A Pan-Cancer Approach to Predict Responsiveness to Immune Checkpoint Inhibitors by Machine Learning
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
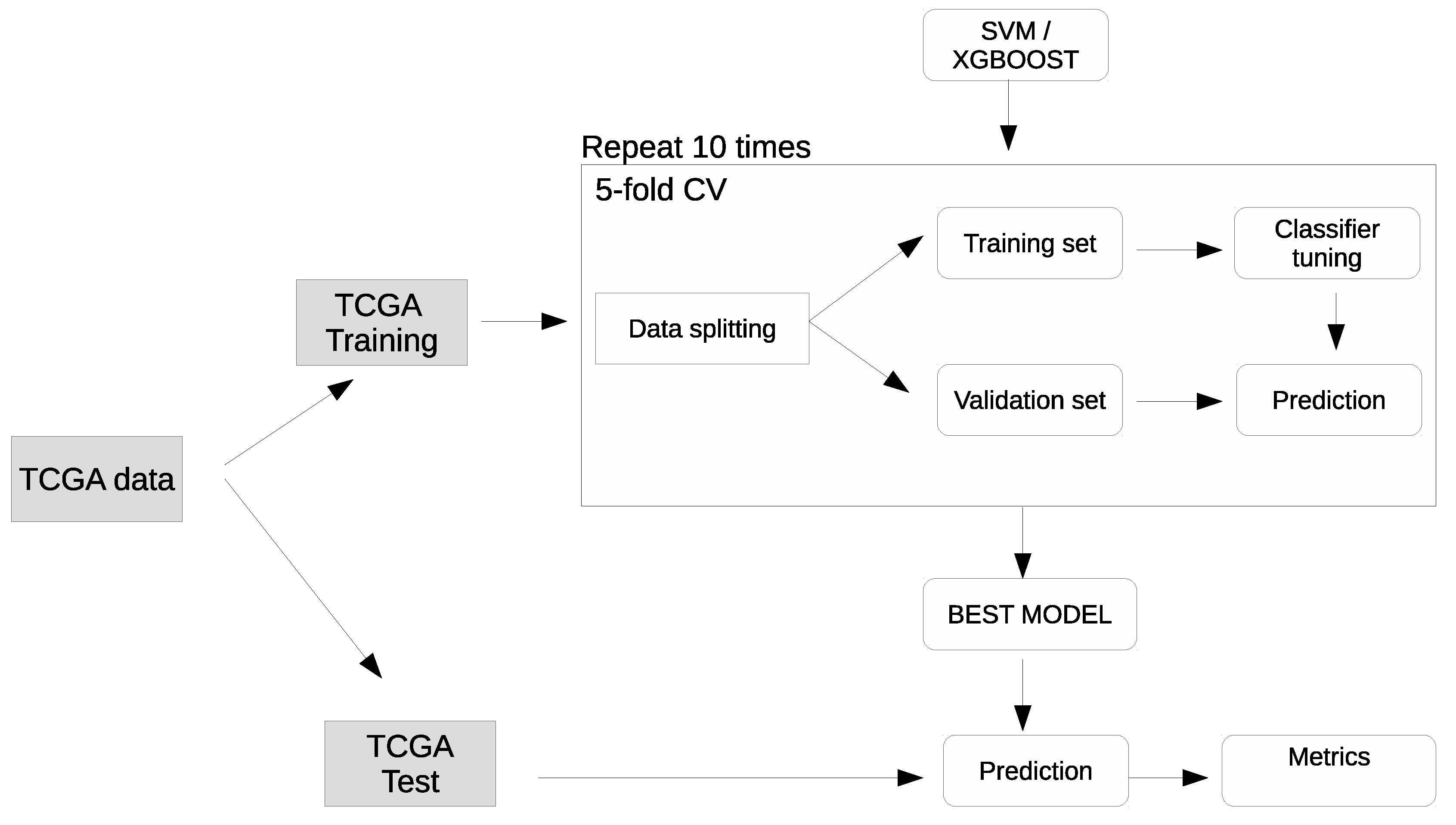
4.1. Datasets
4.2. Machine learning methods
4.3. Computational Details
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ICI | immune checkpoint inhibitors |
| TCGA | The Cancer Genome Atlas |
| XGboost | Extreme distributed gradient boosting library |
| SVM | Support Vector Machine |
| TMB | Tumor Mutational Burden |
| CI | confidence intervals |
| HR | Hazard ratio |
Appendix A


| Cancer Types | Label | Number of Samples | HR | 95% CI for HR | p Value |
|---|---|---|---|---|---|
| UVM | TMB/TGF- score positive | n = 80 | 0.3 | 0.04–2.2 | 0.24 |
| SKCM | TMB/TGF- score positive | n = 103 | 0.45 | 0.11–2 | 0.29 |
| GBM | TMB/TGF- score positive | n = 146 | 0.69 | 0.28–1.7 | 0.42 |
| LIHC | TMB/TGF- score positive | n = 349 | 1 | 0.59–1.8 | 0.91 |
| SARC | TMB/TGF- score positive | n = 201 | 0.85 | 0.41–1.8 | 0.66 |
| PCPG | TMB/TGF- score positive | n = 177 | 4.9 | 0.8–30 | 0.085 |
| TCGT | TMB/TGF- score positive | n = 127 | 1.5 | 0.89–2.6 | 0.12 |
| THCA | TMB/TGF- score positive | n = 481 | 0.87 | 0.11–6.7 | 0.89 |
| PAAD | TMB/TGF- score positive | n = 146 | 1 | 0.53–1.9 | 0.96 |
| PRAD | TMB/TGF- score positive | n = 410 | 3 | 0.74–12 | 0.12 |
| UCEC | TMB/TGF- score positive | n = 542 | 0.25 | 0.092–0.69 | 0.007 |
| CHOL | TMB/TGF- score positive | n = 35 | 2.2 | 0.69–7 | 0.18 |
| KICH | TMB/TGF- score positive | n = 64 | 6.4 | 1.6–26 | 0.0091 |
| BLCA | TMB/TGF- score positive | n = 412 | 0.67 | 0.42–1.1 | 0.096 |
| KIRP | TMB/TGF- score positive | n = 266 | 0.27 | 0.064–1.1 | 0.071 |
| HNSC | TMB/TGF- score positive | n = 488 | 1.1 | 0.8–1.6 | 0.48 |
| CESC | TMB/TGF- score positive | n = 282 | 0.48 | 0.19–1.2 | 0.11 |
| BRCA | TMB/TGF- score positive | n = 1009 | 0.94 | 0.57–1.6 | 0.81 |
| OV | TMB/TGF- score positive | n = 164 | 0.67 | 0.31–1.4 | 0.31 |
| LGG | TMB/TGF- score positive | n = 499 | 0.87 | 0.38–2 | 0.74 |
| LUAD | TMB/TGF- score positive | n = 492 | 0.58 | 0.33–1 | 0.05 |
| ESCA | TMB/TGF- score positive | n = 151 | 1.3 | 0.65–2.7 | 0.45 |
| READ | TMB/TGF- score positive | n = 125 | 0.86 | 0.19–3.9 | 0.85 |
| LUSC | TMB/TGF- score positive | n = 484 | 0.79 | 0.52–1.2 | 0.28 |
| COAD | TMB/TGF- score positive | n = 462 | 0.73 | 0.39–1.4 | 0.33 |
| UCS | TMB/TGF- score positive | n = 56 | 1.1 | 0.4–3.3 | 0.8 |
| MESO | TMB/TGF- score positive | n = 76 | 0.5 | 0.21–1.2 | 0.11 |
| ACC | TMB/TGF- score positive | n = 78 | 6.4 | 2.3–18 | 4 |
| STAD | TMB/TGF- score positive | n = 345 | 0.62 | 0.35–1.1 | 0.088 |
| Cluster Subtype | TMB/TGF- Score Negative | TMB/TGF- Score Positive | Total |
|---|---|---|---|
| 1 | 1957 | 266 | 2223 |
| 2 | 1994 | 379 | 2373 |
| 3 | 1704 | 166 | 1870 |
| 4 | 913 | 150 | 1063 |
| 5 | 334 | 36 | 370 |
| 6 | 149 | 7 | 156 |
References
- Friedrich, M.; Jasinski-Bergner, S.; Lazaridou, M.F.; Subbarayan, K.; Massa, C.; Tretbar, S.; Mueller, A.; Handke, D.; Biehl, K.; Bukur, J.; et al. Tumor-induced escape mechanisms and their association with resistance to checkpoint inhibitor therapy. Cancer Immunol. Immunother. 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Costantini, A.; Takam Kamga, P.; Dumenil, C.; Chinet, T.; Emile, J.F.; Giroux Leprieur, E. Plasma Biomarkers and Immune Checkpoint Inhibitors in Non-Small Cell Lung Cancer: New Tools for Better Patient Selection? Cancers 2019, 11, 1269. [Google Scholar] [CrossRef] [PubMed]
- Havel, J.J.; Chowell, D.; Chan, T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 2019, 19, 133. [Google Scholar] [CrossRef] [PubMed]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 165. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, M. Correlate tumor mutation burden with immune signatures in human cancers. BMC Immunol. 2019, 20, 4. [Google Scholar] [CrossRef]
- Prat, A.; Navarro, A.; Paré, L.; Reguart, N.; Galván, P.; Pascual, T.; Martínez, A.; Nuciforo, P.; Comerma, L.; Alos, L.; et al. Immune-Related Gene Expression Profiling After PD-1 Blockade in Non-Small Cell Lung Carcinoma, Head and Neck Squamous Cell Carcinoma, and Melanoma. Cancer Res. 2017, 77, 3540–3550. [Google Scholar] [CrossRef]
- Zhang, L.; Jones-O’Connor, M.; Awadalla, M.; Zlotoff, D.A.; Thavendiranathan, P.; Groarke, J.D.; Villani, A.C.; Lyon, A.R.; Neilan, T.G. Cardiotoxicity of Immune Checkpoint Inhibitors. Curr. Treat. Options Cardiovasc. Med. 2019, 21, 32. [Google Scholar] [CrossRef]
- Maleki Vareki, S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J. Immunother. Cancer 2018, 6, 157. [Google Scholar] [CrossRef]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef]
- Li, B.; Li, T.; Pignon, J.C.; Wang, B.; Wang, J.; Shukla, S.A.; Dou, R.; Chen, Q.; Hodi, F.S.; Choueiri, T.K.; et al. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat. Genet. 2016, 48, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Schulz, M.; Salamero-Boix, A.; Niesel, K.; Alekseeva, T.; Sevenich, L. Microenvironmental Regulation of Tumor Progression and Therapeutic Response in Brain Metastasis. Front. Immunol. 2019, 10, 1713. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed]
- Khong, H.T.; Restifo, N.P. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat. Immunol. 2002, 3, 999–1005. [Google Scholar] [CrossRef]
- Kather, J.N.; Suarez-Carmona, M.; Charoentong, P.; Weis, C.A.; Hirsch, D.; Bankhead, P.; Horning, M.; Ferber, D.; Kel, I.; Herpel, E.; et al. Topography of cancer-associated immune cells in human solid tumors. eLife 2018, 7, e36967. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Yang, T.H.O.; Porta-Pardo, E.; Gao, G.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef]
- Şenbabaoğlu, Y.; Gejman, R.S.; Winer, A.G.; Liu, M.; Van Allen, E.M.; de Velasco, G.; Miao, D.; Ostrovnaya, I.; Drill, E.; Luna, A.; et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016, 17, 231. [Google Scholar] [CrossRef]
- Danaher, P.; Warren, S.; Lu, R.; Samayoa, J.; Sullivan, A.; Pekker, I.; Wallden, B.; Marincola, F.M.; Cesano, A. Pan-cancer adaptive immune resistance as defined by the Tumor Inflammation Signature (TIS): Results from The Cancer Genome Atlas (TCGA). J. Immunother. Cancer 2018, 6, 63. [Google Scholar] [CrossRef]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFB attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.L. An R package for classification of immune subtypes, in cancer, using gene expression data. 2019; original-date: 2019-05-21T16:04:14Z. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2001. [Google Scholar]
- Matthews, B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Jurman, G.; Riccadonna, S.; Furlanello, C. A comparison of MCC and CEN error measures in multi-class prediction. PLoS ONE 2012, 7, e41882. [Google Scholar] [CrossRef] [PubMed]
- Bonotto, M.; Garattini, S.K.; Basile, D.; Ongaro, E.; Fanotto, V.; Cattaneo, M.; Cortiula, F.; Iacono, D.; Cardellino, G.G.; Pella, N.; et al. Immunotherapy for gastric cancers: Emerging role and future perspectives. Expert Rev. Clin. Pharmacol. 2017, 10, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Visconti, R.; Morra, F.; Guggino, G.; Celetti, A. The between Now and Then of Lung Cancer Chemotherapy and Immunotherapy. Int. J. Mol. Sci. 2017, 18, 1374. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A. Breast Cancer Immunotherapy: Facts and Hopes. Clin. Cancer Res. 2018, 24, 511–520. [Google Scholar] [CrossRef]
- Basile, D.; Garattini, S.K.; Bonotto, M.; Ongaro, E.; Casagrande, M.; Cattaneo, M.; Fanotto, V.; De Carlo, E.; Loupakis, F.; Urbano, F.; et al. Immunotherapy for colorectal cancer: Where are we heading? Expert Opin. Biol. Ther. 2017, 17, 709–721. [Google Scholar] [CrossRef]
- Cattrini, C.; Dellepiane, C.; Cavo, A.; Buzzatti, G.; Tolomeo, F.; Messina, C.; Boccardo, F. Immunotherapy for genitourinary cancer: State of the art and new perspectives. Anticancer Drugs 2016, 27, 585–599. [Google Scholar] [CrossRef]
- Angelova, M.; Charoentong, P.; Hackl, H.; Fischer, M.L.; Snajder, R.; Krogsdam, A.M.; Waldner, M.J.; Bindea, G.; Mlecnik, B.; Galon, J.; et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015, 16, 64. [Google Scholar] [CrossRef]
- Tamborero, D.; Rubio-Perez, C.; Muiños, F.; Sabarinathan, R.; Piulats, J.M.; Muntasell, A.; Dienstmann, R.; Lopez-Bigas, N.; Gonzalez-Perez, A. A Pan-cancer Landscape of Interactions between Solid Tumors and Infiltrating Immune Cell Populations. Clin. Cancer Res. 2018, 24, 3717–3728. [Google Scholar] [CrossRef]
- McGranahan, N.; Furness, A.J.S.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T.; et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Gilligan, B.M.; Yuan, J.; Li, T. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J. Hematol. Oncol. 2016, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Meléndez, B.; Van Campenhout, C.; Rorive, S.; Remmelink, M.; Salmon, I.; D’Haene, N. Methods of measurement for tumor mutational burden in tumor tissue. Transl. Lung Cancer Res. 2018, 7, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.M.; Davis, S.R. GenomicDataCommons R-Package | NCI Genomic Data Commons Access 2019. Available online: https://bioconductor.org/packages/GenomicDataCommons,http://github.com/Bioconductor/GenomicDataCommons (accessed on 31 May 2018).
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; McLellan, M.; et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018, 6, 271–281.e7. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Gibbs, D.L. This Repo Contains the Code Necessary to Reproduce the Clusters Found in “The Immune Landscape of Cancer”. Available online: https://github.com/Gibbsdavidl/Immune-Subtype-Clustering (accessed on 31 May 2018).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]

| Cohort | Cancer Type Full Name | Number of Cases | Percentage of TMB/TGF- Score Positive Cases |
|---|---|---|---|
| HNSC | head and neck squamous cell carcinoma | 488 | 15.57 |
| LUSC | lung squamous cell carcinoma | 476 | 14.71 |
| LIHC | liver hepatocellular carcinoma | 350 | 14.29 |
| UCEC | uterine corpus endometrial carcinoma | 511 | 14.29 |
| CESC | cervical squamous cell carcinoma and endocervical adenocarcinoma | 282 | 14.18 |
| BLCA | bladder urothelial carcinoma | 397 | 14.11 |
| STAD | stomach adenocarcinoma | 349 | 13.75 |
| PRAD | prostate adenocarcinoma | 401 | 13.72 |
| KIRP | kidney renal papillary cell carcinoma | 267 | 13.48 |
| BRCA | breast invasive carcinoma | 970 | 13.30 |
| ESCA | esophageal carcinoma | 151 | 13.25 |
| MESO | mesothelioma | 77 | 12.99 |
| SKCM | skin cutaneous melanoma | 103 | 12.62 |
| UCS | uterine carcinosarcoma | 56 | 12.50 |
| UVM | uveal melanoma | 80 | 12.50 |
| READ | rectum adenocarcinoma | 126 | 11.90 |
| THCA | thyroid carcinoma | 481 | 11.85 |
| COAD | colon adenocarcinoma | 383 | 11.75 |
| PAAD | pancreatic adenocarcinoma | 146 | 11.64 |
| CHOL | cholangiocarcinoma | 35 | 11.43 |
| TGCT | testicular germ cell tumors | 143 | 11.19 |
| PCPG | pheochromocytoma and paraganglioma | 177 | 10.73 |
| LUAD | lung adenocarcinoma | 450 | 10.22 |
| SARC | sarcoma | 201 | 9.95 |
| KICH | kidney chromophobe | 64 | 9.38 |
| LGG | brain lower grade glioma | 501 | 7.98 |
| OV | ovarian serous cystadenocarcinoma | 165 | 7.88 |
| ACC | adrenocortical carcinoma | 78 | 7.69 |
| GBM | glioblastoma multiforme | 147 | 4.08 |
| Endpoint | Status | Number of Samples | HR | 95% CI for HR | p Value |
|---|---|---|---|---|---|
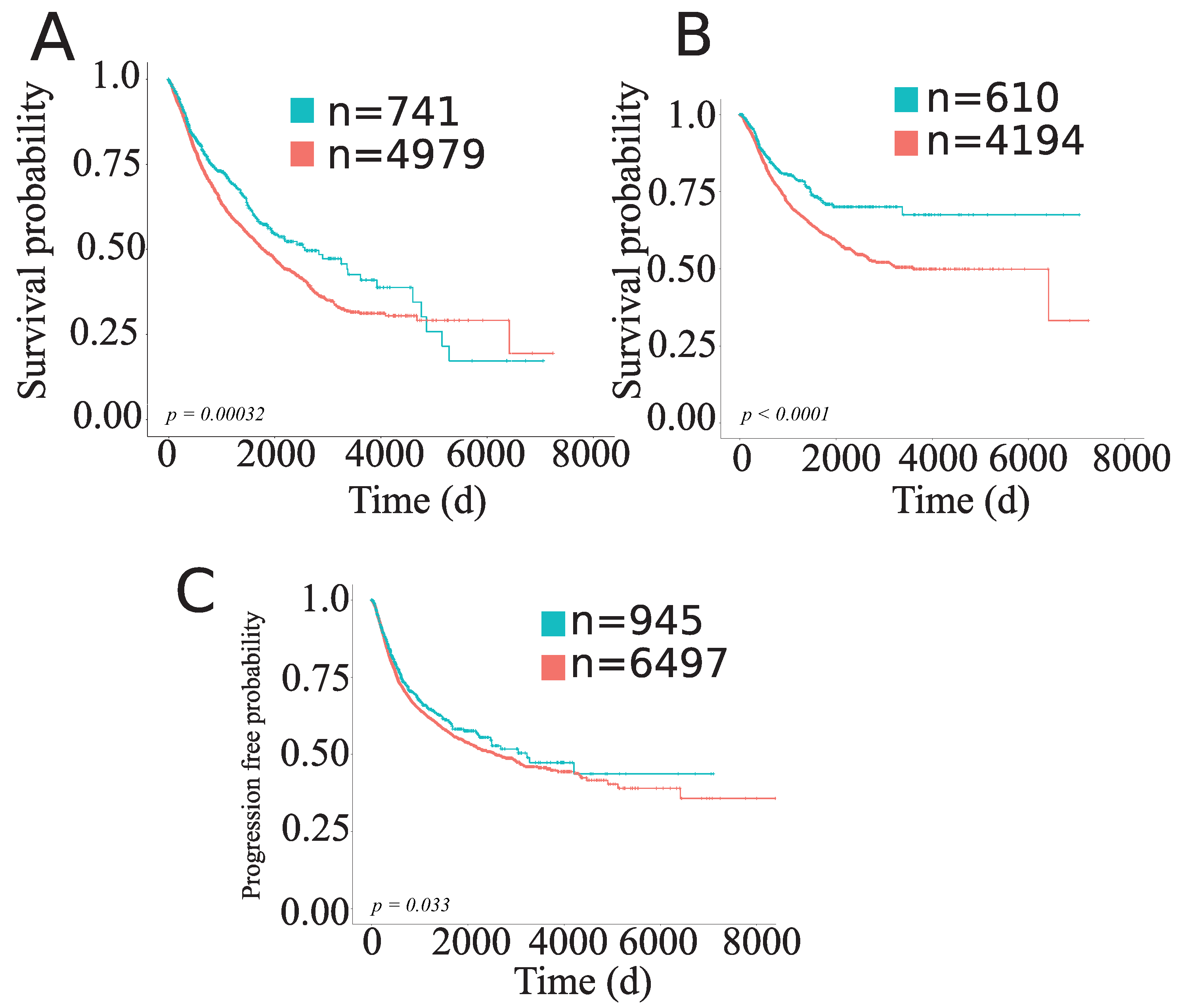
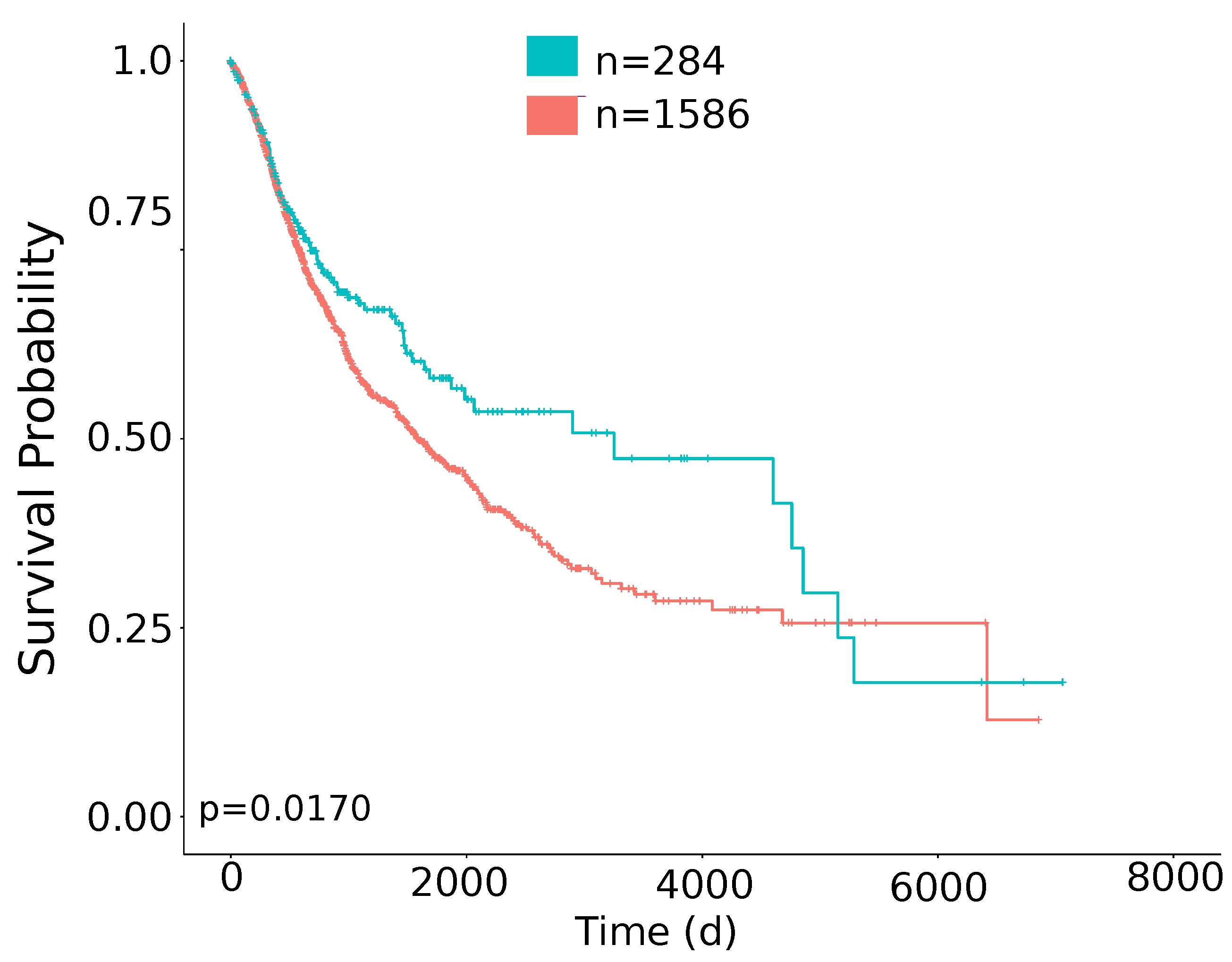
| OS | TMB/TGF- score positive | n = 8007 | 0.86 | 0.75–0.98 | 0.01 |
| DSS | TMB/TGF- score positive | n = 7741 | 0.79 | 0.67–0.93 | 0.0056 |
| PFI | TMB/TGF- score positive | n = 8007 | 0.89 | 0.79–0.99 | 0.059 |
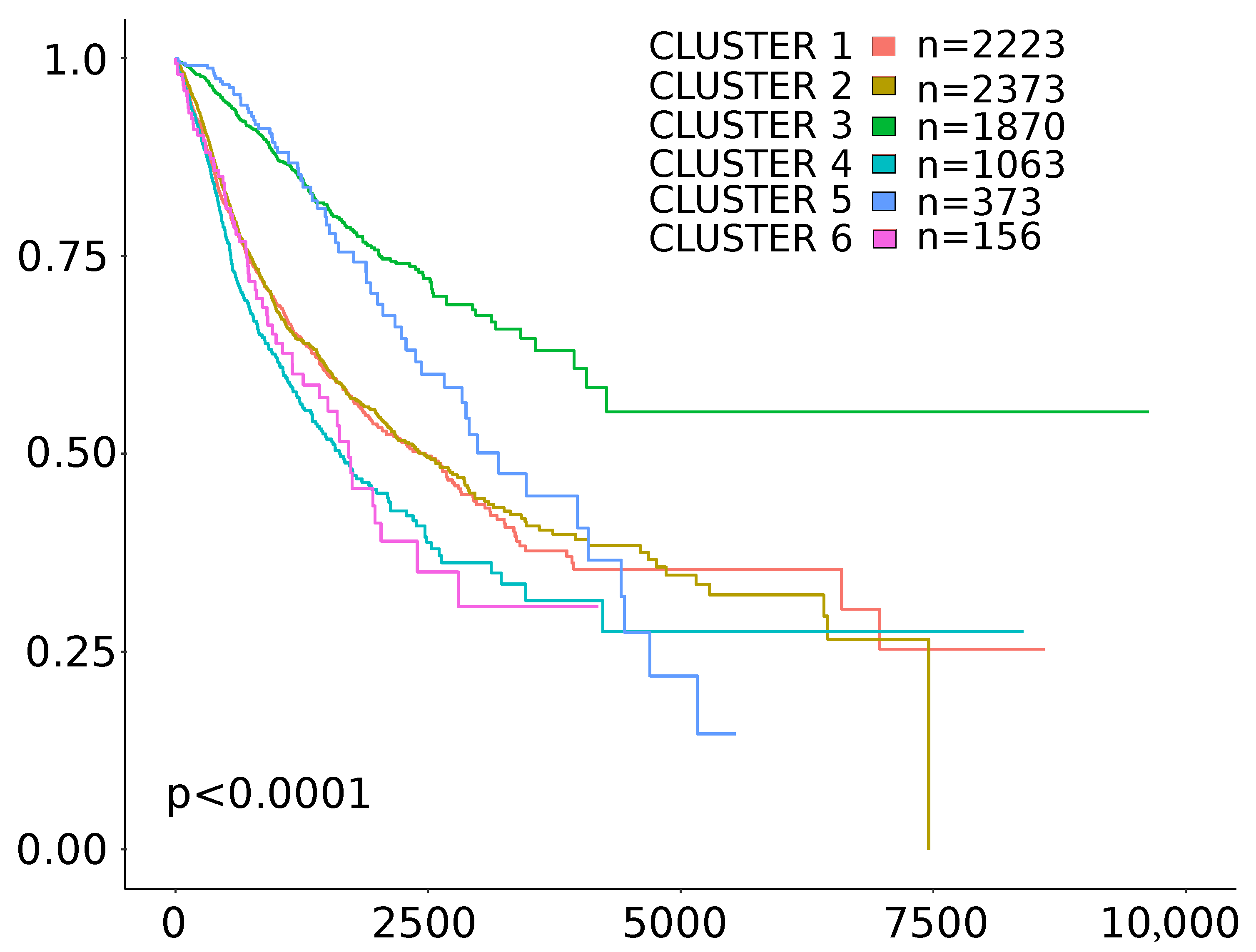
| Cluster | Status | Number of Samples | HR | 95% CI for HR | p Value |
|---|---|---|---|---|---|
| Cluster 1 | TMB/TGF- score positive | n = 2200 | 0.82 | 0.64–1 | 0.11 |
| Cluster 2 | TMB/TGF- score positive | n = 2357 | 0.76 | 0.61–0.93 | 0.0095 |
| Cluster 3 | TMB/TGF- score positive | n = 1867 | 0.84 | 0.53–1.3 | 0.48 |
| Cluster 4 | TMB/TGF- score positive | n = 1061 | 0.72 | 0.52–0.99 | 0.044 |
| Cluster 5 | TMB/TGF- score positive | n = 368 | 1.7 | 0.71–3.9 | 0.24 |
| Cluster 6 | TMB/TGF- score positive | n = 154 | 2.7 | 1.1–6.8 | 0.037 |
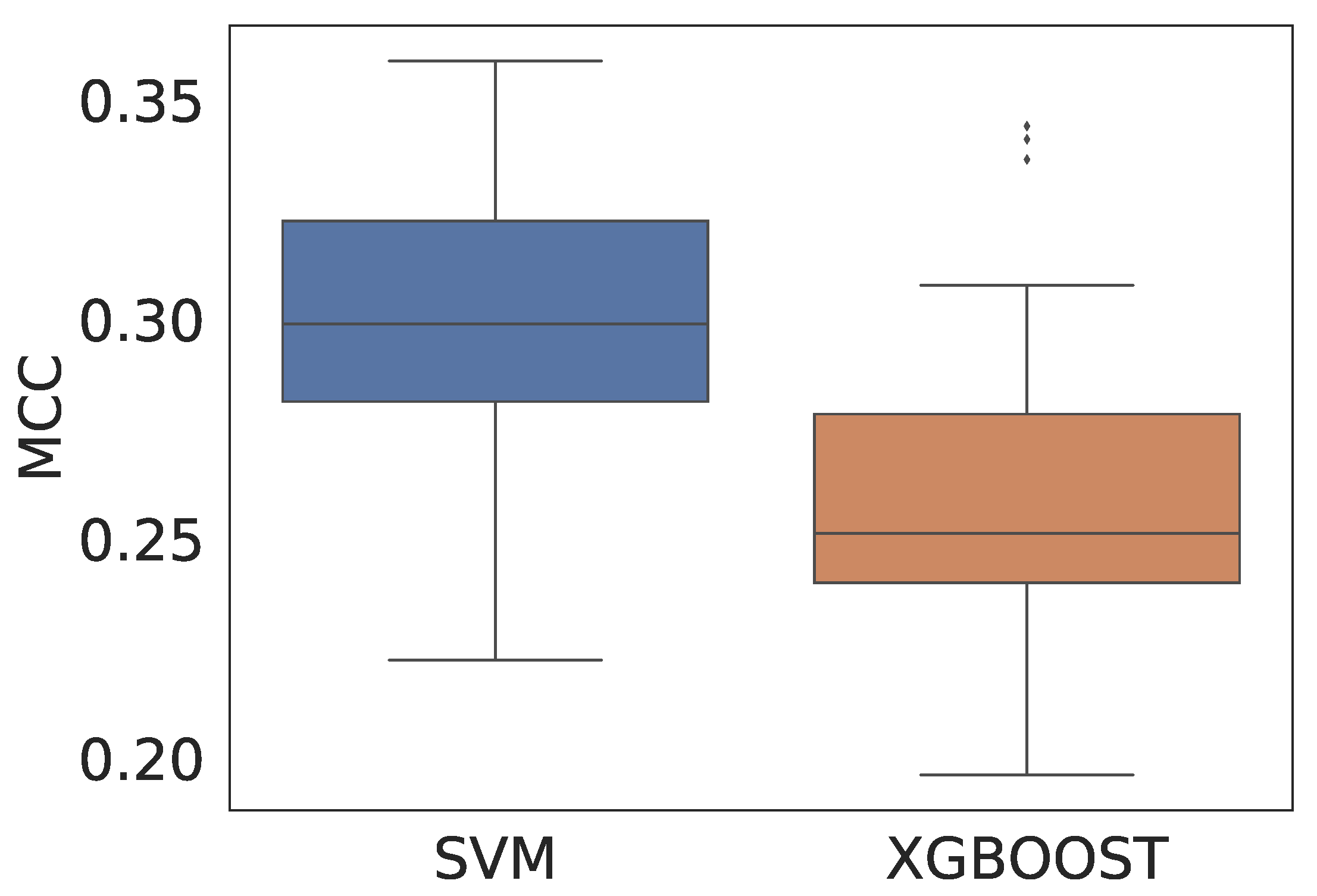
| Model | ACC (CI) | ACC Test | MCC (CI) | MCC Test |
|---|---|---|---|---|
| SVM | 0.879 (0.878–0.881) | 0.877 | 0.296 (0.287–0306) | 0.271 |
| XGBoost | 0.878 (0.877–0.880) | 0.879 | 0.260 (0.250–0.269) | 0.260 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polano, M.; Chierici, M.; Dal Bo, M.; Gentilini, D.; Di Cintio, F.; Baboci, L.; Gibbs, D.L.; Furlanello, C.; Toffoli, G. A Pan-Cancer Approach to Predict Responsiveness to Immune Checkpoint Inhibitors by Machine Learning. Cancers 2019, 11, 1562. https://doi.org/10.3390/cancers11101562
Polano M, Chierici M, Dal Bo M, Gentilini D, Di Cintio F, Baboci L, Gibbs DL, Furlanello C, Toffoli G. A Pan-Cancer Approach to Predict Responsiveness to Immune Checkpoint Inhibitors by Machine Learning. Cancers. 2019; 11(10):1562. https://doi.org/10.3390/cancers11101562
Chicago/Turabian StylePolano, Maurizio, Marco Chierici, Michele Dal Bo, Davide Gentilini, Federica Di Cintio, Lorena Baboci, David L. Gibbs, Cesare Furlanello, and Giuseppe Toffoli. 2019. "A Pan-Cancer Approach to Predict Responsiveness to Immune Checkpoint Inhibitors by Machine Learning" Cancers 11, no. 10: 1562. https://doi.org/10.3390/cancers11101562
APA StylePolano, M., Chierici, M., Dal Bo, M., Gentilini, D., Di Cintio, F., Baboci, L., Gibbs, D. L., Furlanello, C., & Toffoli, G. (2019). A Pan-Cancer Approach to Predict Responsiveness to Immune Checkpoint Inhibitors by Machine Learning. Cancers, 11(10), 1562. https://doi.org/10.3390/cancers11101562






