Classic Chromophobe Renal Cell Carcinoma Incur a Larger Number of Chromosomal Losses Than Seen in the Eosinophilic Subtype
Abstract
1. Introduction
2. Results
2.1. Swiss Cohort
2.2. TCGA Cohort
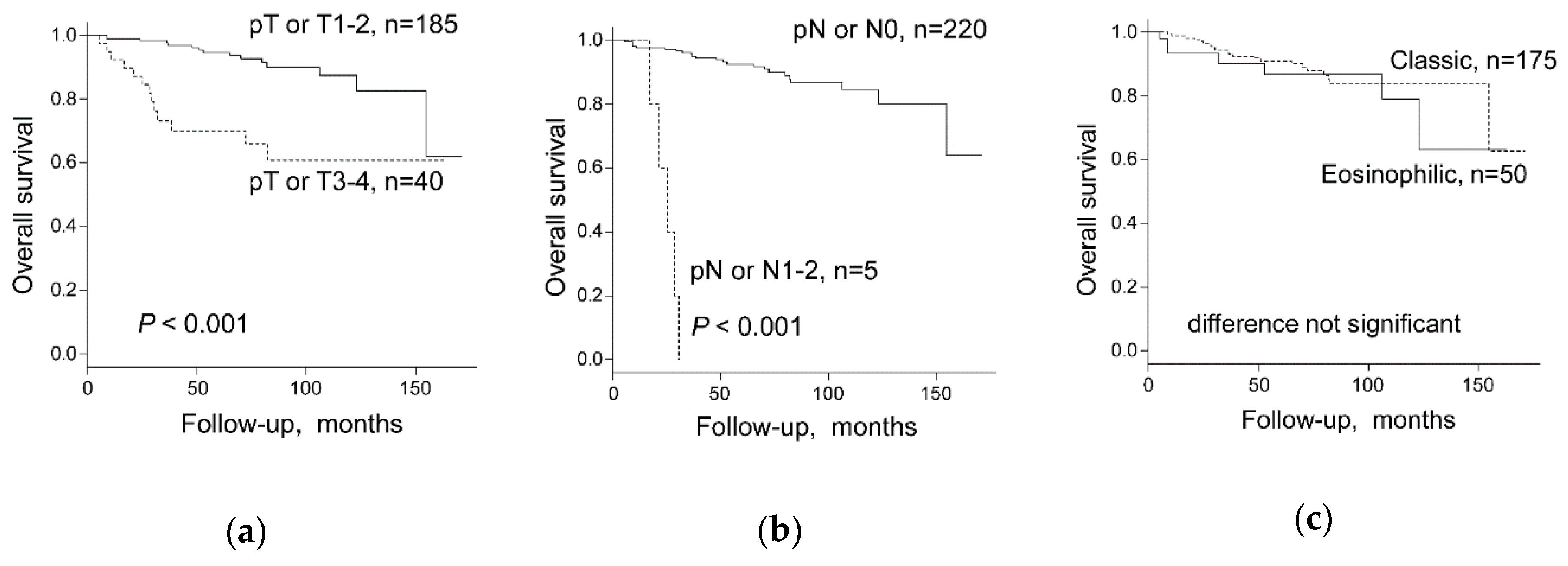
2.3. Chromophobe Renal Cancer Subtype and Survival
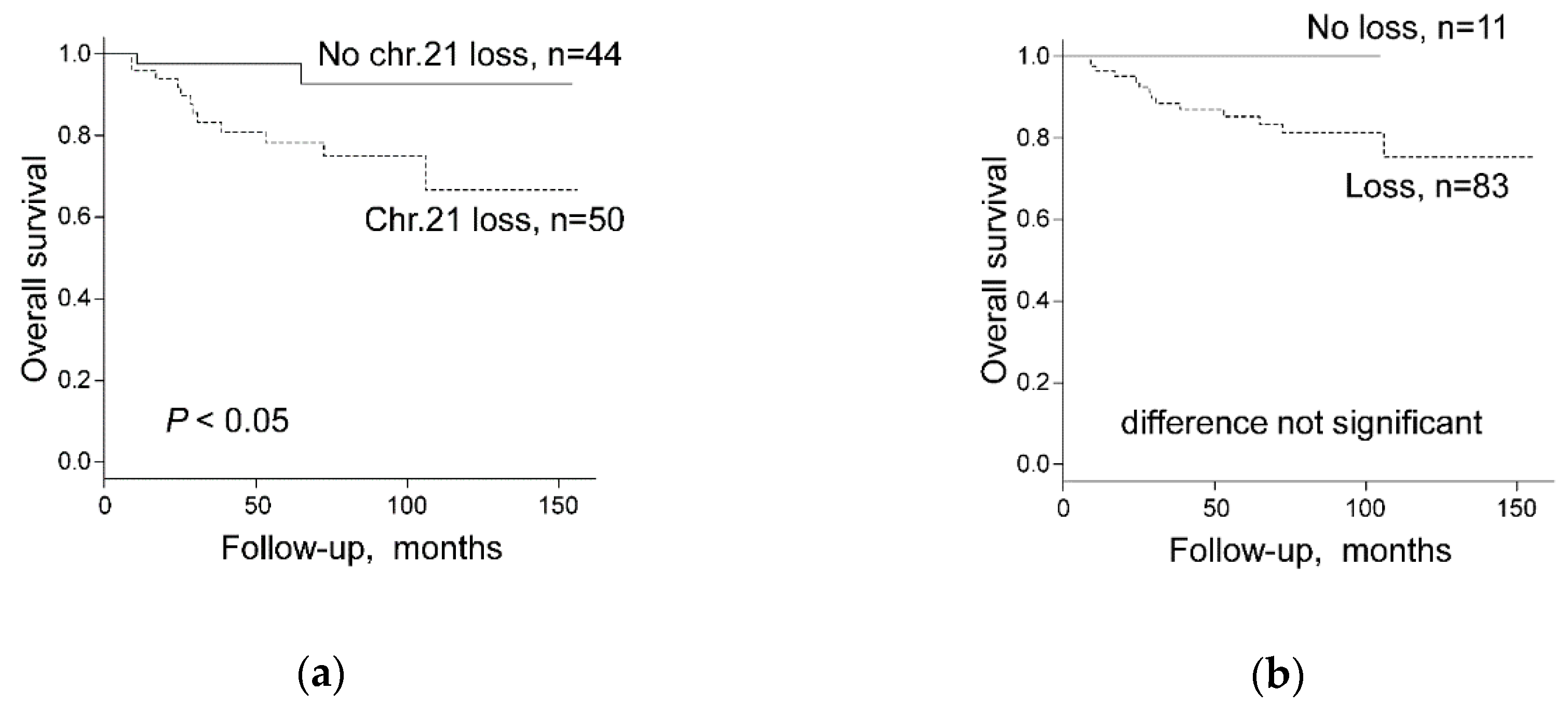
2.4. Chromosomal Copy Number Variation and Survival
3. Discussion
4. Materials and Methods
4.1. Swiss Patients
4.2. The Cancer Genome Atlas (TCGA) Dataset
4.3. Japanese Patients
4.4. OncoScan® CNV Assay of chRCCs
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thoenes, W.; Störkel, S.; Rumpelt, H.J. Human chromophobe cell renal carcinoma. Virchows Arch. B 1985, 48, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Paner, G.; Amin, M.B.; Moch, H.; Störkel, S. Chromophobe renal cell carcinoma. In WHO Classification of Tumours of the Urinary System and Male Genital Organs, 4th ed.; Moch, H., Humphrey, P.A., Ulbright, T.M., Reuter, V.E., Eds.; International Agency for Research on Cancer: Lyon, France, 2016; pp. 27–28. [Google Scholar]
- Thoenes, W.; Störkel, S.; Rumpelt, H.J.; Moll, R.; Baum, H.P.; Werner, S. Chromophobe cell renal carcinoma and its variants—A report on 32 cases. J. Pathol. 1988, 155, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Yap, N.Y.; Rajandram, R.; Ng, K.L.; Pailoor, J.; Fadzli, A.; Gobe, G.C. Genetic and Chromosomal Aberrations and Their Clinical Significance in Renal Neoplasms. Biomed. Res. Int. 2015, 2015, 476508. [Google Scholar] [CrossRef]
- Crotty, T.B.; Farrow, G.M.; Lieber, M.M. Chromophobe cell renal carcinoma: Clinicopathological features of 50 cases. J. Urol. 1995, 154, 964–967. [Google Scholar] [CrossRef]
- Akhtar, M.; Kardar, H.; Linjawi, T.; McClintock, J.; Ali, M.A. Chromophobe cell carcinoma of the kidney. A clinicopathologic study of 21 cases. Am. J. Surg. Pathol. 1995, 19, 1245–1256. [Google Scholar] [CrossRef]
- Latham, B.; Dickersin, G.R.; Oliva, E. Subtypes of chromophobe cell renal carcinoma: An ultrastructural and histochemical study of 13 cases. Am. J. Surg. Pathol. 1999, 23, 530–535. [Google Scholar] [CrossRef]
- Tickoo, S.K.; Lee, M.W.; Eble, J.N.; Amin, M.; Christopherson, T.; Zarbo, R.J.; Amin, M.B. Ultrastructural observations on mitochondria and microvesicles in renal oncocytoma, chromophobe renal cell carcinoma, and eosinophilic variant of conventional (clear cell) renal cell carcinoma. Am. J. Surg. Pathol. 2000, 24, 1247–1256. [Google Scholar] [CrossRef]
- Ng, K.L.; Rajandram, R.; Morais, C.; Yap, N.Y.; Samaratunga, H.; Gobe, G.C.; Wood, S.T. Differentiation of oncocytoma from chromophobe renal cell carcinoma (RCC): Can novel molecular biomarkers help solve an old problem? J. Clin. Pathol. 2014, 67, 97–104. [Google Scholar] [CrossRef]
- Williamson, S.R.; Gadde, R.; Trpkov, K.; Hirsch, M.S.; Srigley, J.R.; Reuter, V.E.; Cheng, L.; Kunju, L.P.; Barod, R.; Rogers, C.G.; et al. Diagnostic criteria for oncocytic renal neoplasms: A survey of urologic pathologists. Hum. Pathol. 2017, 63, 149–156. [Google Scholar] [CrossRef]
- Brunelli, M.; Eble, J.N.; Zhang, S.; Martignoni, G.; Delahunt, B.; Cheng, L. Eosinophilic and classic chromophobe renal cell carcinomas have similar frequent losses of multiple chromosomes from among chromosomes 1, 2, 6, 10 and 17, and this pattern of genetic abnormality is not present in renal oncocytoma. Mod. Pathol. 2005, 18, 161–169. [Google Scholar] [CrossRef]
- Brunelli, M.; Delahunt, B.; Gobbo, S.; Tardanico, R.; Eccher, A.; Bersani, S.; Cossu-Rocca, P.; Parolini, C.; Balzarini, P.; Menestrina, F.; et al. Diagnostic usefulness of fluorescent cytogenetics in differentiating chromophobe renal cell carcinoma from renal oncocytoma: A validation study combining metaphase and interphase analyses. Am. J. Clin. Pathol. 2010, 133, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.F.; Ricketts, C.J.; Wang, M.; Yang, L.; Cherniack, A.D.; Shen, H.; Buhay, C.; Kang, H.; Kim, S.C.; Fahey, C.C.; et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014, 26, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Paolella, B.R.; Gibson, W.J.; Urbanski, L.M.; Alberta, J.A.; Zack, T.I.; Bandopadhayay, P.; Nichols, C.A.; Agarwalla, P.K.; Brown, M.S.; Lamothe, R.; et al. Copy-number and gene dependency analysis reveals partial copy loss of wild-type SF3B1 as a novel cancer vulnerability. Elife 2017, 6, e23268. [Google Scholar] [CrossRef] [PubMed]
- Quddus, M.B.; Pratt, N.; Nabi, G. Chromosomal aberrations in renal cell carcinoma: An overview with implications for clinical practice. Urol. Ann. 2019, 11, 6–14. [Google Scholar] [PubMed]
- Bellanne-Chantelot, C.; Chauveau, D.; Gautier, J.F.; Dubois-Laforgue, D.; Clauin, S.; Beaufils, S.; Wilhelm, J.M.; Boitard, C.; Noël, L.H.; Velho, G.; et al. Clinical spectrum associated with hepatocyte nuclear factor-1beta mutations. Ann. Intern. Med. 2004, 140, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Tong, P.; Kong, W.; Dong, B.; Huang, Y.; Park, I.Y.; Zhou, L.; Liu, X.D.; Ding, Z.; Zhang, X.; et al. HNF1B Loss Exacerbates the Development of Chromophobe Renal Cell Carcinomas. Cancer Res. 2017, 77, 5313–5326. [Google Scholar] [CrossRef]
- Gasparre, G.; Hervouet, E.; de Laplanche, E.; Demont, J.; Pennisi, L.F.; Colombel, M.; Mège-Lechevallier, F.; Scoazec, J.Y.; Bonora, E.; Smeets, R.; et al. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum. Mol. Genet. 2008, 17, 986–995. [Google Scholar] [CrossRef]
- Lang, M.; Vocke, C.D.; Merino, M.J.; Schmidt, L.S.; Linehan, W.M. Mitochondrial DNA mutations distinguish bilateral multifocal renal oncocytomas from familial Birt-Hogg-Dubé tumors. Mod. Pathol. 2015, 28, 1458–1469. [Google Scholar] [CrossRef]
- Nagy, A.; Wilhelm, M.; Sükösd, F.; Ljungberg, B.; Kovacs, G. Somatic mitochondrial DNA mutations in human chromophobe renal cell carcinomas. Genes Chromosomes Cancer 2002, 35, 256–260. [Google Scholar] [CrossRef]
- Digital Slide Archive (DSA). Available online: https://cancer.digitalslidearchive.org/ (accessed on 9 May 2019).
- Trpkov, K.; Williamson, S.R.; Gao, Y.; Martinek, P.; Cheng, L.; Sangoi, A.R.; Yilmaz, A.; Wang, C.; San Miguel Fraile, P.; Perez Montiel, D.M.; et al. Low-grade oncocytic tumour of kidney (CD117-negative, cytokeratin 7-positive): A distinct entity? Histopathology 2019, 75, 174–184. [Google Scholar] [CrossRef]
- Amin, M.B.; Paner, G.P.; Alvarado-Cabrero, I.; Young, A.N.; Stricker, H.J.; Lyles, R.H.; Moch, H. Chromophobe renal cell carcinoma: Histomorphologic characteristics and evaluation of conventional pathologic prognostic parameters in 145 cases. Am. J. Surg. Pathol. 2008, 32, 1822–1834. [Google Scholar] [CrossRef]
- Podduturi, V.; Yourshaw, C.J.; Zhang, H. Eosinophilic variant of chromophobe renal cell carcinoma. Proc. Bayl. Univ. Med. Cent. 2015, 28, 57–58. [Google Scholar] [CrossRef]
- Yip, S.M.; Ruiz Morales, J.M.; Donskov, F.; Fraccon, A.; Basso, U.; Rini, B.I.; Lee, J.L.; Bjarnason, G.A.; Sim, H.W.; Beuselinck, B.; et al. Outcomes of Metastatic Chromophobe Renal Cell Carcinoma (chrRCC) in the Targeted Therapy Era: Results from the International Metastatic Renal Cell Cancer Database Consortium (IMDC). Kidney Cancer 2017, 1, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, R.; Schraml, P.; Batavia, A.; Angori, S.; Simmler, P.; Rupp, N.; Ajioka, Y.; Oliva, E.; Moch, H. Allele Loss and Reduced Expression of CYCLOPS Genes is a Characteristic Feature of Chromophobe Renal Cell Carcinoma. Transl. Oncol. 2019, 12, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Kouba, E.J.; Eble, J.N.; Simper, N.; Grignon, D.J.; Wang, M.; Zhang, S.; Wang, L.; Martignoni, G.; Williamson, S.R.; Brunelli, M.; et al. High fidelity of driver chromosomal alterations among primary and metastatic renal cell carcinomas: Implications for tumor clonal evolution and treatment. Mod. Pathol. 2016, 29, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Algaba, F.; Delahunt, B.; Berney, D.M.; Camparo, P.; Compérat, E.; Griffiths, D.; Kristiansen, G.; Lopez-Beltran, A.; Martignoni, G.; Moch, H.; et al. Handling and reporting of nephrectomy specimens for adult renal tumours: A survey by the European Network of Uropathology. J. Clin. Pathol. 2012, 65, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Trpkov, K.; Grignon, D.J.; Bonsib, S.M.; Amin, M.B.; Billis, A.; Lopez-Beltran, A.; Samaratunga, H.; Tamboli, P.; Delahunt, B.; Egevad, L.; et al. Handling and staging of renal cell carcinoma: The International Society of Urological Pathology Consensus (ISUP) conference recommendations. Am. J. Surg. Pathol. 2013, 37, 1505–1517. [Google Scholar] [CrossRef]
- GDC Data Portal-National Cancer Institute. Available online: https://portal.gdc.cancer.gov/ (accessed on 23 March 2019).
- Broad GDAC Firehose-Broad Institute. Available online: http://gdac.broadinstitute.org/ (accessed on 21 February 2019).
- Deml, K.F.; Schildhaus, H.U.; Compérat, E.; von Teichman, A.; Storz, M.; Schraml, P.; Bonventre, J.V.; Fend, F.; Fleige, B.; Nerlich, A.; et al. Clear cell papillary renal cell carcinoma and renal angiomyoadenomatous tumor: two variants of a morphologic, immunohistochemical, and genetic distinct entity of renal cell carcinoma. Am. J. Surg. Pathol. 2015, 39, 889–901. [Google Scholar] [CrossRef]
- Noske, A.; Brandt, S.; Valtcheva, N.; Wagner, U.; Zhong, Q.; Bellini, E.; Fink, D.; Obermann, E.C.; Moch, H.; Wild, P.J. Detection of CCNE1/URI (19q12) amplification by in situ hybridisation is common in high grade and type II endometrial cancer. Oncotarget 2017, 8, 14794–14805. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the freely-available easy-to-use software “EZR” (Easy R) for medical statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef]
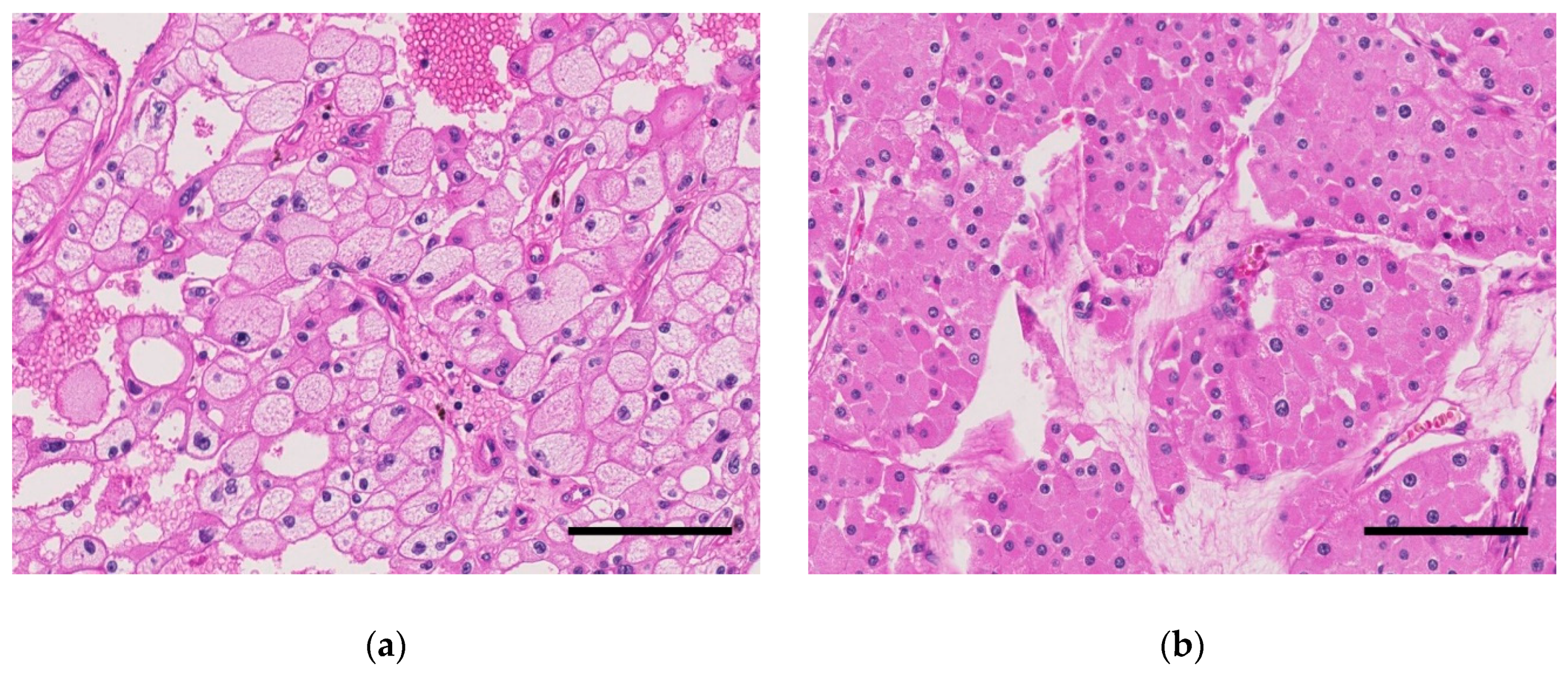

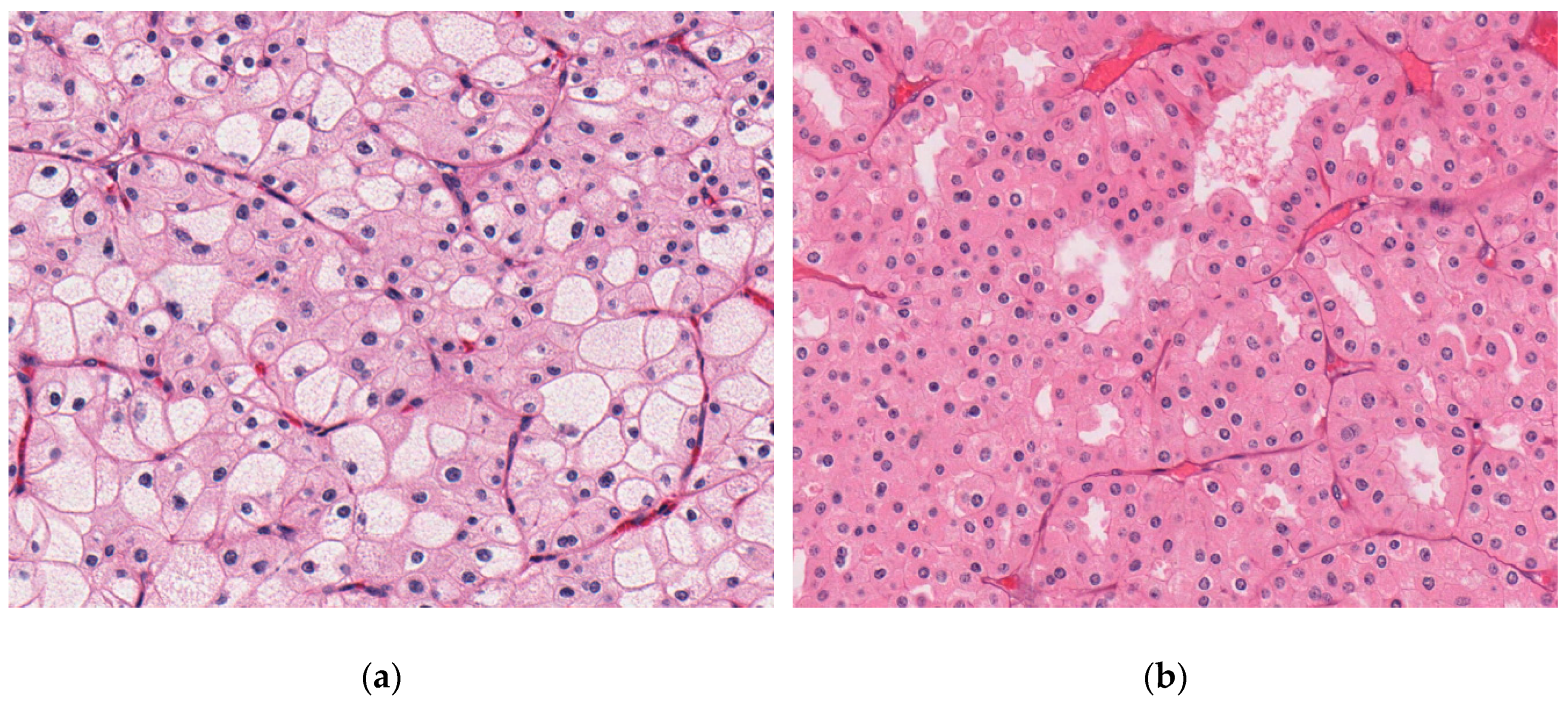
| Characteristics | Swiss Cohort (Total) | TCGA-KICH | Japanese Cohort |
|---|---|---|---|
| Patient number | 42 | 66 | 119 |
| Age (years) | |||
| Range | 18–87 | 17–86 | 26–88 |
| Median | 59 | 50 | 60 |
| Gender | |||
| Female | 13 (31.0%) | 27 (40.9%) | 69 (58.0%) |
| Male | 29 (69.0%) | 39 (59.1%) | 50 (42.0%) |
| pT Stage or T stage, n (%) * | |||
| 1 | 25 (59.5%) | 21 (31.8%) | 85 (71.4%) |
| 2 | 11 (26.2%) | 25 (37.9%) | 19 (16.0%) |
| 3 | 6 (14.3%) | 18 (27.3%) | 14 (11.8%) |
| 4 | 0 (0%) | 2 (3.0%) | 1 (0.8%) |
| Subtype | |||
| classic | 32 (76.2%) | 53 (80.3%) | 90 (75.6%) |
| eosinophilic | 10 (23.8%) | 13 (19.7%) | 29 (24.4%) |
| Cohort | Swiss | Combined * | ||||||
|---|---|---|---|---|---|---|---|---|
| Characteristics | n (%) | Classic chRCC a (%) | Eosinophilic chRCC b (%) | p-Value | n (%) | Classic chRCC a (%) | Eosinophilic chRCC b (%) | p-Value |
| Total | 33 | 22 (66.7) | 11 (33.3) | 99 | 75 (75.8) | 24 (24.2) | ||
| Chr.1 status | ||||||||
| Loss | 32 (97.0) | 22 (100) | 10 (90.9) | n.s. | 87 (87.9) | 70 (93.3) | 17 (70.8) | <0.01 |
| No loss | 1 (3.0) | 0 (0) | 1 (9.1) | 12 (12.1) | 5 (6.7) | 7 (29.2) | ||
| Chr.2 status | ||||||||
| Loss | 24 (72.7) | 19 (86.4) | 5 (45.5) | <0.05 | 73 (73.7) | 63 (84.0) | 10 (41.7) | <0.001 |
| No loss | 9 (27.3) | 3 (13.6) | 6 (54.5) | 26 (26.3) | 12 (16.0) | 14 (58.3) | ||
| Chr.6 status | ||||||||
| Loss | 26 (78.8) | 21 (95.5) | 5 (45.5) | <0.01 | 78 (78.8) | 68 (90.7) | 10 (41.7) | <0.001 |
| No loss | 7 (21.2) | 1 (4.5) | 6 (54.5) | 21 (21.2) | 7 (9.3) | 14 (58.3) | ||
| Chr. 10 status | ||||||||
| Loss | 21 (63.6) | 16 (72.7) | 5 (45.5) | n.s. | 70 (70.7) | 62 (82.7) | 8 (33.3) | <0.001 |
| No loss | 12 (36.4) | 6 (27.3) | 6 (54.5) | 29 (29.3) | 13 (17.3) | 16 (66.7) | ||
| Chr.13 status | ||||||||
| Loss | 23 (69.7) | 17 (77.3) | 6 (54.5) | n.s. | 68 (68.7) | 57 (76.0) | 11 (45.8) | 0.01 |
| No loss | 10 (30.3) | 5 (22.7) | 5 (45.5) | 31 (31.3) | 18 (24.0) | 13 (54.2) | ||
| Chr.17 status | ||||||||
| Loss | 25 (75.8) | 19 (86.4) | 6 (54.5) | n.s. | 75 (75.8) | 64 (85.3) | 11 (45.8) | <0.001 |
| No loss | 8 (24.2) | 3 (13.6) | 5 (45.5) | 24 (24.2) | 11 (14.7) | 13 (54.2) | ||
| Chr.21 status | ||||||||
| Loss | 17 (51.5) | 12 (54.5) | 5 (45.5) | n.s. | 52 (52.5) | 42 (56.0) | 10 (41.7) | n.s. |
| No loss | 16 (48.5) | 10 (45.5) | 6 (54.5) | 47 (47.5) | 33 (44.0) | 14 (58.3) | ||
| Loss of any chromosome c | ||||||||
| present | 29 (87.9) | 21 (95.5) | 8 (72.7) | n.s. | 83 (83.8) | 69 (92.0) | 14 (58.3) | <0.001 |
| absent | 4 (12.1) | 1 (4.5) | 3 (27.3) | 16 (16.2) | 6 (8.0) | 10 (41.7) | ||
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95%CI) | p-Value | HR (95%CI) | p-Value | |
| pT Stage or T stage * (3–4 vs. 1–2) | 4.809 (2.275–10.16) | <0.001 | 3.177 (1.336–7.556) | <0.01 |
| pN Stage or N stage * (1–2 vs. 0) | 42.95 (13.16–140.1) | <0.001 | 21.140 (5.612–79.650) | <0.001 |
| WHO/ISUP grade (Grade 3/4 vs. Grade 2) ** | 1.667 (0.502–5.537) | n.s. | 2.010 (0.594–6.804) | n.s. |
| Subtype (classic vs. eosinophilic) | 0.756 (0.321–1.782) | n.s. | 0.520 (0.207–1.303) | n.s. |
| Cohort | Swiss | Combined | ||||
|---|---|---|---|---|---|---|
| Characteristics | Cases, n (%) | Patient Death, n (%) | p-Value | Cases, n (%) | Patient Death, n (%) | p-Value |
| Total | 30 | 5 (16.7) | 94 | 14 (14.9) | ||
| Chr.1 status | ||||||
| Loss | 30 (100) | 5 (100) | n.s. | 83 (88.3) | 14 (100) | n.s. |
| No loss | 0 (0) | 0 (0) | 11 (11.7) | 0 (0) | ||
| Chr.2 status | ||||||
| Loss | 22 (73.3) | 4 (80.0) | n.s. | 69 (73.4) | 12 (85.7) | n.s. |
| No loss | 8 (26.7) | 1 (20.0) | 25 (26.6) | 2 (14.3) | ||
| Chr.6 status | ||||||
| Loss | 25 (83.3) | 4 (80.0) | n.s. | 75 (79.8) | 12 (85.7) | n.s. |
| No loss | 5 (16.7) | 1 (20.0) | 19 (20.2) | 2 (14.3) | ||
| Chr. 10 status | ||||||
| Loss | 19 (63.3) | 3 (60.0) | n.s. | 67 (71.3) | 12 (85.7) | n.s. |
| No loss | 11 (36.7) | 2 (40.0) | 27 (28.7) | 2 (14.3) | ||
| Chr.13 status | ||||||
| Loss | 21 (70.0) | 4 (80.0) | n.s. | 64 (68.1) | 11 (78.6) | n.s. |
| No loss | 9 (30.0) | 1 (20.0) | 30 (31.9) | 3 (21.4) | ||
| Chr.17 status | ||||||
| Loss | 23 (76.7) | 4 (80.0) | n.s. | 71 (75.5) | 11 (78.6) | n.s. |
| No loss | 7 (23.3) | 1 (20.0) | 23 (24.5) | 3 (21.4) | ||
| Chr.21 status | ||||||
| Loss | 17 (56.7) | 4 (80.0) | n.s. | 50 (53.2) | 12 (85.7) | <0.05 |
| No loss | 13 (43.3) | 1 (20.0) | 44 (46.8) | 2 (14.3) | ||
| Loss of any chromosome * | ||||||
| present | 30 (100) | 5 (100) | n.s. | 83 (88.3) | 14 (100) | n.s. |
| absent | 0 (0) | 0 (0) | 11 (11.7) | 0 (0) | ||
| Variables | Univariate | Multivariate | Multivariate | |||
|---|---|---|---|---|---|---|
| HR (95%CI) | p-Value | HR (95%CI) | p-Value | HR (95%CI) | p-Value | |
| pT Stage or T stage a (3–4 vs. 1–2) | 6.323 (2.078–19.25) | 0.001 | 5.505 (1.935–17.450) | <0.01 | 6.344 (2.010–20.03) | <0.01 |
| Loss of any chromosome b,c (present vs. absent) | 2.503 (0.331–320.637) | n.s. | 1.319 (0.154–172.711) | n.s. | ||
| Loss of chromosome 21 (present vs. absent) | 4.684 (1.047–20.95) | <0.05 | 4.480 (1.000–20.08) | n.s. | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohashi, R.; Schraml, P.; Angori, S.; Batavia, A.A.; Rupp, N.J.; Ohe, C.; Otsuki, Y.; Kawasaki, T.; Kobayashi, H.; Kobayashi, K.; et al. Classic Chromophobe Renal Cell Carcinoma Incur a Larger Number of Chromosomal Losses Than Seen in the Eosinophilic Subtype. Cancers 2019, 11, 1492. https://doi.org/10.3390/cancers11101492
Ohashi R, Schraml P, Angori S, Batavia AA, Rupp NJ, Ohe C, Otsuki Y, Kawasaki T, Kobayashi H, Kobayashi K, et al. Classic Chromophobe Renal Cell Carcinoma Incur a Larger Number of Chromosomal Losses Than Seen in the Eosinophilic Subtype. Cancers. 2019; 11(10):1492. https://doi.org/10.3390/cancers11101492
Chicago/Turabian StyleOhashi, Riuko, Peter Schraml, Silvia Angori, Aashil A. Batavia, Niels J. Rupp, Chisato Ohe, Yoshiro Otsuki, Takashi Kawasaki, Hiroshi Kobayashi, Kazuhiro Kobayashi, and et al. 2019. "Classic Chromophobe Renal Cell Carcinoma Incur a Larger Number of Chromosomal Losses Than Seen in the Eosinophilic Subtype" Cancers 11, no. 10: 1492. https://doi.org/10.3390/cancers11101492
APA StyleOhashi, R., Schraml, P., Angori, S., Batavia, A. A., Rupp, N. J., Ohe, C., Otsuki, Y., Kawasaki, T., Kobayashi, H., Kobayashi, K., Miyazaki, T., Shibuya, H., Usuda, H., Umezu, H., Fujishima, F., Furusato, B., Osakabe, M., Sugai, T., Kuroda, N., ... Moch, H. (2019). Classic Chromophobe Renal Cell Carcinoma Incur a Larger Number of Chromosomal Losses Than Seen in the Eosinophilic Subtype. Cancers, 11(10), 1492. https://doi.org/10.3390/cancers11101492






