MicroRNAs Which Can Prognosticate Aggressiveness of Bladder Cancer
Abstract
1. Introduction
2. Materials and Methods
3. Data Analysis
4. Results
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cumberbatch, M.G.K.; Jubber, I.; Black, P.C.; Esperto, F.; Figueroa, J.D.; Kamat, A.M.; Kiemeney, L.; Lotan, Y.; Pang, K.; Silverman, D.T.; et al. Epidemiology of Bladder Cancer: A Systematic Review and Contemporary Update of Risk Factors in 2018. Eur. Urol. 2018, 74, 784–795. [Google Scholar] [CrossRef]
- Hurst, C.D.; Knowles, M.A. Bladder cancer: Multi-omic profiling refines the molecular view. Nat. Rev. Clin. Oncol. 2018, 15, 203–204. [Google Scholar] [CrossRef]
- Klaassen, Z.; Kamat, A.M.; Kassouf, W.; Gontero, P.; Villavicencio, H.; Bellmunt, J.; Van Rhijn, B.W.; Hartmann, A.; Catto, J.W.; Kulkarni, G.S. Treatment Strategy for Newly Diagnosed T1 High-grade Bladder Urothelial Carcinoma: New Insights and Updated Recommendations. Eur. Urol. 2018, 74, 597–608. [Google Scholar] [CrossRef]
- Lin-Brande, M.; Pearce, S.M.; Ashrafi, A.N.; Nazemi, A.; Burg, M.L.; Ghodoussipour, S.; Miranda, G.; Djaladat, H.; Schuckman, A.; Daneshmand, S. Assessing the Impact of Time to Cystectomy for Variant Histology of Urothelial Bladder Cancer. Urology 2019. [Google Scholar] [CrossRef]
- Psutka, S.P.; Barocas, D.A.; Catto, J.W.F.; Gore, J.L.; Lee, C.T.; Morgan, T.M.; Master, V.A.; Necchi, A.; Rouprêt, M.; Boorjian, S.A. Staging the Host: Personalizing Risk Assessment for Radical Cystectomy Patients. Eur. Urol. Oncol. 2018, 1, 292–304. [Google Scholar] [CrossRef]
- Palou, J.; Brausi, M.; Catto, J.W.F. Management of Patients with Normal Cystoscopy but Positive Cytology or Urine Markers. Eur. Urol. Oncol. 2019. [Google Scholar] [CrossRef]
- Sloan, F.A.; Yashkin, A.P.; Akushevich, I.; Inman, B.A. The Cost to Medicare of Bladder Cancer Care. Eur. Urol. Oncol. 2019. [Google Scholar] [CrossRef]
- Guancial, E.A.; Bellmut, J.; Yeh, S.; Rosenberg, J.E.; Berman, D.M. The evolving understanding of microRNA in bladder cancer. Urol. Oncol. 2014, 32, e31–e41. [Google Scholar] [CrossRef]
- Li, Q.; Wang, H.; Peng, H.; Huang, Q.; Huyan, T.; Huang, Q.; Yang, H.; Shi, J. MicroRNAs: Key Players in Bladder Cancer. Mol. Diagn. Ther. 2019. [Google Scholar] [CrossRef]
- Fang, Z.; Dai, W.; Wang, X.; Chen, W.; Shen, C.; Ye, G.; Li, L. Circulating miR-205: A promissing biomarker for detection and prognosis evaluation of bladder cancer. Tumor. Biol. 2016, 37, 8075–8082. [Google Scholar] [CrossRef]
- Miah, S.; Dudziec, E.; Dryton, R.M.; Zlotta, A.R.; Morgan, S.L.; Rosario, D.J.; Hamdy, F.C.; Catto, J.W.F. An evaluation of urinary micro RNA reveals a high sensitivity for bladder cancer. Br. J. Cancer 2012, 107, 123–128. [Google Scholar] [CrossRef]
- Zhou, B.; Guo, R. Integrative analysis of genomic and clinical data reveals intrinsic characteristics of bladder urothelial carcinoma progression. Genes 2019, 10, 464. [Google Scholar] [CrossRef]
- Jensen, S.G.; Lamy, P.; Rasmussen, M.H.; Ostenfeld, M.S.; Dyrskjøt, L.; Ørntoft, T.F.; Andersen, C.L. Evaluation of two commercial global miRNA expression profiling platforms for detection of less abundant miRNAs. BMC Genom. 2011, 12, 435. [Google Scholar] [CrossRef]
- Enokida, H.; Yoshino, H.; Matsushita, R.; Nakagawa, M. The role of microRNAs in bladder cancer. Investig. Clin. Urol. 2016, 57 (Suppl. 1), S60–S76. [Google Scholar] [CrossRef]
- Lee, J.Y.; Ryu, D.S.; Kim, W.J.; Kim, S.J. Aberrantly expressed microRNAs in the context of bladder tumorigenesis. Investig. Clin. Urol. 2016, 57 (Suppl. 1), S52–S59. [Google Scholar] [CrossRef]
- Sobin, L.H.; Gospodarowicz, M.K.; Wittekind, C.; IUAC (Eds.) TNM Classification of Malignant Tumors, 6th ed.; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Compérat, E.M.; Burger, M.; Gontero, P.; Mostafid, A.H.; Palou, J.; Rouprêt, M.; van Rhijn, B.W.G.; Shariat, S.F.; Sylvester, R.J.; Zigeuner, R.; et al. Grading of Urothelial Carcinoma and The New “World Health Organisation Classification of Tumours of the Urinary System and Male Genital Organs 2016”. Eur. Urol. Focus 2018. [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; Vandesompele, J.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Xiayu, R.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2˅(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Schwarzenbach, H.; da Silva, A.M.; Calin, G.; Pantel, K. Data normalization strategies for microRNA quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef]
- van Kessel, K.E.M.; van der Keur, K.A.; Dyrskjøt, L.; Algaba, F.; Welvaart, N.Y.; Beukers, W.; Segersten, U.; Keck, B.; Maurer, T.; Simic, T.; et al. Molecular Markers Increase Precision of the European Association of Urology Non-Muscle-Invasive Bladder Cancer Progression Risk Groups. Clin. Cancer Res. 2018, 24, 1586–1593. [Google Scholar] [CrossRef]
- Bruchbacher, A.; Soria, F.; Hassler, M.; Shariat, S.F.; D’Andrea, D. Tissue biomarkers in nonmuscle-invasivebladder cancer: Any role in clinical practice? Curr. Opin. Urol. 2018, 28, 584–590. [Google Scholar] [CrossRef]
- Soria, F.; Krabbe, L.M.; Todenhöfer, T.; Dobruch, J.; Mitra, A.P.; Inman, B.A.; Gust, K.M.; Lotan, Y.; Shariat, S.F. Molecular markers in bladder cancer. World J. Urol. 2019, 37, 31–40. [Google Scholar] [CrossRef]
- Miyake, M.; Owari, T.; Hori, S.; Fujimoto, K. Significant lack of urine-based biomarkers to replace cystoscopy for the surveillance of non-muscle invasive bladder cancer. Transl. Androl. Urol. 2019, 8 (Suppl. 3), S332–S334. [Google Scholar] [CrossRef]
- Lee, J.Y.; Yun, S.J.; Jeong, P.; Piao, X.-M.; Kim, Y.-H.; Kim, J.; Subramaniyam, S.; Byun, Y.J.; Kang, H.W.; Seo, S.P.; et al. Identification of differentially expressed miRNAs and miRNA-targeted genes in bladder cancer. Oncotarget 2018, 9, 27656–27766. [Google Scholar] [CrossRef]
- Ratert, N.; Meyer, H.A.; Jung, M.; Lioudmer, P.; Mollenkopf, H.; Wagner, I.; Miller, K.; Kilic, E.; Erbersdobler, A.; Weikert, S.; et al. miRNA profiling identifies candidate miRNAs for bladder cancerdiagnosis and clinical outcome. J. Mol. Diagn. 2013, 15. [Google Scholar] [CrossRef]
- Ratert, N.; Meyer, H.A.; Jung, M.; Mollenkopf, H.-J.; Wagner, I.; Miller, K.; Kilic, E.; Erbersdobler, A.; Weikert, S.; Jung, K. Reference miRNAs for miRNAome analysis of urothelial carcinomas. PLoS ONE 2012, 7, e39309. [Google Scholar] [CrossRef]
- Peltier, H.J.; Latham, G.J. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 2008, 14, 844–852. [Google Scholar] [CrossRef]
- Hofbauer, S.L.; de Martino, M.; Lucca, I.; Haitel, A.; Susani, M.; Shariat, S.F.; Klatte, T. A urinary microRNA (miR) signature for diagnosis of bladder cancer. Urol. Oncol. 2018, 36, 531.e1–531.e8. [Google Scholar] [CrossRef]
- Boisen, M.K.; Dehlendorff, C.; Linnemann, D.; Schultz, N.A.; Jensen, B.V.; Hogdal, E.V.S.; Johansen, J.S. MicroRNA expression in formalin-fixed paraffin-embedded cancer tissue: Identifying reference microRNAs and variability. BMC Cancer 2015, 15, 1024. [Google Scholar] [CrossRef]
- Parvaee, P.; Sarmadian, H.; Khansarinejad, B.; Amini, M.; Mondanizadeh, M. Plasma level of microRNAs, miR-107, miR-194 and miR-210 as potential biomarkers for diagnosis intestinal-type gastric cancer in human. Asian Pac. J. Cancer Prev. 2019, 20, 1421–1426. [Google Scholar] [CrossRef]
- Lenherr, S.; Tsai, S.; Neto, B.S.; Sullivan, T.B.; Cimmino, C.B.; Logvinenko, T.; Gee, J.; Huang, W.; Libertino, J.A.; Summerhayes, I.C.; et al. MicroRNA expression profile identifies high grade, non-muscle-invasive bladder tumors at elevated risk to progress to an invasive phenotype. Genes 2017, 8, 77. [Google Scholar] [CrossRef]
- Dip, N.; Reis, S.T.; Timoszczuk, L.S.; Viana, N.I.; Piantino, C.B.; Morais, D.R.; Moura, C.M.; Abe, D.K.; Silva, I.A.; Srougi, M.; et al. Stage, grade and behavior of bladder urothelial carcinoma defined by the microRNA expression profile. J. Urol. 2012, 188, 1951–1956. [Google Scholar] [CrossRef]
- Ecke, T.H.; Stier, K.; Weickmann, S.; Zhao, Z.; Buckendahl, L.; Stephan, C.; Kilic, E.; Jung, K. miR-199a-3p and miR-214-3p improve the overall survival prediction of muscle-invasive bladder cancer patients after radical cystectomy. Cancer Med. 2017, 6, 2252–2262. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, M.; Liu, Q.; Han, Z.; Zhao, Y.; Ji, S. mir-145-5p inhibits the proliferation and migration of bladder cancer cells by targeting TAGLN2. Oncol. Lett. 2018, 16, 6355–6360. [Google Scholar] [CrossRef]
- Li, D.; Hao, X.; Song, Y. An integrated analysis of key microRNAs, regulatory pathways and clinical relevance in bladder cancer. Onco Targets Ther. 2018, 11, 3075–3085. [Google Scholar] [CrossRef]
- Inamoto, T.; Uehara, H.; Akao, Y.; Ibuki, N.; Komura, K.; Takahara, K.; Takai, T.; Uchimoto, T.; Saito, K.; Tanda, N.; et al. A Panel of MicroRNA Signature as a Tool for Predicting Survival of Patients with Urothelial Carcinoma of the Bladder. Dis. Markers 2018, 2018, 5468672. [Google Scholar] [CrossRef]
- Pignot, G.; Cizeron-Clairac, G.; Vacher, S.; Susini, A.; Tozlu, S.; Vieillefond, A.; Zerbib, M.; Lidereau, R.; Debre, B.; Amsellem-Ouazana, D. microRNA expression profile in a large series of bladder cancer tumors: Identyfication of 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int. J. Cancer 2013, 132, 2479–2491. [Google Scholar] [CrossRef]
- Urquidi, V.; Netherton, M.; Gomes-Giacoia, E.; Serie, D.J.; Eckel-Passow, J.; Rosser, C.J.; Goodison, S. A microRNA biomarker panel for the non-invasive detection of bladder cancer. Oncotarget 2016, 7, 66290–86299. [Google Scholar] [CrossRef]
- Armstromg, D.A.; Green, B.B.; Seigne, J.D.; Schned, J.D.; Marsit, C.J. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol. Cancer 2015, 14, 194. [Google Scholar] [CrossRef]
- Baumgart, S.; Holters, S.; Ohlmann, C.H.; Bohle, R.; Stockle, M.; Ostenfeld, M.S.; Dyrskjøt, L.; Junker, K.; Heinzelmann, J. Exosome of invasive urothelial carcinoma cells are characterized by a specific miRNA expression signature. Oncotarget 2017, 8, 58278–58291. [Google Scholar] [CrossRef]
- Egawa, H.; Jingushi, K.; Hirono, T.; Ueda, Y.; Kitae, K.; Nakata, W.; Fujita, K.; Uemura, M.; Nonomura, N.; Tsujikawa, K. The. miR-130 family promotes cell migration and invasion inbladder cancerthrough FAK and Akt phosphorylation by regulating PTEN. Sci. Rep. 2016, 6, 20574. [Google Scholar] [CrossRef]
- Lv, M.; Zhong, Z.; Chi, H.; Huang, M.; Jiang, R.; Chen, J. Genome-Wide Screen of miRNAs and Targeting mRNAs Reveals the Negatively Regulatory Effect of miR-130b-3p on PTEN by PI3K and Integrin β1 Signaling Pathways in Bladder Carcinoma. Int. J. Mol. Sci. 2016, 18, 78. [Google Scholar] [CrossRef]
- Liu, X.; Kong, C.; Zhang, Z. miR-130bpromotesbladder cancer cell proliferation, migration and invasion by targeting VGLL4. Oncol. Rep. 2018, 39, 2324–2332. [Google Scholar] [CrossRef]


| FCmiR-145 | p-Value | FCmiR-21 | p-Value | FCmiR-182 | p-Value | Abnormal Expression 1 | p-Value | Abnormal Expression 2 | p-Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinicopathological Parameters | HE n (%) | LE n (%) | HE n (%) | LE n (%) | HE n (%) | LE n (%) | Yes n (%) | No n (%) | Yes n (%) | No n (%) | ||||||
| Total | 55 | |||||||||||||||
| Sex | ||||||||||||||||
| Female | 4 (7.27%) | 6 (10.91%) | 3 (5.45%) | 7 (12.73%) | 5 (9.09%) | 5 (9.09%) | 7 (12.73%) | 3 (5.45%) | 3 (5.45%) | 7 (12.73%) | ||||||
| Male | 26 (47.27%) | 19 (34.55%) | 0.503 (Y) | 9 (16.36%) | 36 (65.45%) | 0.787 (Y) | 26 (47.27%) | 19 (34.55%) | 0.923 (Y) | 34 (61.82%) | 11 (20%) | 0.971 (Y) | 20 (36.36%) | 25 (45.45%) | 0.629 (Y) | |
| Age at Diagnosis | ||||||||||||||||
| <60 | 2 (3.64%) | 4 (7.27%) | 1 (1.82%) | 5 (9.09%) | 6 (10.91%) | 0 (0%) | 6 (10.91%) | 0 (0%) | 2 (3.64%) | 4 (7.27%) | ||||||
| >60 | 28 (50.91%) | 21 (38.18%) | 0.502 (Y) | 11 (20%) | 38 (69.09%) | 0.841 (Y) | 25 (45.45%) | 24 (43.64%) | 0.064 (Y) | 35 (63.64%) | 14 (25.45%) | 0.308 (Y) | 21 (38.18%) | 28 (50.91%) | 0.994 (Y) | |
| Smoking Status | ||||||||||||||||
| Yes | 23 (41.82%) | 23 (41.82%) | 9 (16.36%) | 37 (67.27%) | 26 (47.27%) | 20 (36.36%) | 34 (61.82%) | 12 (21.82%) | 17 (30.91%) | 29 (52.73%) | ||||||
| No | 7 (12.73%) | 2 (3.64) | 0.244 (Y) | 3 (5.45%) | 6 (10.91%) | 0.636 (Y) | 5 (9.09%) | 4 (7.27%) | 0.753 (Y) | 7 (12.73%) | 2 (3.64%) | 0.861 (Y) | 6 (10.91%) | 3 (5.45%) | 0.199 (Y) | |
| Occupatinal Exposure | ||||||||||||||||
| Yes | 21 (38.18%) | 19 (34.55%) | 6 (10,91%) | 34 (61.82%) | 21 (38.18%) | 19 (34.55%) | 28 (50.91%) | 12 (21.82%) | 15 (27.27%) | 25 (45.45%) | ||||||
| No | 9 (16.36%) | 6 (10.91%) | 0.622 (V) | 6 (10.91%) | 9 (16.36%) | 0.102 (Y) | 10 (18.18%) | 5 (9.09%) | 0.349 | 13 (23.64%) | 2 (3.64%) | 0.359 (Y) | 8 (14.55%) | 7 (12.73%) | 0.293 (V) | |
| Tumour Stage | ||||||||||||||||
| Ta | 9 (16.36%) | 10 (18.18%) | 1 (1.82%) | 18 (32.73%) | 11 (20%) | 8 (14.55%) | 14 (25.45%) | 5 (9.09%) | 6 (10.91%) | 13 (23.64%) | ||||||
| T1 | 10 (18.18%) | 8 (14.55%) | 6 (10.91%) | 12 (21.82%) | 9 (16.36%) | 9 (16.36%) | 13 (23.64%) | 5 (9.09%) | 8 (14.55%) | 10 (18.18%) | ||||||
| T2 | 11 (20%) | 7 (12.73%) | 0.699 | 5 (9.09%) | 13 (23.64%) | 0.089 | 11 (20%) | 7 (12.73%) | 0.786 | 14 (25.45%) | 4 (7.27%) | 0.924 | 9 (16.36%) | 9 (16.36%) | 0.505 | |
| Grade | ||||||||||||||||
| high grade | 13 (23.64%) | 9 (16.36%) | 5 (9.09%) | 17 (30.91%) | 12 (21.82%) | 10 (18.18%) | 16 (29.09%) | 6 (10.91%) | 11 (20%) | 11 (20%) | ||||||
| low grade | 17 (30.91%) | 16 (29.09) | 0.580 | 7 (12.73%) | 26 (47.27%) | 0.841 (Y) | 19 (34.55%) | 14 (25.45%) | 0.826 (V) | 25 (45.45%) | 8 (14.55%) | 0.802 (V) | 12 (21.82%) | 21 (38,18%) | 0.319 (V) | |
| Recurrence | ||||||||||||||||
| Yes | 13 (23.64%) | 13 (23.64%) | 3 (5.45%) | 23 (41.82%) | 16 (29.09%) | 10 (18.18%) | 21 (38.18%) | 5 (9.09%) | 9 (16.36%) | 17 (30.91%) | ||||||
| No | 17 (30.91%) | 12 (21.82%) | 0.521 | 9 (16.36%) | 20 (36.36%) | 0.083 (V) | 15 (27.27%) | 14 (25.45%) | 0.463 | 20 (36.36%) | 9 (16.36%) | 0.320 (V) | 14 (25.45%) | 15 (27.27%) | 0.305 | |
| Progression | ||||||||||||||||
| Yes | 17 (30.91%) | 13 (23.64%) | 7 (12.73%) | 23 (41.82%) | 16 (29.09%) | 14 (25.45%) | 21 (38.18%) | 9 (16.36%) | 14 (25.45%) | 16 (29.09%) | ||||||
| No | 13 (23.64%) | 12 (21.82%) | 0.729 | 5 (9.09%) | 20 (36.36%) | 0.767 (V) | 15 (27.27%) | 10 (18.18%) | 0.619 | 20 (36.36%) | 5 (9.09%) | 0.401 (V) | 9 (16.36%) | 16 (29.09%) | 0.424 | |
| Death | ||||||||||||||||
| Yes | 10 (18.18%) | 7 (12.73%) | 3 (5.45%) | 14 (25.45%) | 9 (16.36%) | 8 (14.55%) | 11 (20%) | 6 (10.91%) | 9 (16.36%) | 8 (14.55%) | ||||||
| No | 20 (36.36%) | 18 (32.73%) | 0.673 (V) | 9 (16.36%) | 29 (52.73%) | 0.882 (Y) | 22 (40%) | 16 (29.09%) | 0.734 | 30 (54.55%) | 8 (14.55%) | 0.432 (Y) | 14 (25.45%) | 24 (43.64%) | 0.267 (V) | |
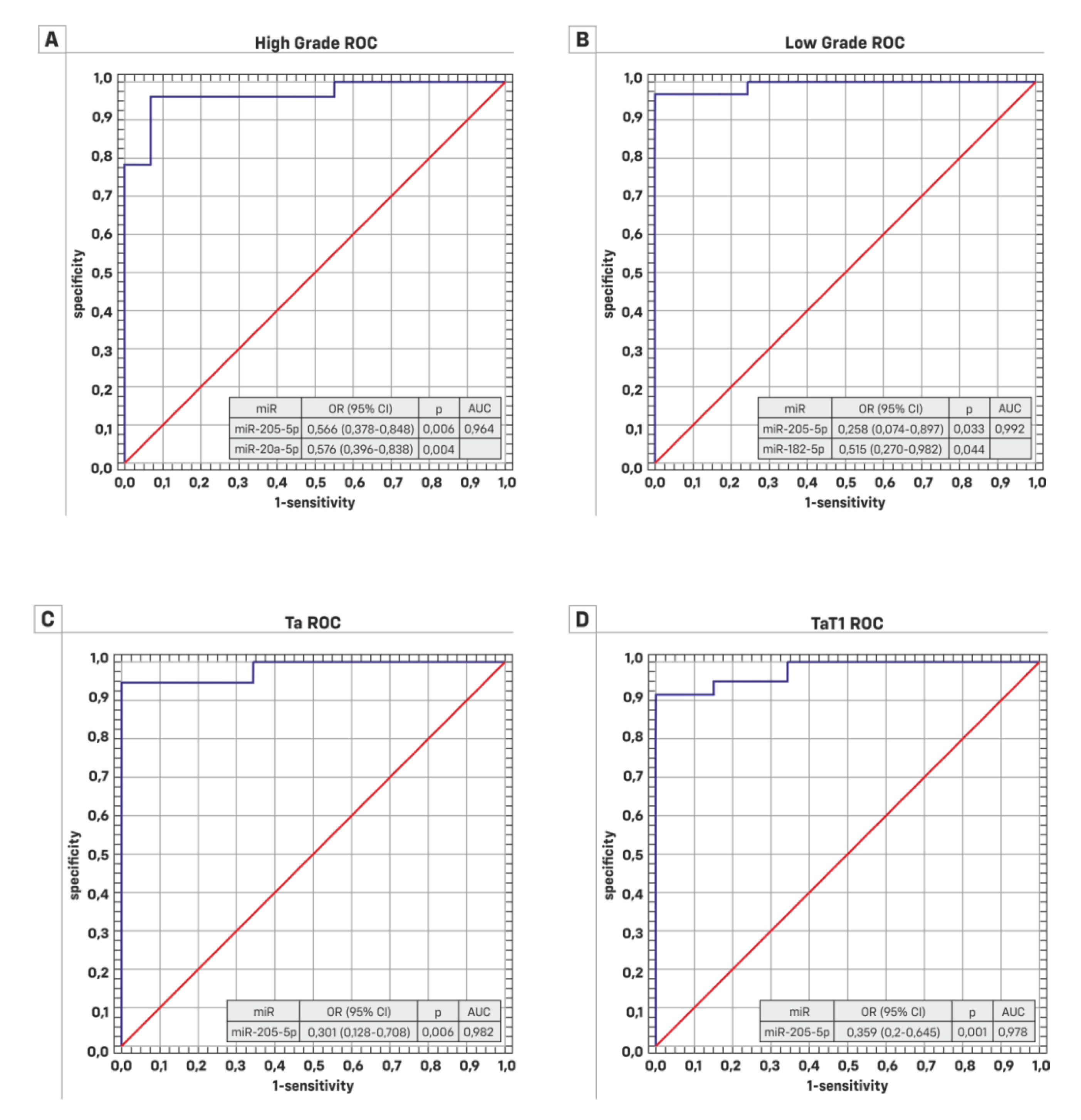
| A) | TaT1 p-value | T2 p-value |
| miR-145-5p | 0.4357505 * | 0.055556 |
| miR-205-5p | 0.440646 | 0.929801 |
| miR-130b-3p | 0.001136 * | 0.2648165 * |
| miR-21-5p | 0.421321 | 0.724233 |
| miR-20a-5p | 0.115487 | 0.1028555 * |
| miR-182-5p | 0.126511 * | 0.269855 * |
| miR-10a-5p | 0.3987205 * | 0.2946955 * |
| B) | HG p-value | LG p-value |
| miR-145-5p | 0.132994 | 0.336568 * |
| miR-205-5p | 0.065169 | 0.030956 * |
| miR-130b-3p | 0.00531 * | 0.138824 * |
| miR-21-5p | 0.606318 | 0.141797 * |
| miR-20a-5p | 0.019231 | 0.038561 |
| miR-182-5p | 0.037793 * | 0.015572 * |
| miR-10a-5p | 0.06102 * | 0.081524 * |
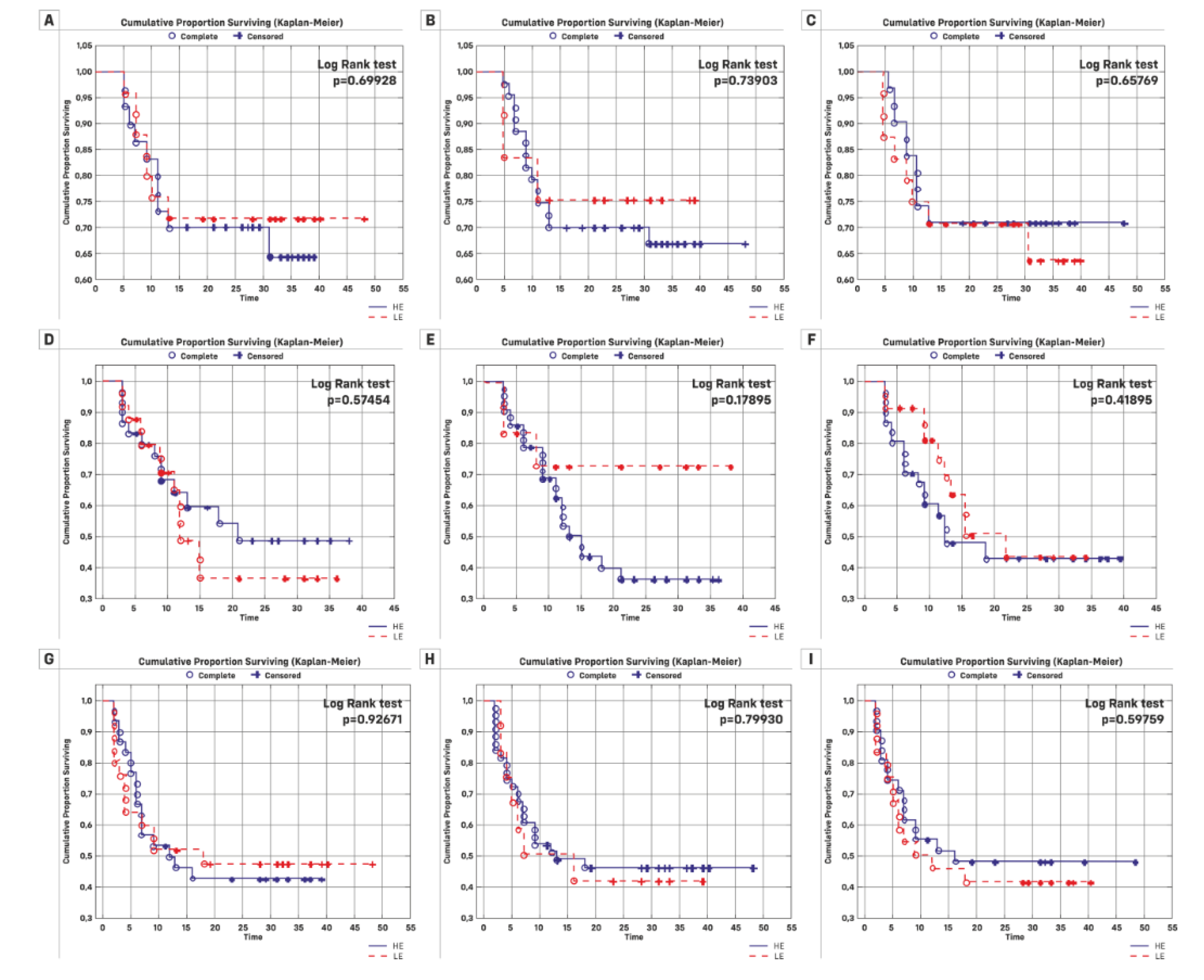
| Kaplan-Meier Analysis | |||||||
|---|---|---|---|---|---|---|---|
| Overall Survival | Recurrence | Progression | |||||
| Overall n (%) | Rate | Log-Rank Value | Rate | Log-Rank Value | Rate | Log-Rank Value | |
| Total | 55 | ||||||
| FCmiR-145 | |||||||
| HE | 30 | 10 | 13 | 17 | |||
| LE | 25 | 7 | 0.6992 | 13 | 0.5745 | 13 | 0.9267 |
| FCmiR-21 | |||||||
| HE | 12 | 3 | 3 | 7 | |||
| LE | 43 | 14 | 0.7390 | 23 | 0.1789 | 7 | 0.7993 |
| FCmiR-182 | |||||||
| HE | 31 | 9 | 16 | 16 | |||
| LE | 24 | 8 | 0.6576 | 10 | 0.4189 | 14 | 0.5976 |
| Total | 55 | ||||||
| Abnormal expression 1 | |||||||
| Yes | 41 | 11 | 21 | 21 | |||
| No | 14 | 6 | 0.2875 | 5 | 0.3499 | 9 | 0.2847 |
| Abnormal expression 2 | |||||||
| Yes | 23 | 9 | 9 | 14 | |||
| No | 32 | 8 | 0.2551 | 17 | 0.6881 | 16 | 0.5205 |
| Overall Survival | Time to Recurrence | Time to Progression | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | HR (95% CI) | p-value | p-value for Chi2 | Beta | HR (95% CI) | p-value | p-value for Chi2 | Beta | HR (95% CI) | p-value | p-value for Chi2 | |
| Gender | −0.57 | 0.56 (0.13–2.47) | 0.448 | 0.415 | −0.816 | 0.44 (0.13–1.47) | 0.184 | 0.142 | 0.029 | 1.03 (0.42–2.42) | 0.948 | 0.948 |
| Age at diagnosis | 0.266 | 1.30 (1.17-1.45) | 0.000 | 0.000 | -0.002 | 0.997 (0.99-1.005) | 0.526 | 0.524 | 0.068 | 1.07 (1.02-1.12) | 0.0034 | 0.003 |
| Stage | ||||||||||||
| Ta–T1&T2 | −0.013 | 0.98 (0.36–2.67) | 0.97 | 0.98 | 0.738 | 2.09 (0.97–4.53) | 0.0607 | 0.061 | −1.388 | 0.25 (0.09–0.65) | 0.005 | 0.0013 |
| Ta&T1–T2 | 1.821 | 6.17 (2.25–16.89) | 0.0004 | 0.00028 | −0.766 | 0.46 (0.16–1.35) | 0.159 | 0.126 | 1.109 | 3.03 (1.46–6.26) | 0.0027 | 0.0034 |
| Occupatinal Exposure | 1.12 | 3.08 (0.7–13.47) | 0.135 | 0.087 | −0.669 | 0.51 (0.23–1.13) | 0.097 | 0.108 | 0.874 | 2.39 (0.91–6.28) | 0.075 | 0.052 |
| Grade | 2.85 | 17.36 (3.89–77.41) | 0.00018 | 0.000 | −1.615 | 0.19 (0.06–0.66) | 0.008 | 0.001 | 1.775 | 5.89 (2.59–13.38) | 0.00002 | 0.00001 |
| Smoking Status | 0.37 | 1.45 (0.33–6.37) | 0.619 | 0.603 | 0.101 | 1.11 (0.38–3.21) | 0.852 | 0.85 | −0.07 | 0.93 (0.36–2.43) | 0.885 | 0.886 |
| Recurrence | −1.28 | 0.28 (0.09–0.86) | 0.026 | 0.015 | −2.229 | 0.107 (0.04–0.28) | 0.000008 | 0.00000 | ||||
| Progression | 2.16 | 8.67 (1.97–8.13) | 0.004 | 0.00031 | −1.717 | 0.18 (0.07–0.48) | 0.0006 | 0.00008 | ||||
| FCmiR-145 | 0.0003 | 1.0003 (1.00009–1.0006) | 0.0069 | 0.038 | −0.019 | 0.98 (0.93–1.03) | 0.393 | 0.099 | 0.0001 | 1.0001 (0.99–1.0003) | 0.243 | 0.321 |
| FCmiR-205 | 0.12 | 1.13 (1.03–1.24) | 0.0089 | 0.045 | −0.167 | 0.85 (0.36–1.96) | 0.697 | 0.521 | 0.046 | 1.05 (0.97–1.13) | 0.233 | 0.311 |
| FCmiR-130b | 0.0003 | 0.99 (0.99–1.00) | 0.466 | 0.398 | 0.0003 | 1.0003 (0.99–1.0007) | 0.131 | 0.176 | −0.0002 | 0.99 (0.99–1.00) | 0.484 | 0.437 |
| FCmiR-21 | 0.00009 | 1.00009 (1.000025–1.00015) | 0.0069 | 0.038 | 0.0004 | 1.0000006 (0.98–1.006) | 0.145 | 0.156 | 0.00003 | 1.00003 (0.99–1.00008) | 0.259 | 0.336 |
| FCmiR-20a | −0.00013 | 0.999 (0.999–1.0) | 0.412 | 0.177 | 0.000002 | 1.000002 (1.0–1.000003) | 0.031 | 0.097 | −0.00013 | 0.999 (0.999–1.0) | 0.412 | 0.177 |
| FCmiR-182 | −0.034 | 0.966 (0.87–1.07) | 0.529 | 0.172 | 0.0006 | 1.0006 (0.00004–1.001) | 0.035 | 0.104 | −0.0009 | 0.999 (0.995–1.002) | 0.599 | 0.243 |
| FCmiR-10a | −0.0004 | 0.999 (0.997–1.001) | 0.672 | 0.47 | −0.0004 | 0.999 (0.998–1.0007) | 0.505 | 0.301 | 0.0003 | 1.0003 (0.999–1.0006) | 0.129 | 0.218 |
| Abnormal Expression 1 | −0.5328 | 0.587 (0.217–1.588) | 0.294 | 0.309 | 0.4376 | 1.549 (0.583–4.11) | 0.379 | 0.358 | −0.4315 | 0.649 (0.297–1.42) | 0.279 | 0.295 |
| Abnormal Expression 2 | 0.5419 | 1.719 (0.663–4.459) | 0.265 | 0.265 | −0.1626 | 0.85 (0.378–1.91) | 0.694 | 0.691 | 0.2274 | 1.255 (0.612–2.575) | 0.535 | 0.536 |
| Mann Whitney U Test | BC Group | Subgroups | |||
|---|---|---|---|---|---|
| p-value | HG p-value | LG p-value | Ta p-value | TaT1 p-value | |
| miR-145-5p | 0.000005 | 0.003612 | 0.000002 | 0.000026 | 0.000001 |
| miR-205-5p | 0.000000 | 0.00000 | 0.00000 | 0.000000 | 0.000000 |
| miR-130b-3p | 0.073733 | 0.770102 | 0.011493 | 0.257699 | 0.479923 |
| miR-21-5p | 0.000000 | 0.000004 | 0.000024 | 0.000000 | 0.000000 |
| miR-20-5p | 0.000000 | 0.000001 | 0.000001 | 0.000003 | 0.000001 |
| miR-182-5p | 0.000000 | 0.000009 | 0.00000 | 0.000001 | 0.000000 |
| miR-10a-5p | 0.000048 | 0.014889 | 0.000016 | 0.000009 | 0.000004 |
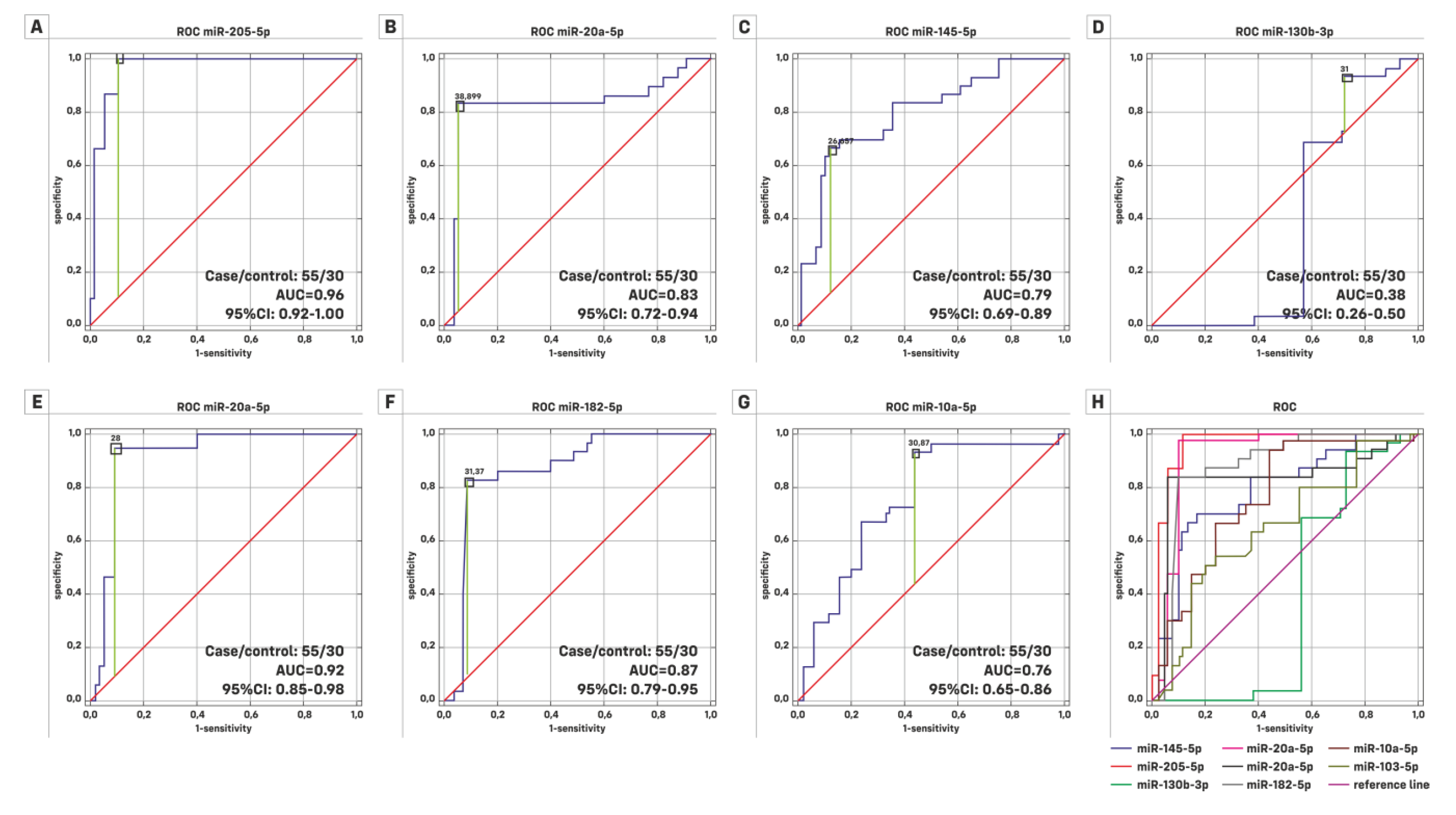
| HG (Case/Control = 22/30) | LG (Case/Control = 33/30) | |||||
|---|---|---|---|---|---|---|
| ROC Characteristics | AUC | 95% Cl | Significance p | AUC | 95% Cl | Significance p |
| miR-145-5p | 0.732 | 0.591–0.873 | 0.0013 | 0.833 | 0.731–0.936 | 0.0001 |
| miR-205-5p | 0.941 | 0.860–1.000 | 0.0001 | 0.981 | 0.955–1.000 | 0.0001 |
| miR-130b-3p | 0.475 | 0.287–0.663 | 0.7964 | 0.313 | 0.167–0.458 | 0.0115 |
| miR-21-5p | 0.851 | 0.717–0.984 | 0.0001 | 0.936 | 0.866–1.000 | 0.0001 |
| miR-20a-5p | 0.87 | 0.761–0.978 | 0.0001 | 0.801 | 0.675–0.927 | 0.0001 |
| miR-182-5p | 0.841 | 0.703–0.976 | 0.0001 | 0.895 | 0.809–0.980 | 0.0001 |
| miR-10a-5p | 0.696 | 0.545–0.846 | 0.0109 | 0.807 | 0.698–0.916 | 0.0001 |
| Ta (case/control = 19/30) | TaT1 (case/control = 37/30) | |||||
| ROC Characteristics | AUC | 95% Cl | Significance p | AUC | 95% Cl | Significance p |
| miR-145-5p | 0.842 | 0.734–0.950 | 0.0001 | 0.83 | 0.728–0.932 | 0.0001 |
| miR-205-5p | 0.982 | 0.947–1.000 | 0.0001 | 0.978 | 0.950–1.000 | 0.0001 |
| miR-130b-3p | 0.401 | 0.205–0.597 | 0.3236 | 0.448 | 0.300–0.596 | 0.493 |
| miR-21-5p | 0.939 | 0.837–1.000 | 0.0001 | 0.925 | 0.849–1.000 | 0.0001 |
| miR-20a-5p | 0.872 | 0.764–0.980 | 0.0001 | 0.83 | 0.713–0.946 | 0.0001 |
| miR-182-5p | 0.888 | 0.777–0.998 | 0.0001 | 0.902 | 0.821–0.983 | 0.0001 |
| miR-10a-5p | 0.858 | 0.738–0.978 | 0.0001 | 0.817 | 0.714–0.920 | 0.0001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borkowska, E.M.; Konecki, T.; Pietrusiński, M.; Borowiec, M.; Jabłonowski, Z. MicroRNAs Which Can Prognosticate Aggressiveness of Bladder Cancer. Cancers 2019, 11, 1551. https://doi.org/10.3390/cancers11101551
Borkowska EM, Konecki T, Pietrusiński M, Borowiec M, Jabłonowski Z. MicroRNAs Which Can Prognosticate Aggressiveness of Bladder Cancer. Cancers. 2019; 11(10):1551. https://doi.org/10.3390/cancers11101551
Chicago/Turabian StyleBorkowska, Edyta Marta, Tomasz Konecki, Michał Pietrusiński, Maciej Borowiec, and Zbigniew Jabłonowski. 2019. "MicroRNAs Which Can Prognosticate Aggressiveness of Bladder Cancer" Cancers 11, no. 10: 1551. https://doi.org/10.3390/cancers11101551
APA StyleBorkowska, E. M., Konecki, T., Pietrusiński, M., Borowiec, M., & Jabłonowski, Z. (2019). MicroRNAs Which Can Prognosticate Aggressiveness of Bladder Cancer. Cancers, 11(10), 1551. https://doi.org/10.3390/cancers11101551






