Tumor BRCA Test for Patients with Epithelial Ovarian Cancer: The Role of Molecular Pathology in the Era of PARP Inhibitor Therapy
Abstract
:1. Introduction
2. Results
2.1. Tumor BRCA Test Performance
2.2. Tumor BRCA1 and BRCA2 Alterations Distribution
2.3. Tumor BRCA1/2 Status According to Clinicopathological Characteristics
2.4. Tumor and Germline BRCA Tests Concordance
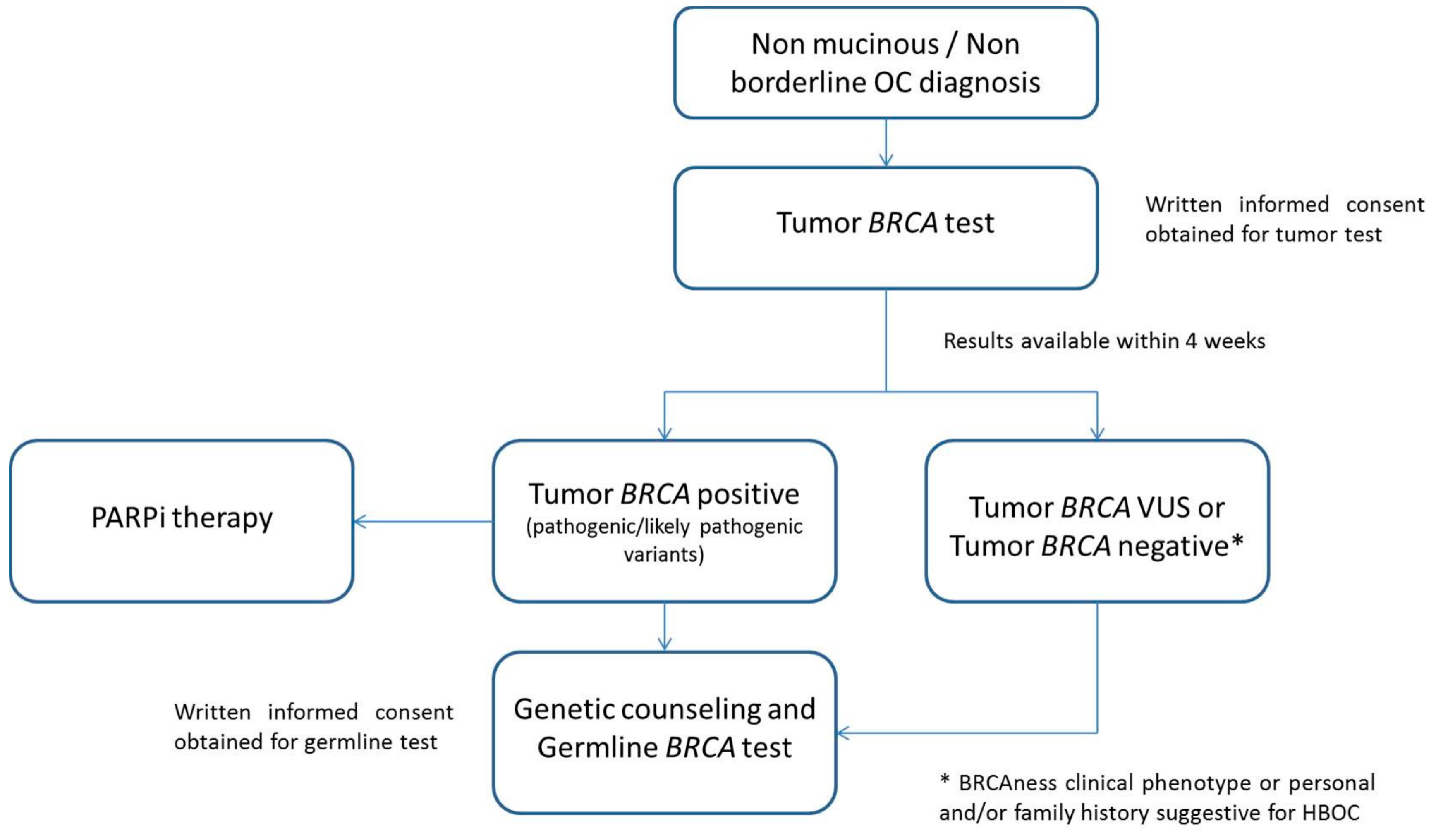
2.5. Tumor BRCA Test Workflow in Clinical Diagnostic Setting
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Next-Generation Sequencing Analysis of BRCA Status in Tumor Samples
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- SEER Cancer Statistics Factsheets: Ovary Cancer. National Cancer Institute. Bethesda. Available online: https://seer.cancer.gov/statfacts/html/ovary.html (accessed on 11 February 2019).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Du Bois, A.; Reuss, A.; Pujade-Lauraine, E.; Harter, P.; Ray-Coquard, I.; Pfisterer, J. Role of surgical outcome as prognostic factor in advanced epithelial ovarian cancer: A combined exploratory analysis of 3 prospectively randomized phase 3 multicenter trials: By the Arbeitsgemeinschaft Gynaekologische Onkologie Studiengruppe Ovarialkarzinom (AGO-OVAR) and the Groupe d’Investigateurs Nationaux Pour les Etudes des Cancers de l’Ovaire (GINECO). Cancer 2009, 115, 1234–1244. [Google Scholar] [CrossRef] [PubMed]
- AstraZeneca. Lynparza Summary of Product Characteristics 2014. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_Product_Information/human/003726/WC500180151.pdf (accessed on 28 June 2019).
- Tesaro. Zejula Summary of Product Characteristics 2017. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_Product_Information/human/004249/WC500239289.pdf (accessed on 28 June 2019).
- Clovis Oncology UK. Rubraca Summary of Product Characteristics 2018. Available online: https://www.ema.europa.eu/documents/product-information/rubraca-epar-product-information_en.pdf (accessed on 28 June 2019).
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Capoluongo, E.; Ellison, G.; López-Guerrero, J.A.; Penault-Llorca, F.; Ligtenberg, M.J.L.; Banerjee, S.; Singer, C.; Friedman, E.; Markiefka, B.; Schirmacher, P.; et al. Guidance Statement on BRCA1/2 Tumor Testing in Ovarian Cancer Patients. Semin. Oncol. 2017, 44, 187–197. [Google Scholar] [CrossRef]
- Wallace, A.J. New challenges for BRCA testing: A view from the diagnostic laboratory. Eur. J. Hum. Genet. 2016, 24, S10–S18. [Google Scholar] [CrossRef]
- Koczkowska, M.; Zuk, M.; Gorczynski, A.; Ratajska, M.; Lewandowska, M.; Biernat, W.; Limon, J.; Wasag, B. Detection of somatic BRCA1/2 mutations in ovarian cancer-next-generation sequencing analysis of 100 cases. Cancer Med. 2016, 5, 1640–1646. [Google Scholar] [CrossRef]
- Mafficini, A.; Simbolo, M.; Parisi, A.; Rusev, B.; Luchini, C.; Cataldo, I.; Piazzola, E.; Sperandio, N.; Turri, G.; Franchi, M.; et al. BRCA somatic and germline mutation detection in paraffin embedded ovarian cancers by next-generation sequencing. Oncotarget 2016, 7, 1076–1083. [Google Scholar] [CrossRef] [Green Version]
- De Jonge, M.M.; Ruano, D.; van Eijk, R.; van der Stoep, N.; Nielsen, M.; Wijnen, J.T.; Ter Haar, N.T.; Baalbergen, A.; Bos, M.E.M.M.; Kagie, M.J.; et al. Validation and Implementation of BRCA1/2 Variant Screening in Ovarian Tumor Tissue. J. Mol. Diagn. 2018, 20, 600–611. [Google Scholar] [CrossRef]
- Guidelines for BRCA Test Implementation: “Raccomandazioni per L’implementazione Del Test BRCA Nelle Pazienti Con Carcinoma Ovarico E Nei Familiari a Rischio Elevato Di Neoplasia”. A Cura del Gruppo di Lavoro AIOM-SIGU-SIBIOC-SIAPEC-IAP, 31 Ottobre 2018. Available online: https://www.aiom.it/pubblicazioni/raccomandazioni-position-paper/raccomandazioni-per-limplementazione-del-test-brca-nelle-pazienti-con-carcinoma-ovarico-e-nei-familiari-a-rischio-elevato-di-neoplasia/ (accessed on 11 February 2019).
- Rigakos, G.; Razis, E. BRCAness: Finding the Achilles heel in ovarian cancer. Oncologist 2012, 17, 956–962. [Google Scholar] [CrossRef]
- AIFA Registry of Lynparza 2016. Available online: http://www.aifa.gov.it/content/pubblicazione-schede-di-monitoraggio-registro-lynparza-27042016 (accessed on 11 February 2019).
- Wong, S.Q.; Li, J.; Tan, A.Y.; Vedururu, R.; Pang, J.M.; Do, H.; Ellul, J.; Doig, K.; Bell, A.; MacArthur, G.A.; et al. Sequence artefacts in a prospective series of Formalin-Fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med. Genom. 2014, 7, 23. [Google Scholar] [CrossRef]
- Shin, S.; Kim, Y.; Chul Oh, S.; Yu, N.; Lee, S.-T.; Choi, J.R.; Lee, K.-A. Validation and optimization of the Ion Torrent S5 XL sequencer and Oncomine workflow for BRCA1 and BRCA2 genetic testing. Oncotarget 2017, 23, 34858–34866. [Google Scholar] [CrossRef] [PubMed]
- Pennington, K.P.; Walsh, T.; Harrell, M.I.; Lee, M.K.; Pennil, C.C.; Rendi, M.H.; Thornton, A.; Norquist, B.M.; Casadei, S.; Nord, A.S.; et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin. Cancer Res. 2014, 20, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, B.T.; Timms, K.M.; Carey, M.S.; Gutin, A.; Meyer, L.A.; Flake, D.D., 2nd; Abkevich, V.; Potter, J.; Pruss, D.; Glenn, P.; et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J. Clin. Oncol. 2010, 28, 3570–3576. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, J.M.; Cicek, M.S.; Larson, N.B.; Davila, J.; Wang, C.; Larson, M.C.; Song, H.; Dicks, E.M.; Harrington, P.; Wick, M.; et al. Clinical characteristics of ovarian cancer classified by BRCA1, BRCA2, and RAD51C status. Sci. Rep. 2014, 4, 4026. [Google Scholar] [CrossRef]
- Moschetta, M.; George, A.; Kaye, S.B.; Banerjee, S. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann. Oncol. 2016, 27, 1449–1455. [Google Scholar] [CrossRef]
- Kwon, J. Scientific Plenary session: Costs and benefits of tumor testing for BRCA mutations in high-grade serous ovarian cancer as a triage for confirmatory genetic testing. In Proceedings of the SGO 50th Annual Meeting on Womens Cancers, Honolulu, HI, USA, 16–19 March 2019. [Google Scholar]
- Arts-de Jong, M.; de Bock, G.H.; van Asperen, C.J.; Mourits, M.J.; de Hullu, J.A.; Kets, C.M. Germline BRCA1/2 mutation testing is indicated in every patient with epithelial ovarian cancer: A systematic review. Eur. J. Cancer 2016, 61, 137–145. [Google Scholar] [CrossRef]
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Aranda, V.; Bardelli, A.; Blanpain, C.; Bock, C.; Borowski, C.; Caldas, C.; Califano, A.; Doherty, M.; Elsner, M.; et al. Toward understanding and exploiting tumor heterogeneity. Nat. Med. 2015, 21, 846–853. [Google Scholar] [CrossRef]
- Patch, A.-M.; The Australian Ovarian Cancer Study Group; Christie, E.L.; Etemadmoghadam, D.; Garsed, D.W.; George, J.; Fereday, S.; Nones, K.; Cowin, P.; Alsop, K.; et al. Whole-Genome characterization of chemoresistant ovarian cancer. Nature 2015, 521, 489–494. [Google Scholar] [CrossRef]
- Norquist, B.; Wurz, K.A.; Pennil, C.C.; Garcia, R.; Gross, J.; Sakai, W.; Karlan, B.Y.; Taniguchi, T.; Swisher, E.M. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J. Clin. Oncol. 2011, 29, 3008–3015. [Google Scholar] [CrossRef] [PubMed]
- Swisher, E.M.; Sakai, W.; Karlan, B.Y.; Wurz, K.; Urban, N.; Taniguchi, T. Secondary BRCA1 mutations in BRCA1-Mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008, 68, 2581–2586. [Google Scholar] [CrossRef] [PubMed]
- Loman, N.J.; Misra, R.V.; Dallman, T.J.; Constantinidou, C.; E Gharbia, S.; Wain, J.; Pallen, M.J. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012, 30, 434–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bragg, L.M.; Stone, G.; Butler, M.K.; Hugenholtz, P.; Tyson, G.W. Shining a light on dark sequencing: Characterising errors in Ion Torrent PGM data. PLoS Comput. Biol. 2013, 9, e1003031. [Google Scholar] [CrossRef] [PubMed]
- Yeo, Z.X.; Wong, J.C.; Rozen, S.G.; Lee, A.S.G. Evaluation and optimization of indel detection workflows for ion torrent sequecing of the BRCA1 and BRCA2 gene. BMC Genom. 2014, 15, 516. [Google Scholar] [CrossRef] [PubMed]
- American Joint Committee on Cancer (AJCC). AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017. [Google Scholar]
- Kurman, R.J.; Carcangiu, M.L.; Herrington, C.S.; Young, R.H. WHO Classification of Tumours of Female Reproductive Organs, 4th ed.; IARC Press: Lyon, France, 2014. [Google Scholar]
- Fumagalli, C.; Catania, C.; Ranghiero, A.; Bosi, C.; Viale, G.; de Marinis, F.; Barberis, M.; Guerini-Rocco, E. Molecular Profile of Advanced Non-Small Cell Lung Cancers in Octogenarians: The Door to Precision Medicine in Elderly Patients. J. Clin. Med. 2019, 8, 112. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, C.; Vacirca, D.; Rappa, A.; Passaro, A.; Guarize, J.; Raviele, P.R.; De Marinis, F.; Spaggiari, L.; Casadio, C.; Viale, G.; et al. The long tail of molecular alterations in non-small cell lung cancer: A single-institution experience of next-generation sequencing in clinical molecular diagnostics. J. Clin. Pathol. 2018, 71, 767–773. [Google Scholar] [CrossRef]
- ENIGMA BRCA1/2 Gene Variant Classification Criteria. Available online: https://enigmaconsortium.org/wp-content/uploads/2018/10/ENIGMA_Rules_2017-06-29-v2.5.1.pdf (accessed on 11 February 2019).
- BRCA Exchange Database. Available online: https://brcaexchange.org/ (accessed on 11 February 2019).
- ClinVar Database. Available online: https://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 11 February 2019).
- Leiden Open Source Variation Database. Available online: https://databases.lovd.nl/shared/variants/BRCA1/unique (accessed on 11 February 2019).


| Clinicopathological Features | BRCA1 | BRCA2 | ||||
|---|---|---|---|---|---|---|
| Pathogenic/Likely Pathogenic Mutations | VUS | WT | Pathogenic/Likely Pathogenic Mutations | VUS | WT | |
| Tumor Site | ||||||
| Primary tumors (n = 200) | 46 (23.0%) | 13 (6.5%) | 141 (70.5%) | 14 (7.0%) | 7 (3.5%) | 179 (89.5%) |
| Metastases/Recurrences (n = 21) | 1 (4.8%) | 3 (14.3%) | 17 (81.0%) | 1 (4.8%) | 2 (9.5%) | 18 (85.7%) |
| Histologycal Subtype | ||||||
| High-grade serous carcinoma (n = 194) | 47 (24.2%) | 16 (8.2%) | 131 (67.5%) | 14 (7.2%) | 7 (3.6%) | 173 (89.2%) |
| Non-serous carcinoma/subtype not defined (n = 27) | 0 | 0 | 27 (100%) | 1 (3.7%) | 2 (7.4%) | 24 (88.9%) |
| Pathological Staging | ||||||
| T1 (n = 15) | 2 (13.3%) | 0 | 13 (86.7%) | 2 (13.3%) | 2 (13.3%) | 11 (73.3%) |
| T2 (n = 29) | 6 (20.7%) | 1 (3.4%) | 22 (75.9%) | 3 (10.3%) | 0 | 26 (89.7%) |
| T3 (n = 116) | 25 (21.6%) | 8 (6.9%) | 83 (71.6%) | 4 (3.4%) | 3 (2.6%) | 109 (94.0%) |
| NA (n = 61) | 14 (23.0%) | 7 (11.5%) | 40 (65.6%) | 6 (9.8%) | 4 (6.6%) | 51 (83.6%) |
| N0 (n = 39) | 5 (12.8%) | 0 | 34 (87.2%) | 3 (7.7%) | 4 (10.3%) | 32 (82.1%) |
| N1 (n = 65) | 18 (2.7%) | 6 (9.2%) | 41 (63.1%) | 2 (3.1%) | 1 (1.5%) | 62 (95.4%) |
| NX (n = 56) | 10 (17.9%) | 3 (5.4%) | 43 (76.8%) | 4 (7.1%) | 0 | 52 (92.9%) |
| NA (n = 61) | 14 (23.0%) | 7 (11.5%) | 40 (65.6%) | 6 (9.8%) | 4 (6.6%) | 51 (83.6%) |
| Family History | ||||||
| Positive (n = 93) | 24 (25.8%) | 8 (8.6%) | 61 (65.6%) | 7 (7.5%) | 6 (6.5%) | 80 (86.0%) |
| Negative (n = 101) | 16 (15.8%) | 7 (6.9%) | 78 (77.2%) | 7 (6.9%) | 2 (2.0%) | 92 (91.1%) |
| NA (n = 27) | 7 (25.9%) | 1 (3.7%) | 19 (70.4%) | 1 (3.7%) | 1 (3.7%) | 25 (92.6%) |
| Time of Test Request | ||||||
| At pathological diagnosis (n = 123) | 28 (22.8%) | 8 (6.5%) | 87 (70.7%) | 8 (6.5%) | 5 (4.1%) | 110 (89.4%) |
| At least 6 months after pathological diagnosis without relapse (n = 35) | 9 (25.7%) | 2 (5.7%) | 24 (68.6%) | 3 (8.6%) | 3 (8.6%) | 29 (82.9%) |
| At relapse (n = 37) | 4 (10.8%) | 5 (13.5%) | 28 (75.7%) | 2 (5.4%) | 1 (2.7%) | 34 (91.9%) |
| NA (n = 26) | 6 (23.1%) | 1 (3.8%) | 19 (73.1%) | 2 (7.7%) | 0 | 24 (92.3%) |
| Clinicopathological Features | BRCA MUTATED (Phatogenic/Likely Pathogenic Mutations) | BRCA WT * | p Value ** |
|---|---|---|---|
| Tumor Site | |||
| Primary tumors (n = 200) | 62 (31.0%) | 138 (69.0%) | 0.07 |
| Metastases/Recurrences (n = 21) | 2 (9.5%) | 19 (90.5%) | |
| Histologycal Subtype | |||
| High-grade serous carcinoma (n = 194) | 61 (31.4%) | 133 (68.6%) | 0.005 |
| Non-serous carcinoma/subtype not defined (n = 27) | 1 (3.7%) | 26 (96.3%) | |
| Pathological Staging | |||
| T1 (n = 15) | 4 (26.7%) | 11 (73.3%) | 0.716 |
| T2 (n = 29) | 9 (31.0%) | 20 (69.0%) | |
| T3 (n = 116) | 29 (25.0%) | 87 (75.0%) | |
| NA (n = 61) | 20 (32.8%) | 41 (67.2%) | |
| N0 (n = 39) | 8 (20.5%) | 31 (79.5%) | 0.518 |
| N1 (n = 65) | 20 (30.8%) | 45 (69.2%) | |
| NX (n = 56) | 14 (25.0%) | 42 (75.0%) | |
| NA (n = 61) | 20 (32.8%) | 41 (67.2%) | |
| Family History | |||
| Positive (n = 93) | 31 (33.3%) | 62 (66.7%) | 0.258 |
| Negative (n = 101) | 23 (22.8%) | 78 (77.2%) | |
| NA (n = 27) | 8 (29.6%) | 19 (70.4%) | |
| Time of Test Request | |||
| At pathological diagnosis (n = 123) | 36 (29.3%) | 87 (70.7%) | 0.33 |
| At least 6 months after pathological diagnosis without relapse (n = 35) | 12 (34.3%) | 23 (65.7%) | |
| At relapse (n = 37) | 6 (16.2%) | 31 (83.8%) | |
| NA (n = 26) | 8 (30.8%) | 18 (69.2%) | |
| Tumor BRCA Test | Germline BRCA Test | |||
|---|---|---|---|---|
| Positive | Negative | |||
| Pathogenic/Likely Pathogenic | VUS | |||
| Positive | Pathogenic/Likely Pathogenic | 18 (78.3%) | / | 5 (21.7%) |
| VUS | / | 2 (40%) | 3 (60%) | |
| Negative | / | / | 34 (100%) | |
| Clinicopathological Features | n (%) |
|---|---|
| Time of Test Request | |
| At pathological diagnosis | 123 (55.2%) |
| At least 6 months after pathological diagnosis without relapse | 35 (15.7%) |
| At relapse | 37 (16.6%) |
| NA | 28 (12.6%) |
| Histological Subtype | |
| High-grade serous carcinoma | 195 (87.4%) |
| Endometroid carcinoma | 16 (7.2%) |
| Clear cell carcinoma | 9 (4%) |
| Carcinoma—subtype not defined | 3 (1.3%) |
| TN Pathological Staging | |
| T1 | 15 (6.7%) |
| T2 | 29 (13%) |
| T3 | 116 (52%) |
| NA | 63 (28.3%) |
| N0 | 39 (17.5%) |
| N1 | 65 (29.1%) |
| NX | 56 (25.1%) |
| NA | 63 (28.3%) |
| Specimen Analyzed | |
| Primary Tumor | 201 (90.1%) |
| Ovary localization | 169 (75.8%) |
| Extra-ovary localization | 32 (14.3%) |
| Pelvic localization | 14 (43.8%) |
| Extra-Pelvic localization | 12 (37.5%) |
| NA | 6 (18.7%) |
| Metastasis/recurrence | 22 (9.9%) |
| Lymph nodal metastasis | 3 (13.6%) |
| Extra lymph nodal metastasis | 19 (86.4%) |
| Neoadjuvant Treatment | |
| Yes | 46 (20.6%) |
| No | 114 (51.1%) |
| NA | 63 (28.3%) |
| Surgery | |
| R0 - no residual disease | 135 (60.5%) |
| R+ < 1 cm | 42 (18.8%) |
| R+ > 1 cm | 19 (8.5%) |
| NA | 27 (12.1%) |
| Family History | |
| Positive | 93 (41.7%) |
| Negative | 101 (45.3%) |
| NA | 29 (13%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fumagalli, C.; Tomao, F.; Betella, I.; Rappa, A.; Calvello, M.; Bonanni, B.; Bernard, L.; Peccatori, F.; Colombo, N.; Viale, G.; et al. Tumor BRCA Test for Patients with Epithelial Ovarian Cancer: The Role of Molecular Pathology in the Era of PARP Inhibitor Therapy. Cancers 2019, 11, 1641. https://doi.org/10.3390/cancers11111641
Fumagalli C, Tomao F, Betella I, Rappa A, Calvello M, Bonanni B, Bernard L, Peccatori F, Colombo N, Viale G, et al. Tumor BRCA Test for Patients with Epithelial Ovarian Cancer: The Role of Molecular Pathology in the Era of PARP Inhibitor Therapy. Cancers. 2019; 11(11):1641. https://doi.org/10.3390/cancers11111641
Chicago/Turabian StyleFumagalli, Caterina, Federica Tomao, Ilaria Betella, Alessandra Rappa, Mariarosaria Calvello, Bernardo Bonanni, Loris Bernard, Fedro Peccatori, Nicoletta Colombo, Giuseppe Viale, and et al. 2019. "Tumor BRCA Test for Patients with Epithelial Ovarian Cancer: The Role of Molecular Pathology in the Era of PARP Inhibitor Therapy" Cancers 11, no. 11: 1641. https://doi.org/10.3390/cancers11111641
APA StyleFumagalli, C., Tomao, F., Betella, I., Rappa, A., Calvello, M., Bonanni, B., Bernard, L., Peccatori, F., Colombo, N., Viale, G., Barberis, M., & Guerini-Rocco, E. (2019). Tumor BRCA Test for Patients with Epithelial Ovarian Cancer: The Role of Molecular Pathology in the Era of PARP Inhibitor Therapy. Cancers, 11(11), 1641. https://doi.org/10.3390/cancers11111641







