FOXC1 Regulation of miR-31-5p Confers Oxaliplatin Resistance by Targeting LATS2 in Colorectal Cancer
Abstract
1. Introduction
2. Results
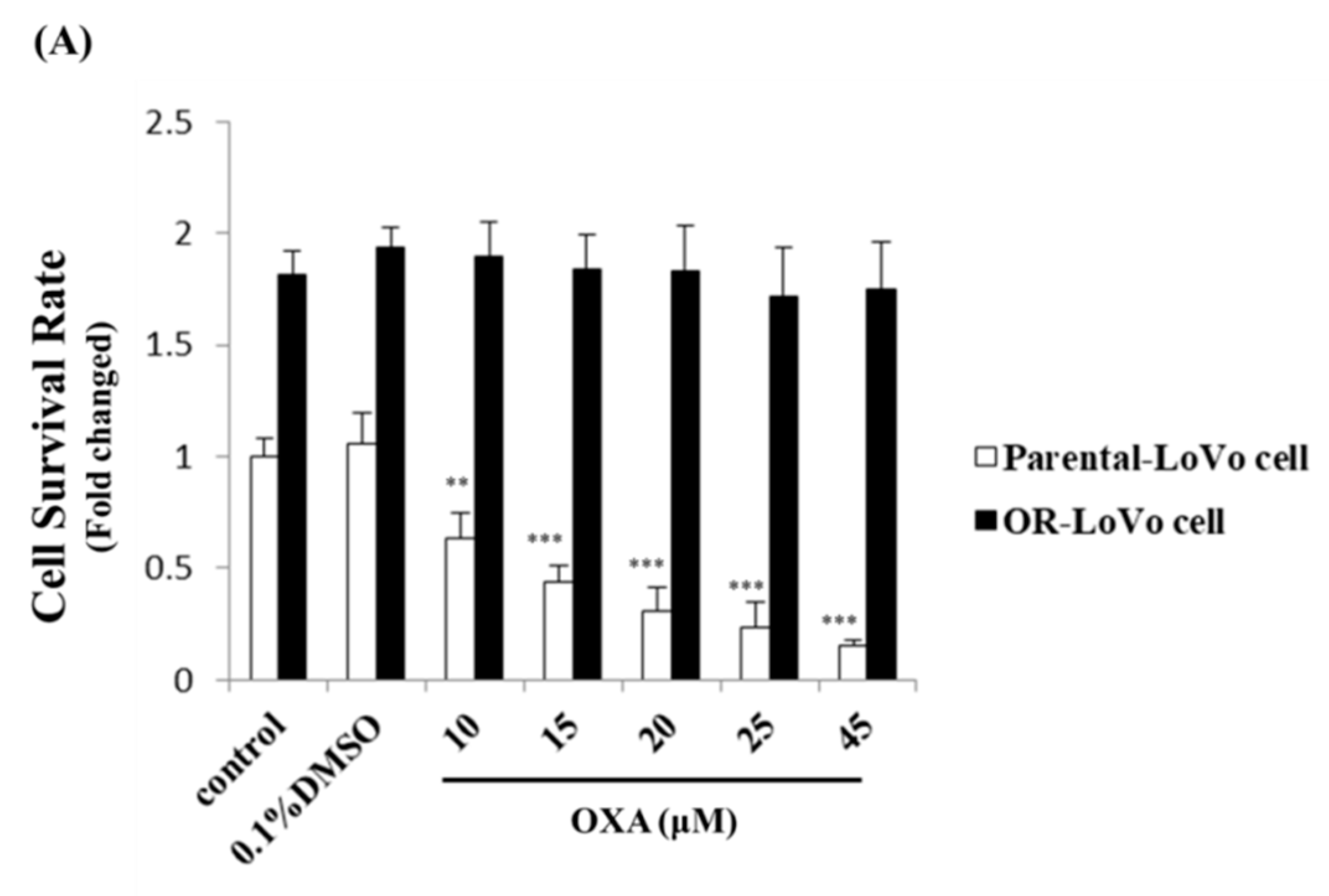
2.1. Evaluation of the Cell Properties of LoVo and OR-LoVo Cells
2.2. MicroRNA Expression Differed between LoVo and OR-LoVo Cancer Cells
2.3. MiR-31-5p Regulates Cell Survival and Cell Death in LoVo and OR-LoVo Cells in Vitro
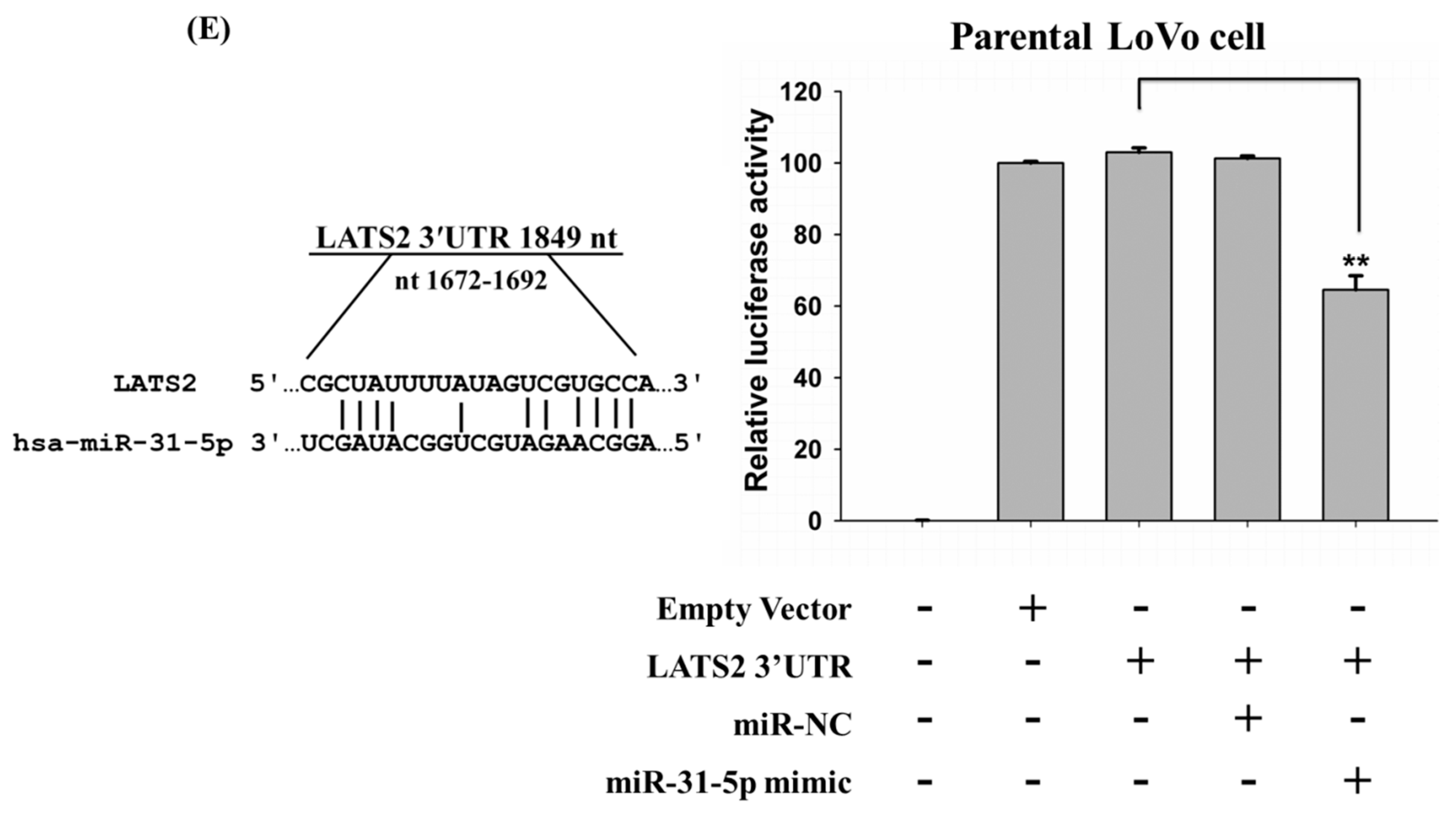
2.4. LATS2 is a Direct Target of miR-31-5p in Colorectal Cancer Cells
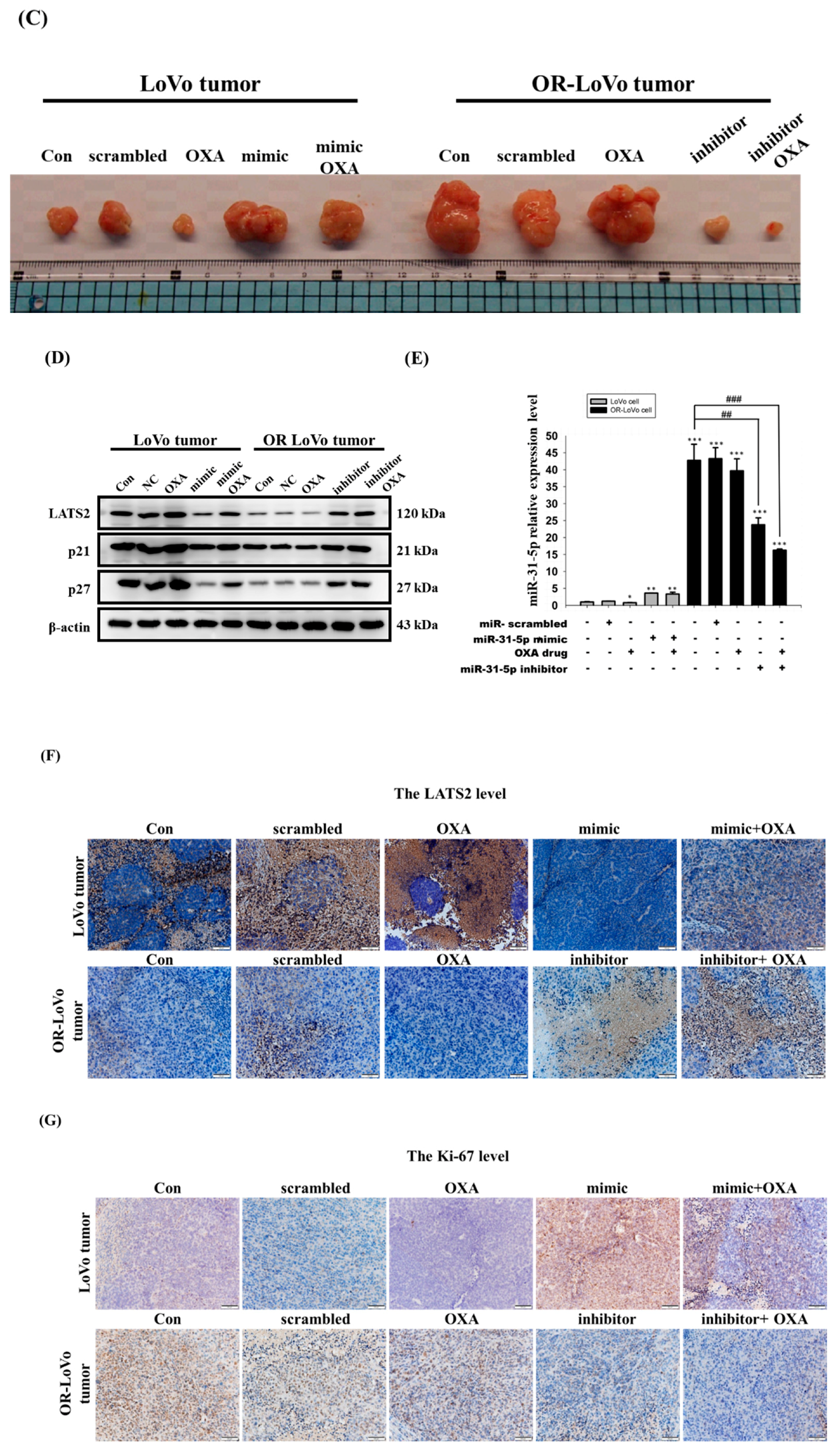
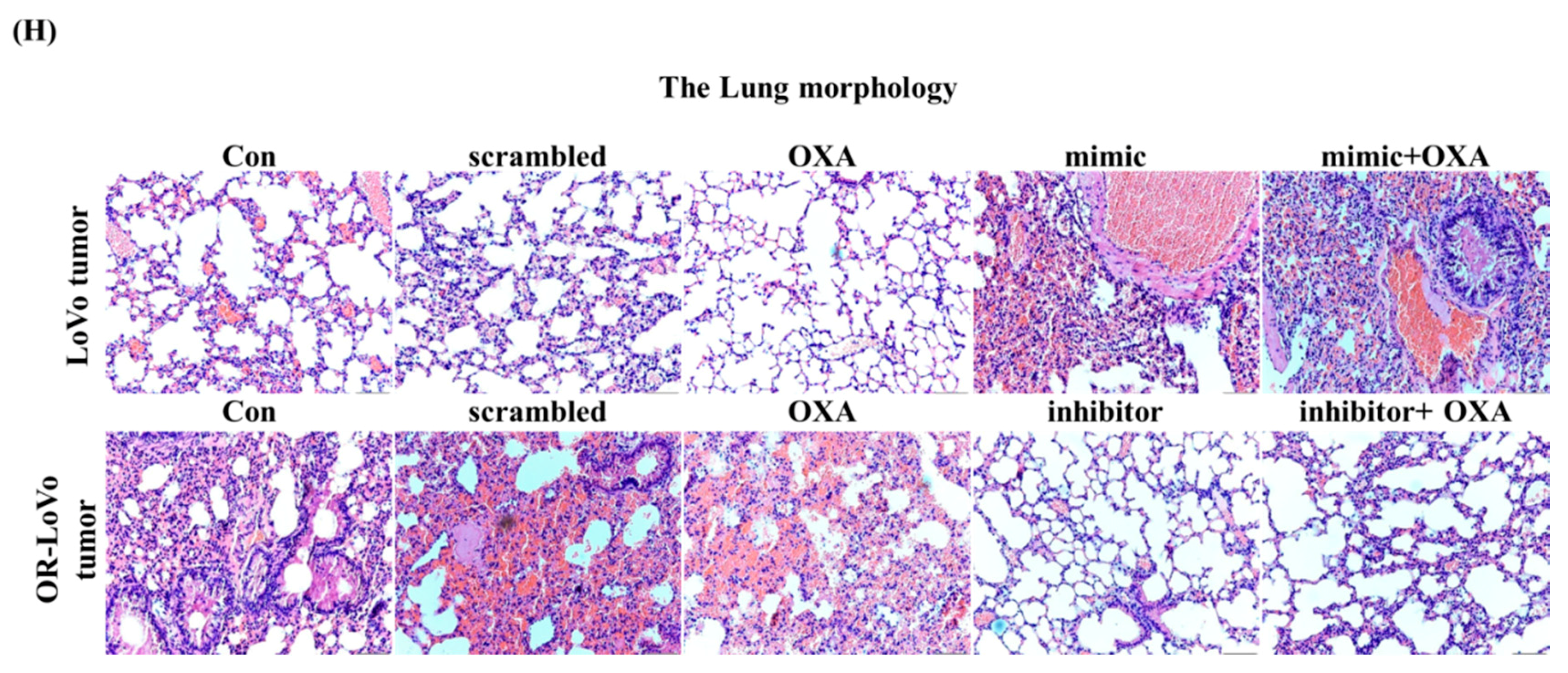
2.5. MiR-31-5p Regulates Tumorigenesis and Chemosensitivity by Targeting LATS2 in A Xenograft Tumor Model
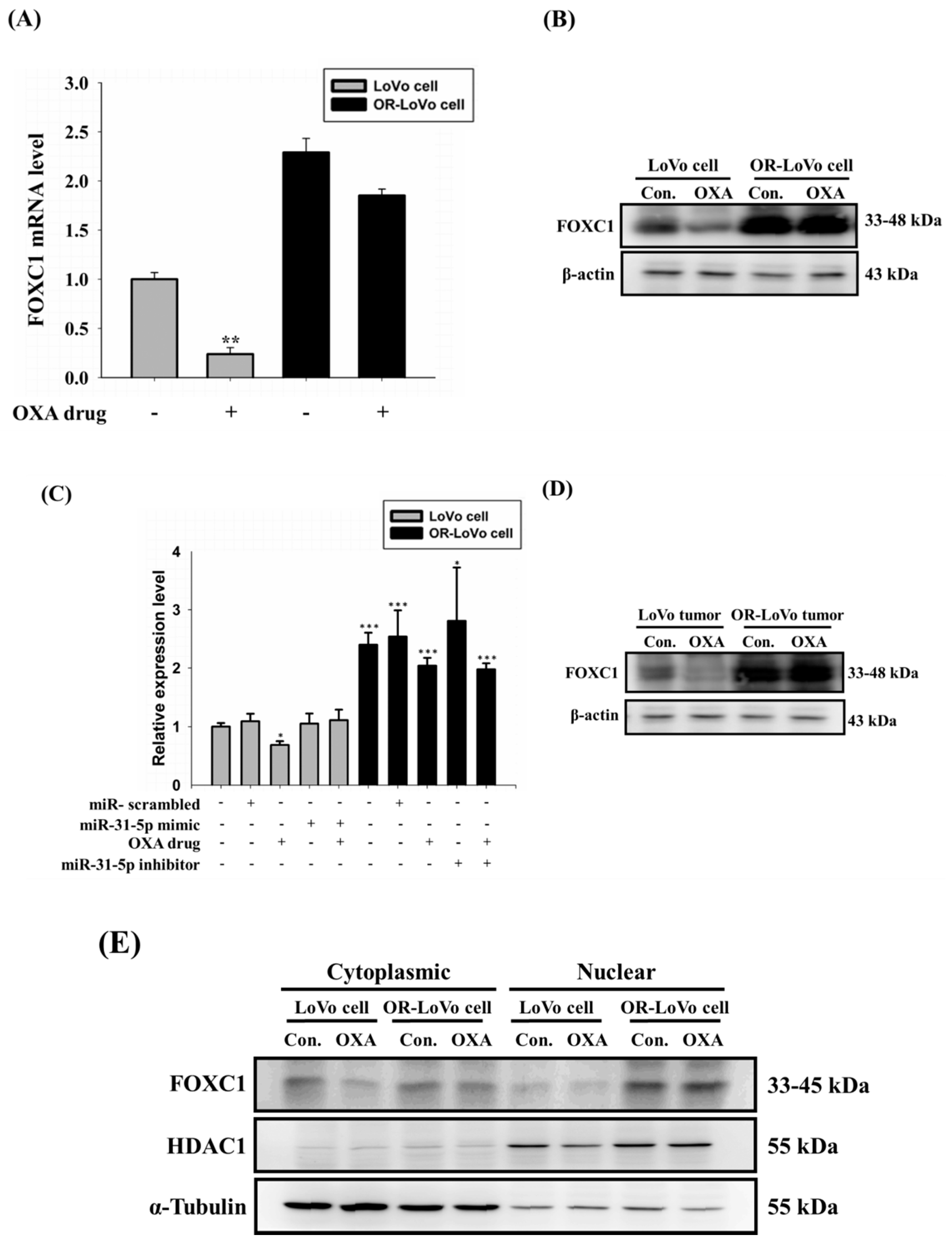
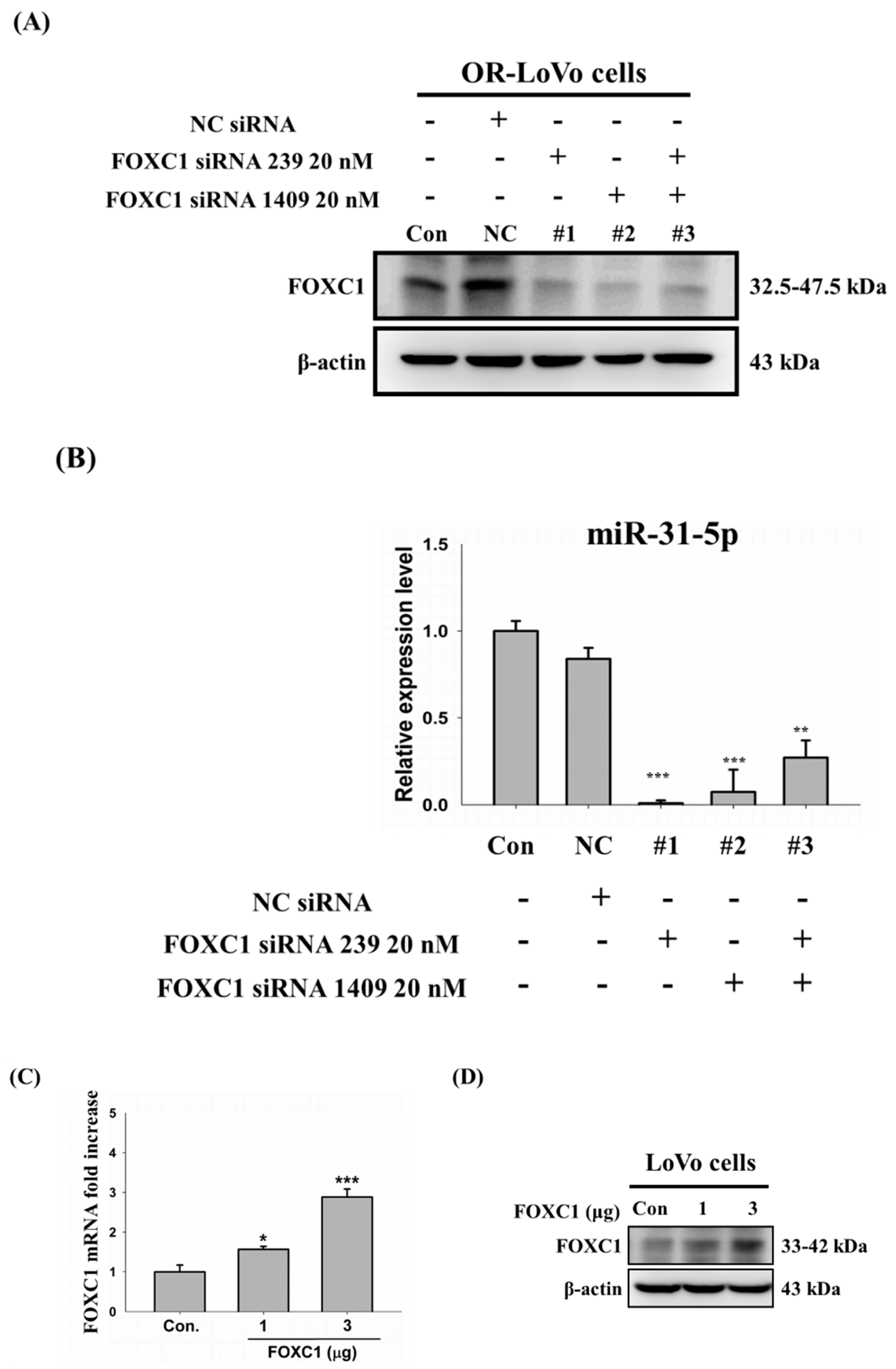
2.6. FOXC1 mRNA and Protein Expression in Colorectal Cancer
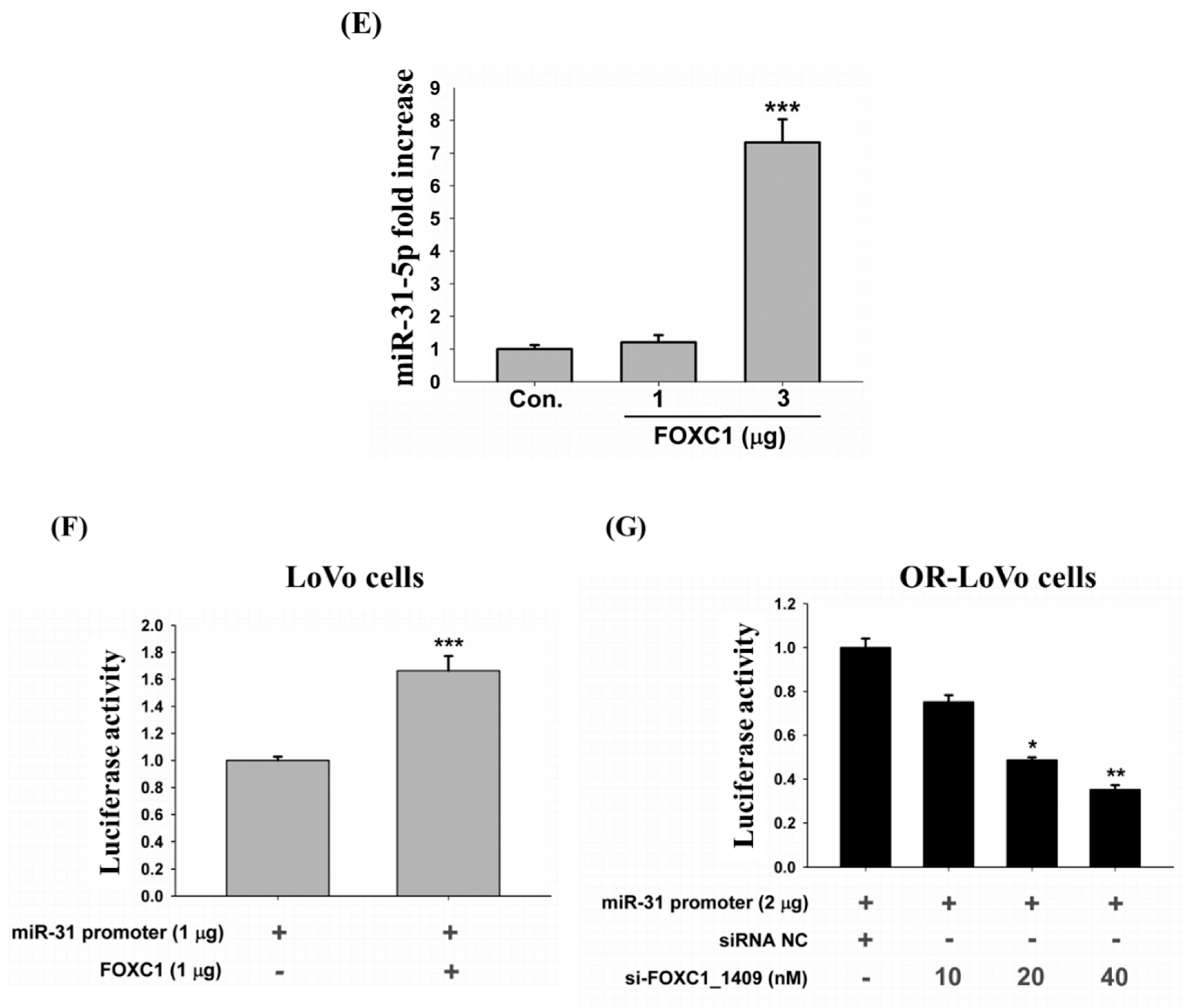
2.7. Binding of the FOXC1 Transcription Factor to the miR-31 Promoter Induces High miR-31-5p Expression
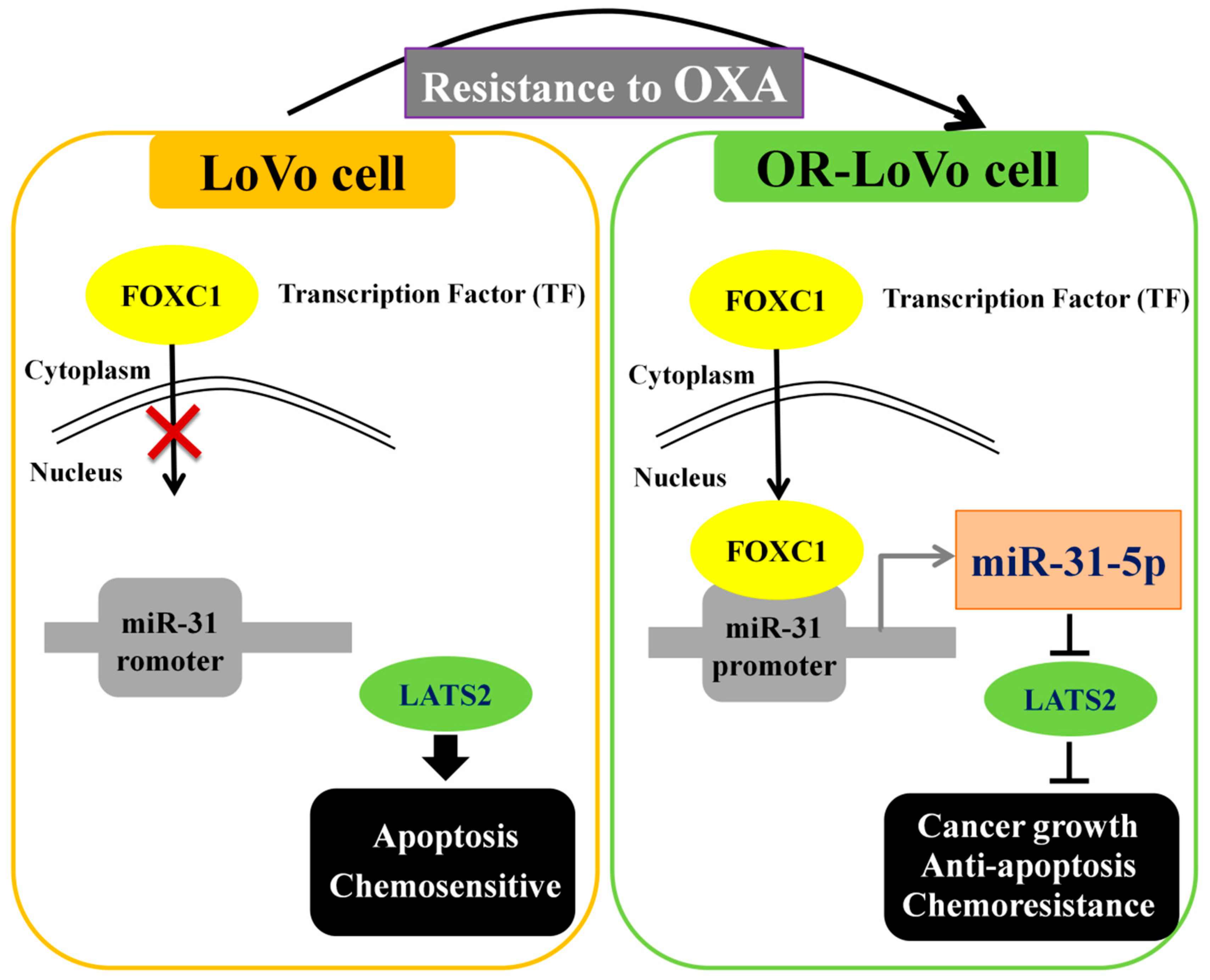
3. Discussion
4. Materials and Methods
4.1. Cell Culture and OXA-Resistant Cell Establishment
4.2. MTT (Thiazolyl Blue Tetrazolium Bromide) Assay
4.3. Microarray Array Analysis
4.4. Prediction of mRNA Targets
4.5. Plasmid, miR-31-5p Overexpression (Mimic) or Knockdown (Inhibitor), Scrambled miRNA (Mimic NC or Inhibitor NC), and siRNA by Transfection
4.6. Western Blot Analysis
4.7. RNA Preparation and Quantitative RT-PCR Analysis
4.8. Luciferase Reporter Assay
4.9. TUNEL Assay
4.10. Subcutaneous Tumor Xenograft Model
4.11. Immunohistochemistry (IHC) Staining
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ali, A.S.; Ajaz, A. The role of mucin-educated platelet activation in tumor invasiveness: An unfolding concern in the realm of cancer biology. Biomedicine 2017, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Gulei, D.; Magdo, L.; Jurj, A.; Raduly, L.; Cojocneanu-Petric, R.; Moldovan, A.; Moldovan, C.; Florea, A.; Paşca, S.; Pop, L.-A.; et al. The silent healer: miR-205-5p up-regulation inhibits epithelial to mesenchymal transition in colon cancer cells by indirectly up-regulating E-cadherin expression. Cell Death Dis. 2018, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Yu, F.; Ma, Y.; Zhao, R.; Chen, X.; Zhu, J.; Zhang, C.Y.; Chen, J.; Zhang, J. Microrna-31 activates the ras pathway and functions as an oncogenic microrna in human colorectal cancer by repressing ras p21 gtpase activating protein 1 (rasa1). J. Biol. Chem. 2013, 288, 9508–9518. [Google Scholar] [CrossRef] [PubMed]
- De Krijger, I.; Mekenkamp, L.J.; Punt, C.J.; Nagtegaal, I.D. MicroRNAs in colorectal cancer metastasis. J. Pathol. 2011, 224, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Su, S.-Y.; Huang, J.-Y.; Jian, Z.-H.; Ho, C.-C.; Lung, C.-C.; Liaw, Y.-P. Mortality of colorectal cancer in Taiwan, 1971–2010: Temporal changes and age–period–cohort analysis. Int. J. Color. Dis. 2012, 27, 1665–1672. [Google Scholar] [CrossRef] [PubMed]
- Selvachandran, S.N.; Hodder, R.J.; Ballal, M.S.; Jones, P.; Cade, D. Prediction of colorectal cancer by a patient consultation questionnaire and scoring system: A prospective study. Lancet 2002, 360, 278–283. [Google Scholar] [CrossRef]
- Chung, T.K.; Lau, T.S.; Cheung, T.H.; Yim, S.F.; Lo, K.W.; Siu, N.S.; Chan, L.K.; Yu, M.Y.; Kwong, J.; Doran, G.; et al. Dysregulation of microrna-204 mediates migration and invasion of endometrial cancer by regulating foxc1. Int. J. Cancer 2012, 130, 1036–1045. [Google Scholar] [CrossRef]
- Smith, D.; Ballal, M.; Hodder, R.; Soin, G.; Selvachandran, S.; Cade, D. Symptomatic Presentation of Early Colorectal Cancer. Ann. R. Coll. Surg. Engl. 2006, 88, 185–190. [Google Scholar] [CrossRef]
- Xu, Z.; Chi, P.; Pan, J.; Shen, S.; Sun, Y.; Wang, X.; Lu, X. Knockdown of KLK11 inhibits cell proliferation and increases oxaliplatin sensitivity in human colorectal cancer. Exp. Ther. Med. 2016, 12, 2855–2860. [Google Scholar] [CrossRef][Green Version]
- Baskaran, R.; Day, C.H.; Lin, Y.-J.; Padma, V.V.; Huang, C.-Y.; Hsu, H.-H.; Chen, M.-C.; Lin, Y.-M.; Tu, C.-C.; Kuo, W.-W.; et al. Oxaliplatin resistance in colorectal cancer cells is mediated via activation of ABCG2 to alleviate ER stress induced apoptosis. J. Cell. Physiol. 2018, 233, 5458–5467. [Google Scholar]
- Lai, H.-H.; Lin, L.-J.; Hung, L.-Y.; Chen, P.-S. Role of Dicer in regulating oxaliplatin resistance of colon cancer cells. Biochem. Biophys. Res. Commun. 2018, 506, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Jeng, L.-B.; Velmurugan, B.K.; Chen, M.-C.; Hsu, H.-H.; Ho, T.-J.; Day, C.-H.; Lin, Y.-M.; Padma, V.V.; Tu, C.-C.; Huang, C.-Y. Fisetin mediated apoptotic cell death in parental and Oxaliplatin/irinotecan resistant colorectal cancer cells in vitro and in vivo. J. Cell. Physiol. 2018, 233, 7134–7142. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-C.; Hsu, H.-H.; Chu, Y.-Y.; Cheng, S.-F.; Shen, C.-Y.; Lin, Y.-J.; Chen, R.-J.; Viswanadha, V.P.; Lin, Y.-M.; Huang, C.-Y. Lupeol alters ER stress-signaling pathway by downregulating ABCG2 expression to induce Oxaliplatin-resistant LoVo colorectal cancer cell apoptosis. Environ. Toxicol. 2018, 33, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Siomi, H.; Siomi, M.C. Posttranscriptional Regulation of MicroRNA Biogenesis in Animals. Mol. Cell 2010, 38, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Wery, M.; Kwapisz, M.; Morillon, A. Noncoding rnas in gene regulation. Wiley interdisciplinary reviews. Syst. Biol. Med. 2011, 3, 728–738. [Google Scholar]
- Fleming, J.L.; Gable, D.L.; Samadzadeh-Tarighat, S.; Cheng, L.; Yu, L.; Gillespie, J.L.; Toland, A.E. Differential expression of mir-1, a putative tumor suppressing microrna, in cancer resistant and cancer susceptible mice. PeerJ 2013, 1, e68. [Google Scholar] [CrossRef][Green Version]
- Reddy, K.B. Microrna (mirna) in cancer. Cancer Cell Int. 2015, 15, 38. [Google Scholar] [CrossRef]
- Paladini, L.; Fabris, L.; Bottai, G.; Raschioni, C.; Calin, G.A.; Santarpia, L. Targeting microRNAs as key modulators of tumor immune response. J. Exp. Clin. Cancer Res. 2016, 35, 103. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Bottai, G.; Nuciforo, P.G.; Di Tommaso, L.; Giovannetti, E.; Peg, V.; Losurdo, A.; Perez-Garcia, J.; Masci, G.; Corsi, F.; et al. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget 2015, 6, 37269–37280. [Google Scholar] [CrossRef]
- Ganju, A.; Khan, S.; Hafeez, B.B.; Behrman, S.W.; Yallapu, M.M.; Chauhan, S.C.; Jaggi, M. Mirna nanotherapeutics for cancer. Drug Discov. Today 2017, 22, 424–432. [Google Scholar] [CrossRef]
- Shah, M.Y.; Calin, G.A. MicroRNAs as therapeutic targets in human cancers. Wiley Interdiscip. Rev. RNA 2014, 5, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sempere, L.F.; Ouyang, H.; Memoli, V.A.; Andrew, A.S.; Luo, Y.; Demidenko, E.; Korc, M.; Shi, W.; Preis, M.; et al. Microrna-31 functions as an oncogenic microrna in mouse and human lung cancer cells by repressing specific tumor suppressors. J. Clin. Investig. 2010, 120, 1298–1309. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yi, J.; Zhang, K.; Bai, F.; Feng, B.; Wang, R.; Chu, X.; Chen, L.; Song, H. Downregulation of MiR-31 stimulates expression of LATS2 via the hippo pathway and promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 161. [Google Scholar] [CrossRef] [PubMed]
- Bandrés, E.; Cubedo, E.; Agirre, X.; Malumbres, R.; Zarate, R.; Ramirez, N.; Abajo, A.; Navarro, A.; Moreno, I.; Monzó, M.; et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 2006, 5, 29. [Google Scholar] [CrossRef]
- Augoff, K.; McCue, B.; Plow, E.F.; Sossey-Alaoui, K. Mir-31 and its host gene lncrna loc554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol. Cancer 2012, 11, 5. [Google Scholar] [CrossRef]
- Asangani, I.A.; Harms, P.W.; Dodson, L.; Pandhi, M.; Kunju, L.P.; Maher, C.A.; Fullen, D.R.; Johnson, T.M.; Giordano, T.J.; Palanisamy, N.; et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget 2012, 3, 1011–1025. [Google Scholar] [CrossRef]
- Creighton, C.J.; Fountain, M.D.; Yu, Z.; Nagaraja, A.K.; Zhu, H.; Khan, M.; Olokpa, E.; Zariff, A.; Gunaratne, P.H.; Matzuk, M.M.; et al. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010, 70, 1906–1915. [Google Scholar] [CrossRef]
- Ye, Y.; Zhuang, J.; Wang, G.; He, S.; Ni, J.; Xia, W.; Wang, J. microRNA-605 promotes cell proliferation, migration and invasion in non-small cell lung cancer by directly targeting LATS2. Exp. Ther. Med. 2017, 14, 867–873. [Google Scholar] [CrossRef]
- He, Y.; Wang, J.; Wang, J.; Yung, V.Y.-W.; Hsu, E.; Li, A.; Kang, Q.; Ma, J.; Han, Q.; Jin, P.; et al. MicroRNA-135b regulates apoptosis and chemoresistance in colorectal cancer by targeting large tumor suppressor kinase 2. Am. J. Cancer Res. 2015, 5, 1382–1395. [Google Scholar]
- Guan, X.; Zong, Z.-H.; Chen, S.; Sang, X.-B.; Wu, D.-D.; Wang, L.-L.; Liu, Y.; Zhao, Y. The role of miR-372 in ovarian carcinoma cell proliferation. Gene 2017, 624, 14–20. [Google Scholar] [CrossRef]
- Lit, L.C.; Scott, S.; Zhang, H.; Stebbing, J.; Photiou, A.; Giamas, G. LATS2 is a modulator of estrogen receptor alpha. Anticancer. Res. 2013, 33, 53–63. [Google Scholar] [PubMed]
- Mitamura, T.; Watari, H.; Wang, L.; Kanno, H.; Kitagawa, M.; Hassan, M.K.; Kimura, T.; Tanino, M.; Nishihara, H.; Tanaka, S.; et al. microRNA 31 functions as an endometrial cancer oncogene by suppressing Hippo tumor suppressor pathway. Mol. Cancer 2014, 13, 97. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, L.; Wang, L.; Wang, C.-Y.; Yu, J.; Guan, K.-L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012, 26, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-R.; Lin, C.; Lu, C.-C.; Kuo, S.-C.; Tsao, J.-W.; Juan, Y.-N.; Chiu, H.-Y.; Lee, F.-Y.; Yang, J.-S.; Tsai, F.-J. YC-1 induces G0/G1 phase arrest and mitochondria-dependent apoptosis in cisplatin-resistant human oral cancer CAR cells. BioMedicine 2017, 7, 12. [Google Scholar] [CrossRef]
- Karamitopoulou, E.; Haemmig, S.; Baumgartner, U.; Schlup, C.; Wartenberg, M.; Vassella, E. MicroRNA dysregulation in the tumor microenvironment influences the phenotype of pancreatic cancer. Mod. Pathol. 2017, 30, 1116–1125. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, L.; Liu, Y. Targeting thyroid receptor interacting protein 6 by microrna-589-5p inhibits cell proliferation, migration, and invasion in endometrial carcinoma. Cancer Biother. Radiopharm. 2019, 34, 529–536. [Google Scholar] [CrossRef]
- Song, T.; Hou, X.; Lin, B. Microrna-758 inhibits cervical cancer cell proliferation and metastasis by targeting hmgb3 through the wnt/beta-catenin signaling pathway. Oncol. Lett. 2019, 18, 1786–1792. [Google Scholar]
- Han, B.; Bhowmick, N.; Qu, Y.; Chung, S.; Giuliano, A.E.; Cui, X. FOXC1: An emerging marker and therapeutic target for cancer. Oncogene 2017, 36, 3957–3963. [Google Scholar] [CrossRef]
- Igarashi, H.; Kurihara, H.; Mitsuhashi, K.; Ito, M.; Okuda, H.; Kanno, S.; Naito, T.; Yoshii, S.; Takahashi, H.; Kusumi, T.; et al. Association of microrna-31-5p with clinical efficacy of anti-egfr therapy in patients with metastatic colorectal cancer. Ann. Surg. Oncol. 2015, 22, 2640–2648. [Google Scholar] [CrossRef]
- Dong, Y.; Wu, W.K.; Wu, C.W.; Sung, J.J.Y.; Yu, J.; Ng, S.S.M. MicroRNA dysregulation in colorectal cancer: A clinical perspective. Br. J. Cancer 2011, 104, 893–898. [Google Scholar] [CrossRef]
- Yang, X.; Xu, X.; Zhu, J.; Zhang, S.; Wu, Y.; Wu, Y.; Zhao, K.; Xing, C.; Cao, J.; Zhu, H.; et al. miR-31 affects colorectal cancer cells by inhibiting autophagy in cancer-associated fibroblasts. Oncotarget 2016, 7, 79617–79628. [Google Scholar] [CrossRef]
- Ye, J.-J.; Cao, J. MicroRNAs in colorectal cancer as markers and targets: Recent advances. World J. Gastroenterol. 2014, 20, 4288–4299. [Google Scholar] [CrossRef]
- Valastyan, S.; Reinhardt, F.; Benaich, N.; Calogrias, D.; Szasz, A.M.; Wang, Z.C.; Brock, J.E.; Richardson, A.L.; Weinberg, R.A. A pleiotropically acting microrna, mir-31, inhibits breast cancer metastasis. Cell 2009, 137, 1032–1046. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, J.; Li, D.; Xiao, B.; Miao, Y.; Jiang, Z.; Zhuo, H. Down-regulation of mir-31 expression in gastric cancer tissues and its clinical significance. Med. Oncol. 2010, 27, 685–689. [Google Scholar] [CrossRef]
- Feng, S.; Pan, W.; Jin, Y.; Zheng, J. MiR-25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumor Biol. 2014, 35, 12339–12344. [Google Scholar] [CrossRef]
- Lee, K.H.; Goan, Y.G.; Hsiao, M.; Lee, C.H.; Jian, S.H.; Lin, J.T.; Chen, Y.L.; Lu, P.J. Microrna-373 (mir-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (lats2) and stimulates proliferation in human esophageal cancer. Exp. Cell Res. 2009, 315, 2529–2538. [Google Scholar] [CrossRef]
- Xia, Y.; Gao, Y. MicroRNA-181b promotes ovarian cancer cell growth and invasion by targeting LATS2. Biochem. Biophys. Res. Commun. 2014, 447, 446–451. [Google Scholar] [CrossRef]
- Fang, L.; Du, W.W.; Yang, W.; Rutnam, Z.J.; Peng, C.; Li, H.; O’Malley, Y.Q.; Askeland, R.W.; Sugg, S.; Liu, M.; et al. MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle 2012, 11, 4352–4365. [Google Scholar] [CrossRef]
- Yi, J.; Lu, L.; Yanger, K.; Wang, W.; Sohn, B.H.; Stanger, B.Z.; Zhang, M.; Martin, J.F.; Ajani, J.A.; Chen, J.; et al. Large tumor suppressor homologs 1 and 2 regulate mouse liver progenitor cell proliferation and maturation through antagonism of the coactivators YAP and TAZ. Hepatology 2016, 64, 1757–1772. [Google Scholar] [CrossRef]
- Yu, C.; Wang, M.; Li, Z.; Xiao, J.; Peng, F.; Guo, X.; Deng, Y.; Jiang, J.; Sun, C. MicroRNA-138-5p regulates pancreatic cancer cell growth through targeting FOXC1. Cell. Oncol. 2015, 38, 173–181. [Google Scholar] [CrossRef]
- Ray, P.S.; Bagaria, S.P.; Wang, J.; Shamonki, J.M.; Ye, X.; Sim, M.-S.; Steen, S.; Qu, Y.; Cui, X.; Giuliano, A.E. Basal-Like Breast Cancer Defined by FOXC1 Expression Offers Superior Prognostic Value: A Retrospective Immunohistochemical Study. Ann. Surg. Oncol. 2011, 18, 3839–3847. [Google Scholar] [CrossRef]
- Ray, P.S.; Wang, J.; Qu, Y.; Sim, M.-S.; Shamonki, J.; Bagaria, S.P.; Ye, X.; Liu, B.; Elashoff, D.; Hoon, D.S.; et al. FOXC1 Is a Potential Prognostic Biomarker with Functional Significance in Basal-like Breast Cancer. Cancer Res. 2010, 70, 3870–3876. [Google Scholar] [CrossRef]
- Wang, J.; Ray, P.S.; Sim, M.S.; Zhou, X.Z.; Lu, K.P.; Lee, A.V.; Lin, X.; Bagaria, S.P.; Giuliano, A.E.; Cui, X. Foxc1 regulates the functions of human basal-like breast cancer cells by activating nf-kappab signaling. Oncogene 2012, 31, 4798–4802. [Google Scholar] [CrossRef]
- Xu, Y.; Shao, Q.-S.; Yao, H.-B.; Jin, Y.; Ma, Y.-Y.; Jia, L.-H. Overexpression of FOXC1 correlates with poor prognosis in gastric cancer patients. Histopathology 2014, 64, 963–970. [Google Scholar] [CrossRef]
- Wang, L.; Gu, F.; Liu, C.Y.; Wang, R.J.; Li, J.; Xu, J.Y. High level of foxc1 expression is associated with poor prognosis in pancreatic ductal adenocarcinoma. Tumour Biol. 2013, 34, 853–858. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, H.-H.; Kuo, W.-W.; Shih, H.-N.; Cheng, S.-F.; Yang, C.-K.; Chen, M.-C.; Tu, C.-C.; Viswanadha, V.P.; Liao, P.-H.; Huang, C.-Y. FOXC1 Regulation of miR-31-5p Confers Oxaliplatin Resistance by Targeting LATS2 in Colorectal Cancer. Cancers 2019, 11, 1576. https://doi.org/10.3390/cancers11101576
Hsu H-H, Kuo W-W, Shih H-N, Cheng S-F, Yang C-K, Chen M-C, Tu C-C, Viswanadha VP, Liao P-H, Huang C-Y. FOXC1 Regulation of miR-31-5p Confers Oxaliplatin Resistance by Targeting LATS2 in Colorectal Cancer. Cancers. 2019; 11(10):1576. https://doi.org/10.3390/cancers11101576
Chicago/Turabian StyleHsu, Hsi-Hsien, Wei-Wen Kuo, Hui-Nung Shih, Sue-Fei Cheng, Ching-Kuo Yang, Ming-Cheng Chen, Chuan-Chou Tu, Vijaya Padma Viswanadha, Po-Hsiang Liao, and Chih-Yang Huang. 2019. "FOXC1 Regulation of miR-31-5p Confers Oxaliplatin Resistance by Targeting LATS2 in Colorectal Cancer" Cancers 11, no. 10: 1576. https://doi.org/10.3390/cancers11101576
APA StyleHsu, H.-H., Kuo, W.-W., Shih, H.-N., Cheng, S.-F., Yang, C.-K., Chen, M.-C., Tu, C.-C., Viswanadha, V. P., Liao, P.-H., & Huang, C.-Y. (2019). FOXC1 Regulation of miR-31-5p Confers Oxaliplatin Resistance by Targeting LATS2 in Colorectal Cancer. Cancers, 11(10), 1576. https://doi.org/10.3390/cancers11101576




