A Sexually Dimorphic Role for STAT3 in Sonic Hedgehog Medulloblastoma
Abstract
:1. Introduction
2. Results
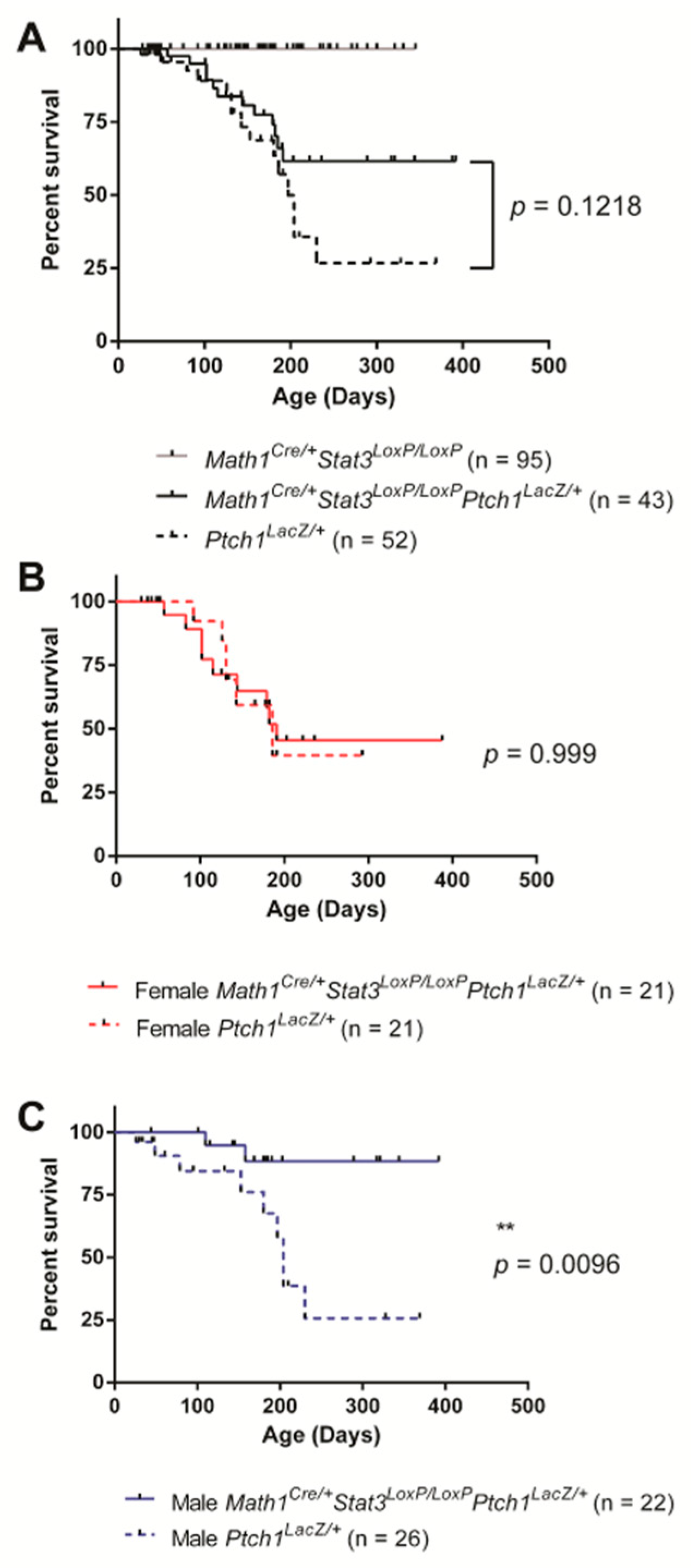
2.1. STAT3 Expression in SHH MB Contributes to Disease Onset in Male but Not Female Mice
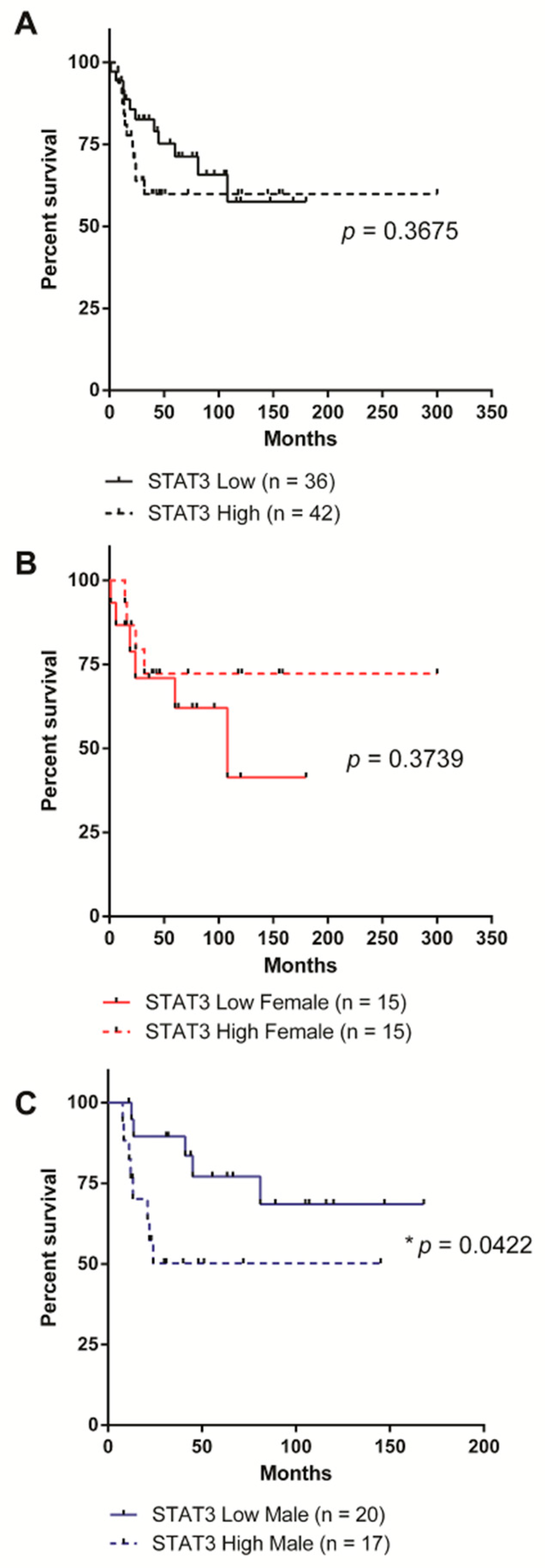
2.2. High STAT3 Expression Correlates with Worse Overall Survival in Male SHH MB Patients
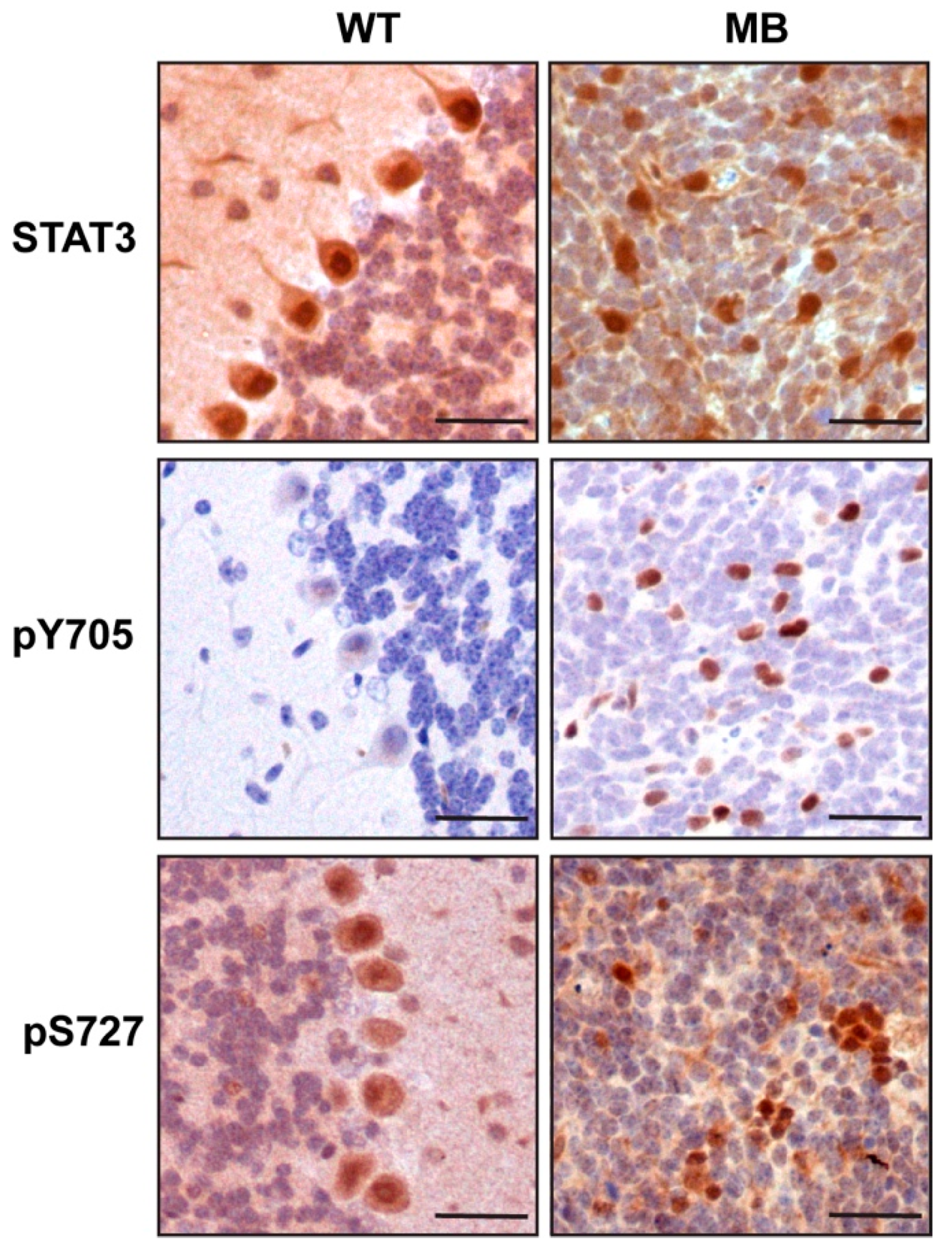
2.3. STAT3 Expression Has No Impact on Proliferation, Cell Death or Differentiation
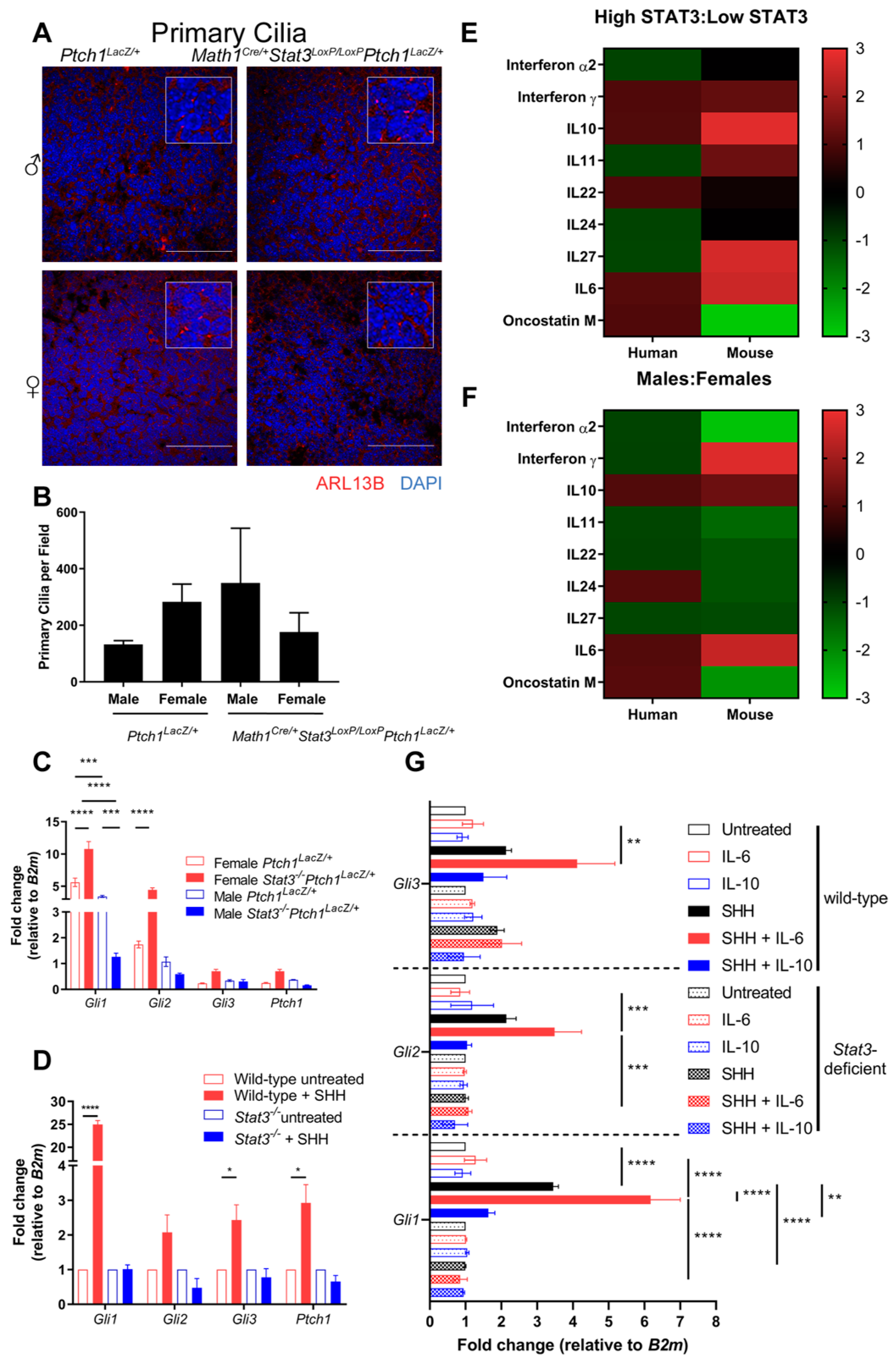
2.4. STAT3 Is Required for SHH Signaling
2.5. IL-6 Expression Is Elevated in Males and Augments SHH Signaling
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Bioinformatic Analysis of Human Tissue Samples
4.3. Cell Culture
4.4. Immunohistochemistry
4.5. PCR Array
4.6. RNA Isolation and Gene Expression Analyses
4.7. Mouse Genotyping PCR
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Northcott, P.A.; Korshunov, A.; Witt, H.; Hielscher, T.; Eberhart, C.G.; Mack, S.; Bouffet, E.; Clifford, S.C.; Hawkins, C.E.; French, P.; et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 2011, 29, 1408–1414. [Google Scholar] [CrossRef] [PubMed]
- Moxon-Emre, I.; Taylor, M.D.; Bouffet, E.; Hardy, K.; Campen, C.J.; Malkin, D.; Hawkins, C.; Laperriere, N.; Ramaswamy, V.; Bartels, U.; et al. Intellectual Outcome in Molecular Subgroups of Medulloblastoma. J. Clin. Oncol. 2016, 34, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Jager, N.; Kool, M.; Zichner, T.; Hutter, B.; Sultan, M.; Cho, Y.J.; Pugh, T.J.; Hovestadt, V.; Stutz, A.M.; et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 2012, 488, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Pugh, T.J.; Weeraratne, S.D.; Archer, T.C.; Pomeranz Krummel, D.A.; Auclair, D.; Bochicchio, J.; Carneiro, M.O.; Carter, S.L.; Cibulskis, K.; Erlich, R.L.; et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 2012, 488, 106–110. [Google Scholar] [CrossRef]
- Robinson, G.; Parker, M.; Kranenburg, T.A.; Lu, C.; Chen, X.; Ding, L.; Phoenix, T.N.; Hedlund, E.; Wei, L.; Zhu, X.; et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012, 488, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Archer, T.C.; Mahoney, E.L.; Pomeroy, S.L. Medulloblastoma: Molecular Classification-Based Personal Therapeutics. Neurotherapeutics 2017, 14, 265–273. [Google Scholar] [CrossRef] [Green Version]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Jones, D.T.W.; Banito, A.; Grunewald, T.G.P.; Haber, M.; Jager, N.; Kool, M.; Milde, T.; Molenaar, J.J.; Nabbi, A.; Pugh, T.J.; et al. Molecular characteristics and therapeutic vulnerabilities across paediatric solid tumours. Nat. Rev. Cancer 2019, 19, 420–438. [Google Scholar] [CrossRef]
- Northcott, P.A.; Korshunov, A.; Pfister, S.M.; Taylor, M.D. The clinical implications of medulloblastoma subgroups. Nat. Rev. Neurol. 2012, 8, 340–351. [Google Scholar] [CrossRef]
- Dressler, E.V.; Dolecek, T.A.; Liu, M.; Villano, J.L. Demographics, patterns of care, and survival in pediatric medulloblastoma. J. Neurooncol. 2017, 132, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Kerr, I.M.; Williams, B.R.; Silverman, R.H.; Schreiber, R.D. How cells respond to interferons. Annu. Rev. Biochem. 1998, 67, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Nichane, M.; Ren, X.; Bellefroid, E.J. Self-regulation of Stat3 activity coordinates cell-cycle progression and neural crest specification. EMBO J. 2010, 29, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Hata, R.; Ma, Y.J.; Tanaka, J.; Mitsuda, N.; Kumon, Y.; Hanakawa, Y.; Hashimoto, K.; Nakajima, K.; Sakanaka, M. Suppression of Stat3 promotes neurogenesis in cultured neural stem cells. J. Neurosci. Res. 2005, 81, 163–171. [Google Scholar] [CrossRef]
- Hong, S.; Song, M.R. Signal transducer and activator of transcription-3 maintains the stemness of radial glia at mid-neurogenesis. J. Neurosci. 2015, 35, 1011–1023. [Google Scholar] [CrossRef]
- Yoshimatsu, T.; Kawaguchi, D.; Oishi, K.; Takeda, K.; Akira, S.; Masuyama, N.; Gotoh, Y. Non-cell-autonomous action of STAT3 in maintenance of neural precursor cells in the mouse neocortex. Development 2006, 133, 2553–2563. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Ren, W.; Ren, B. Expression of signal transducers and activator of transcription 3 (STAT3) determines differentiation of olfactory bulb cells. Mol. Cell. Biochem. 2009, 320, 101–108. [Google Scholar] [CrossRef]
- Bonni, A.; Sun, Y.; Nadal-Vicens, M.; Bhatt, A.; Frank, D.A.; Rozovsky, I.; Stahl, N.; Yancopoulos, G.D.; Greenberg, M.E. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 1997, 278, 477–483. [Google Scholar] [CrossRef]
- Lee, H.S.; Han, J.; Lee, S.H.; Park, J.A.; Kim, K.W. Meteorin promotes the formation of GFAP-positive glia via activation of the Jak-STAT3 pathway. J. Cell Sci. 2010, 123, 1959–1968. [Google Scholar] [CrossRef] [Green Version]
- Schaefer, L.K.; Ren, Z.; Fuller, G.N.; Schaefer, T.S. Constitutive activation of Stat3alpha in brain tumors: Localization to tumor endothelial cells and activation by the endothelial tyrosine kinase receptor (VEGFR-2). Oncogene 2002, 21, 2058–2065. [Google Scholar] [CrossRef]
- Birner, P.; Toumangelova-Uzeir, K.; Natchev, S.; Guentchev, M. STAT3 tyrosine phosphorylation influences survival in glioblastoma. J. Neurooncol. 2010, 100, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Garg, N.; Bakhshinyan, D.; Venugopal, C.; Mahendram, S.; Rosa, D.A.; Vijayakumar, T.; Manoranjan, B.; Hallett, R.; McFarlane, N.; Delaney, K.H.; et al. CD133+ brain tumor-initiating cells are dependent on STAT3 signaling to drive medulloblastoma recurrence. Oncogene 2016, 36, 606. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Fan, Q.; Zhang, X.; Xie, J. A role for transcription factor STAT3 signaling in oncogene smoothened-driven carcinogenesis. J. Biol. Chem. 2012, 287, 38356–38366. [Google Scholar] [CrossRef] [PubMed]
- Chedotal, A. Should I stay or should I go? Becoming a granule cell. Trends Neurosci. 2010, 33, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, L.V.; Milenkovic, L.; Higgins, K.M.; Scott, M.P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 1997, 277, 1109–1113. [Google Scholar] [CrossRef]
- Huynh, J.; Chand, A.; Gough, D.; Ernst, M. Therapeutically exploiting STAT3 activity in cancer—using tissue repair as a road map. Nat. Rev. Cancer 2019, 19, 82–96. [Google Scholar] [CrossRef]
- Matei, V.; Pauley, S.; Kaing, S.; Rowitch, D.; Beisel, K.W.; Morris, K.; Feng, F.; Jones, K.; Lee, J.; Fritzsch, B. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 2005, 234, 633–650. [Google Scholar] [CrossRef]
- Vanner, R.J.; Remke, M.; Gallo, M.; Selvadurai, H.J.; Coutinho, F.; Lee, L.; Kushida, M.; Head, R.; Morrissy, S.; Zhu, X.; et al. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell 2014, 26, 33–47. [Google Scholar] [CrossRef]
- Levy, D.E.; Lee, C.K. What does Stat3 do? J. Clin. Investig. 2002, 109, 1143–1148. [Google Scholar] [CrossRef]
- Huangfu, D.; Liu, A.; Rakeman, A.S.; Murcia, N.S.; Niswander, L.; Anderson, K.V. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 2003, 426, 83–87. [Google Scholar] [CrossRef]
- Bay, S.N.; Long, A.B.; Caspary, T. Disruption of the ciliary GTPase Arl13b suppresses Sonic hedgehog overactivation and inhibits medulloblastoma formation. Proc. Natl. Acad. Sci. USA 2018, 115, 1570–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciucci, A.; Meco, D.; De Stefano, I.; Travaglia, D.; Zannoni, G.F.; Scambia, G.; Riccardi, R.; Saran, A.; Mancuso, M.; Gallo, D. Gender effect in experimental models of human medulloblastoma: Does the estrogen receptor beta signaling play a role? PLoS ONE 2014, 9, e101623. [Google Scholar] [CrossRef] [PubMed]
- Zannoni, G.F.; Ciucci, A.; Marucci, G.; Travaglia, D.; Stigliano, E.; Foschini, M.P.; Scambia, G.; Gallo, D. Sexual dimorphism in medulloblastoma features. Histopathology 2016, 68, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Zhang, W.; Chen, N.H. Sexual dimorphism in cerebral ischemia injury. Eur. J. Pharmacol. 2013, 711, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, M.T.; Iulita, M.F.; Cavedo, E.; Chiesa, P.A.; Schumacher Dimech, A.; Santuccione Chadha, A.; Baracchi, F.; Girouard, H.; Misoch, S.; Giacobini, E.; et al. Sex differences in Alzheimer disease—The gateway to precision medicine. Nat. Rev. Neurol. 2018, 14, 457–469. [Google Scholar] [CrossRef]
- Robison, L.S.; Gannon, O.J.; Salinero, A.E.; Zuloaga, K.L. Contributions of sex to cerebrovascular function and pathology. Brain Res. 2019, 1710, 43–60. [Google Scholar] [CrossRef]
- Dziennis, S.; Jia, T.; Ronnekleiv, O.K.; Hurn, P.D.; Alkayed, N.J. Role of signal transducer and activator of transcription-3 in estradiol-mediated neuroprotection. J. Neurosci. 2007, 27, 7268–7274. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Abarbanell, A.; Tan, J.; Weil, B.; Herrmann, J.; Meldrum, D.R. Both endogenous and exogenous testosterone decrease myocardial STAT3 activation and SOCS3 expression after acute ischemia and reperfusion. Surgery 2009, 146, 138–144. [Google Scholar] [CrossRef]
- Kriz, J.; Lalancette-Hebert, M. Inflammation, plasticity and real-time imaging after cerebral ischemia. Acta Neuropathol. 2009, 117, 497–509. [Google Scholar] [CrossRef] [Green Version]
- Di Domenico, F.; Casalena, G.; Sultana, R.; Cai, J.; Pierce, W.M.; Perluigi, M.; Cini, C.; Baracca, A.; Solaini, G.; Lenaz, G.; et al. Involvement of Stat3 in mouse brain development and sexual dimorphism: A proteomics approach. Brain Res. 2010, 1362, 1–12. [Google Scholar] [CrossRef]
- Gough, D.J.; Corlett, A.; Schlessinger, K.; Wegrzyn, J.; Larner, A.C.; Levy, D.E. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 2009, 324, 1713–1716. [Google Scholar] [CrossRef] [PubMed]
- Wegrzyn, J.; Potla, R.; Chwae, Y.J.; Sepuri, N.B.; Zhang, Q.; Koeck, T.; Derecka, M.; Szczepanek, K.; Szelag, M.; Gornicka, A.; et al. Function of mitochondrial Stat3 in cellular respiration. Science 2009, 323, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Raz, R.; Lee, C.K.; Cannizzaro, L.A.; d’Eustachio, P.; Levy, D.E. Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 1999, 96, 2846–2851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
White, C.L.; Jayasekara, W.S.N.; Picard, D.; Chen, J.; Watkins, D.N.; Cain, J.E.; Remke, M.; Gough, D.J. A Sexually Dimorphic Role for STAT3 in Sonic Hedgehog Medulloblastoma. Cancers 2019, 11, 1702. https://doi.org/10.3390/cancers11111702
White CL, Jayasekara WSN, Picard D, Chen J, Watkins DN, Cain JE, Remke M, Gough DJ. A Sexually Dimorphic Role for STAT3 in Sonic Hedgehog Medulloblastoma. Cancers. 2019; 11(11):1702. https://doi.org/10.3390/cancers11111702
Chicago/Turabian StyleWhite, Christine L., W. Samantha N. Jayasekara, Daniel Picard, Jasmine Chen, D. Neil Watkins, Jason E. Cain, Marc Remke, and Daniel J. Gough. 2019. "A Sexually Dimorphic Role for STAT3 in Sonic Hedgehog Medulloblastoma" Cancers 11, no. 11: 1702. https://doi.org/10.3390/cancers11111702



