Circular RNAs in Clear Cell Renal Cell Carcinoma: Their Microarray-Based Identification, Analytical Validation, and Potential Use in a Clinico-Genomic Model to Improve Prognostic Accuracy
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics and Study Design
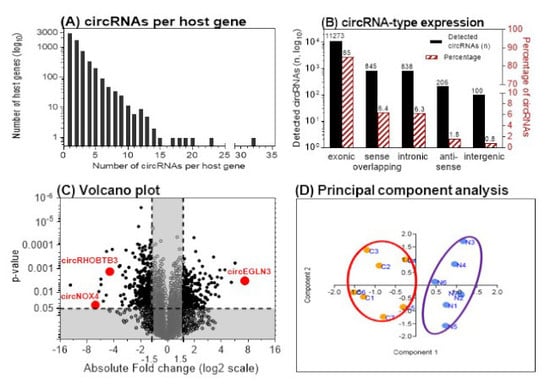
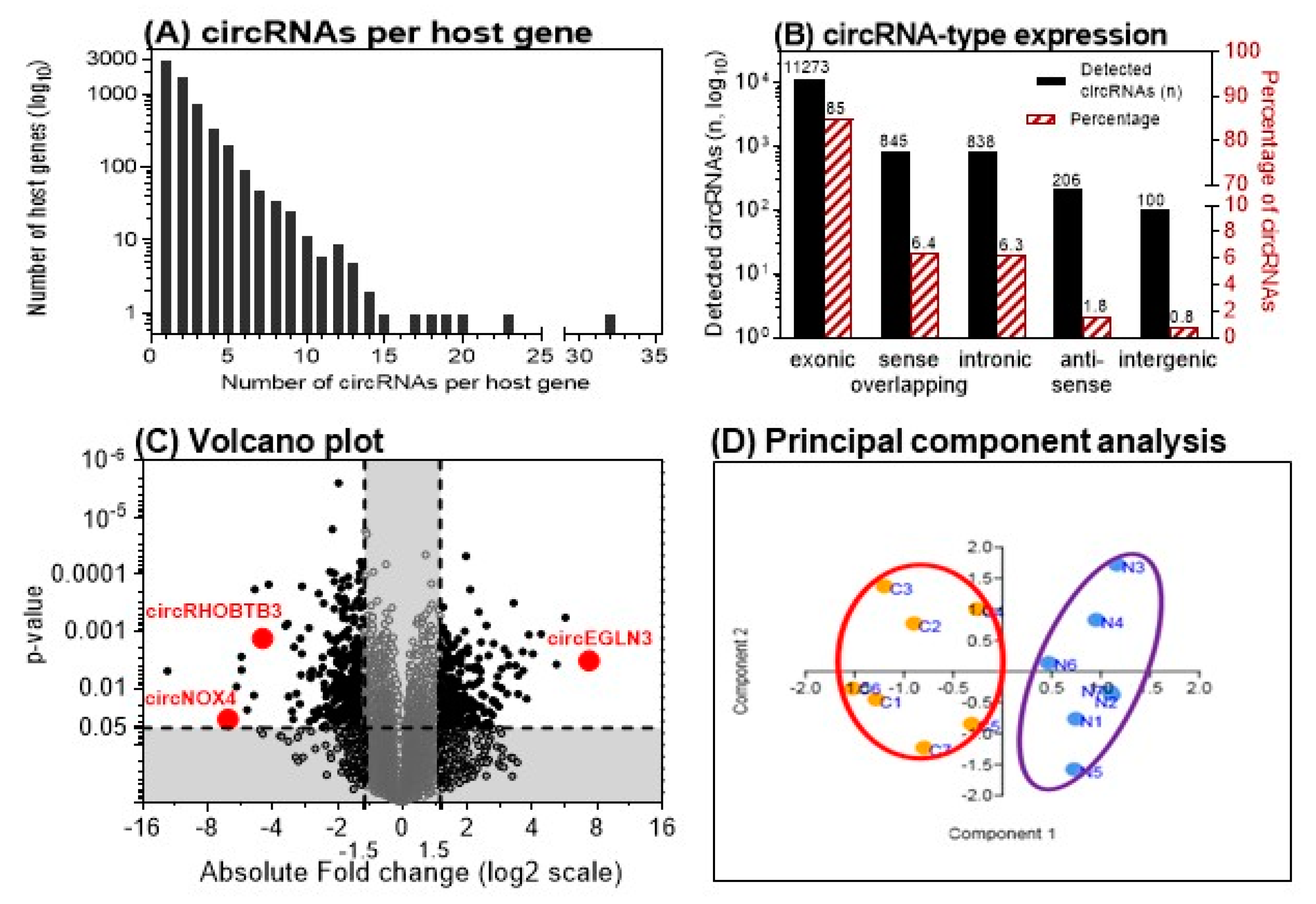
2.2. Discovery of circRNAs in ccRCC Tissue Using Microarray Analysis
2.2.1. Identification of Differentially Expressed circRNAs
2.2.2. Selection of Three circRNAs for Further Evaluation
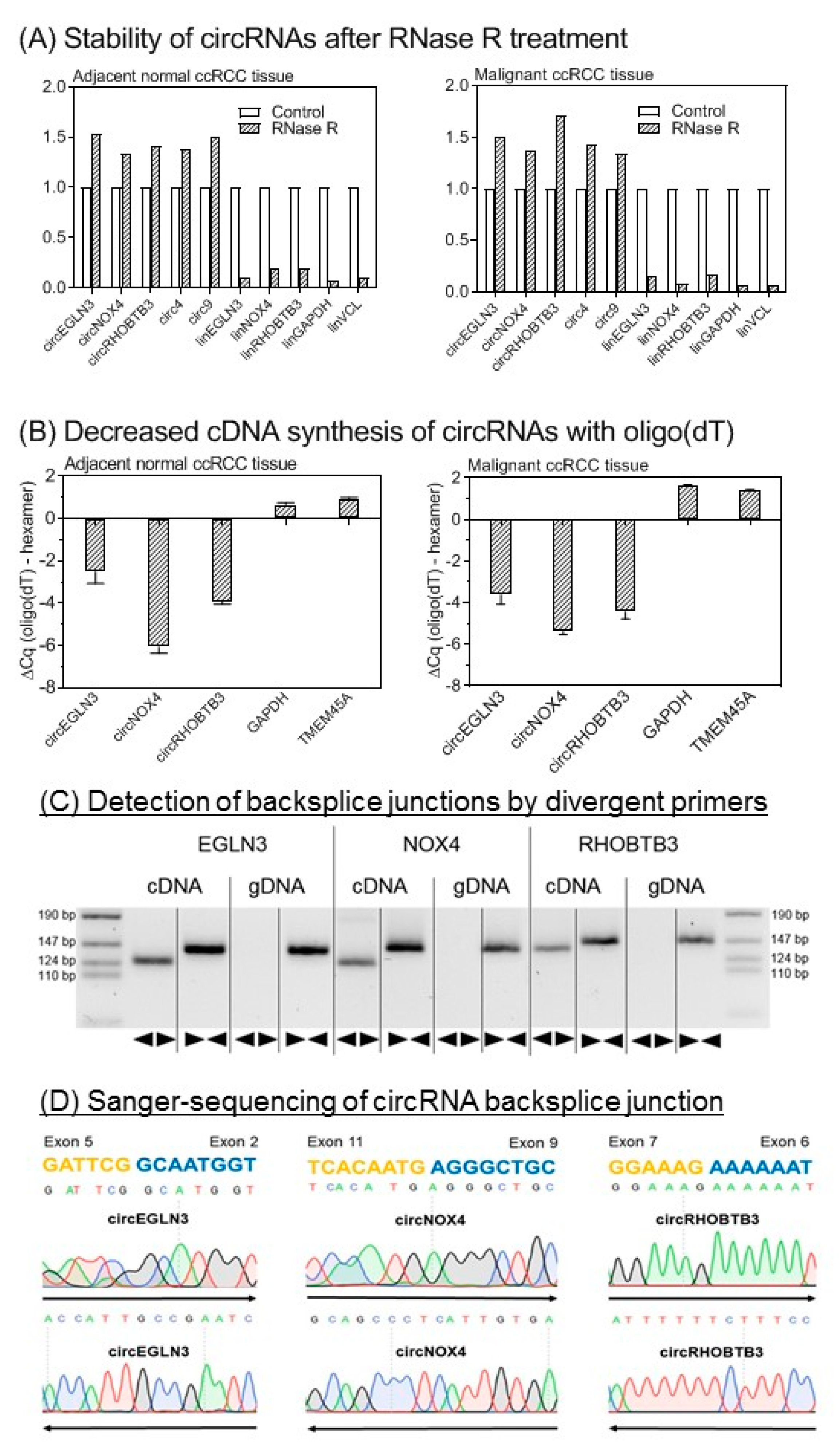
2.3. Analytical Validation of Selected circRNAs
2.3.1. Experimental Confirmation of the Circularity of Transcripts
2.3.2. Analytical Performance of RT-qPCR Assays
2.4. Clinical Assessment
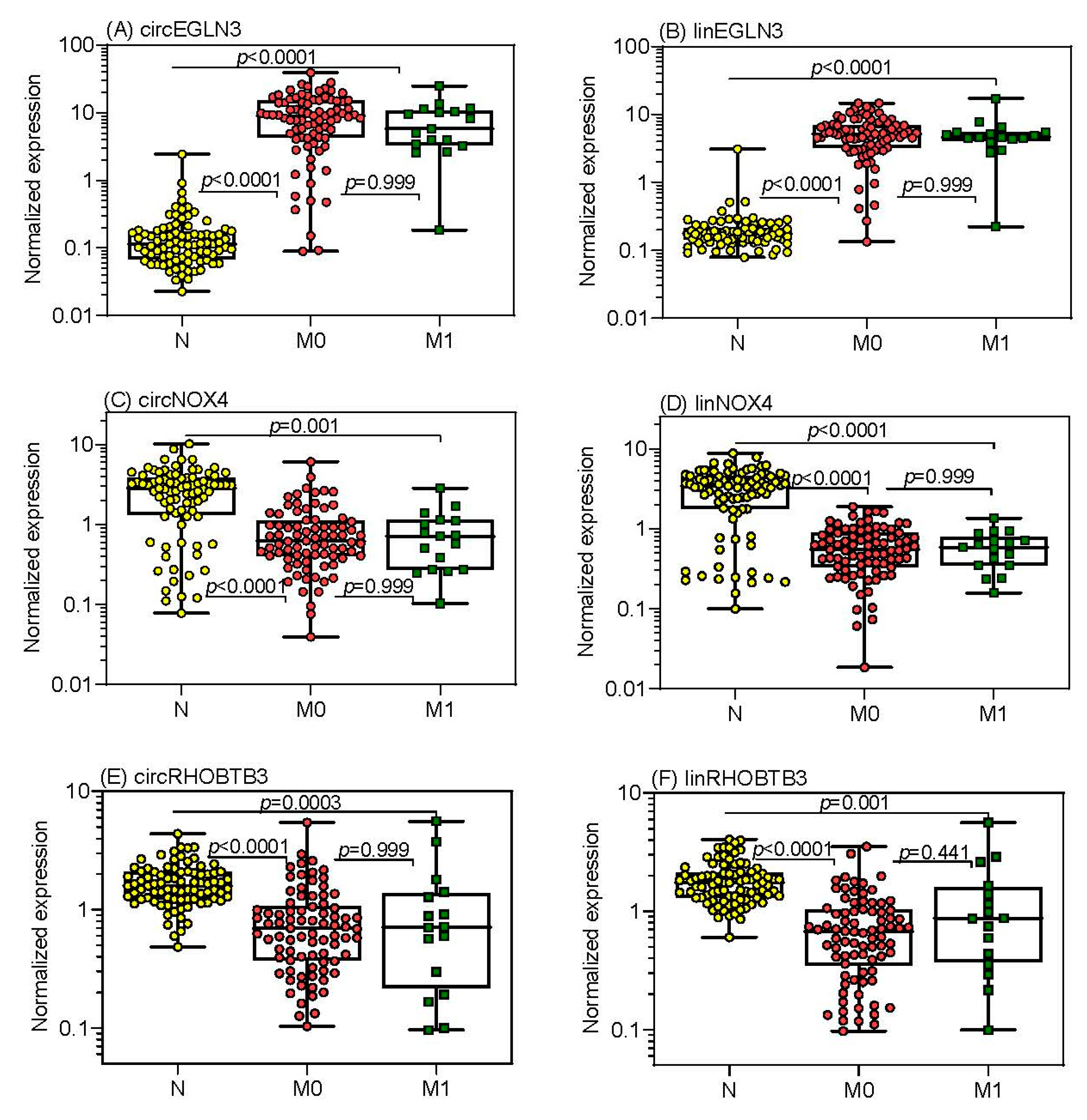
2.4.1. Differential Expression of circRNAs in Relation to Clinicopathological Factors
2.4.2. CircRNAs as Prognostic Markers and Elaboration of RNA Signatures
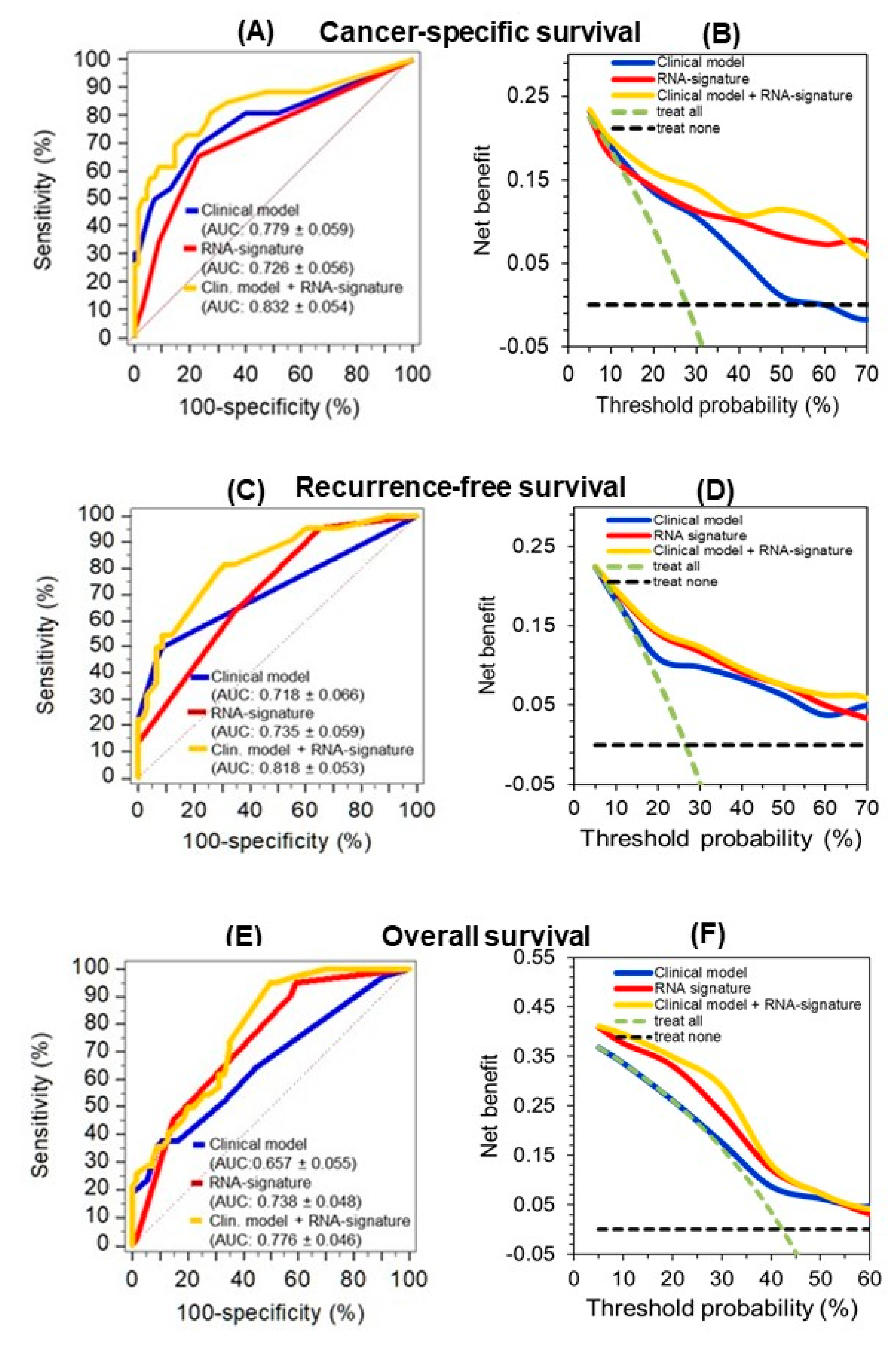
2.4.3. A circRNA-Based Predictive Clinico-Genomic Model to Improve Prognostic Accuracy
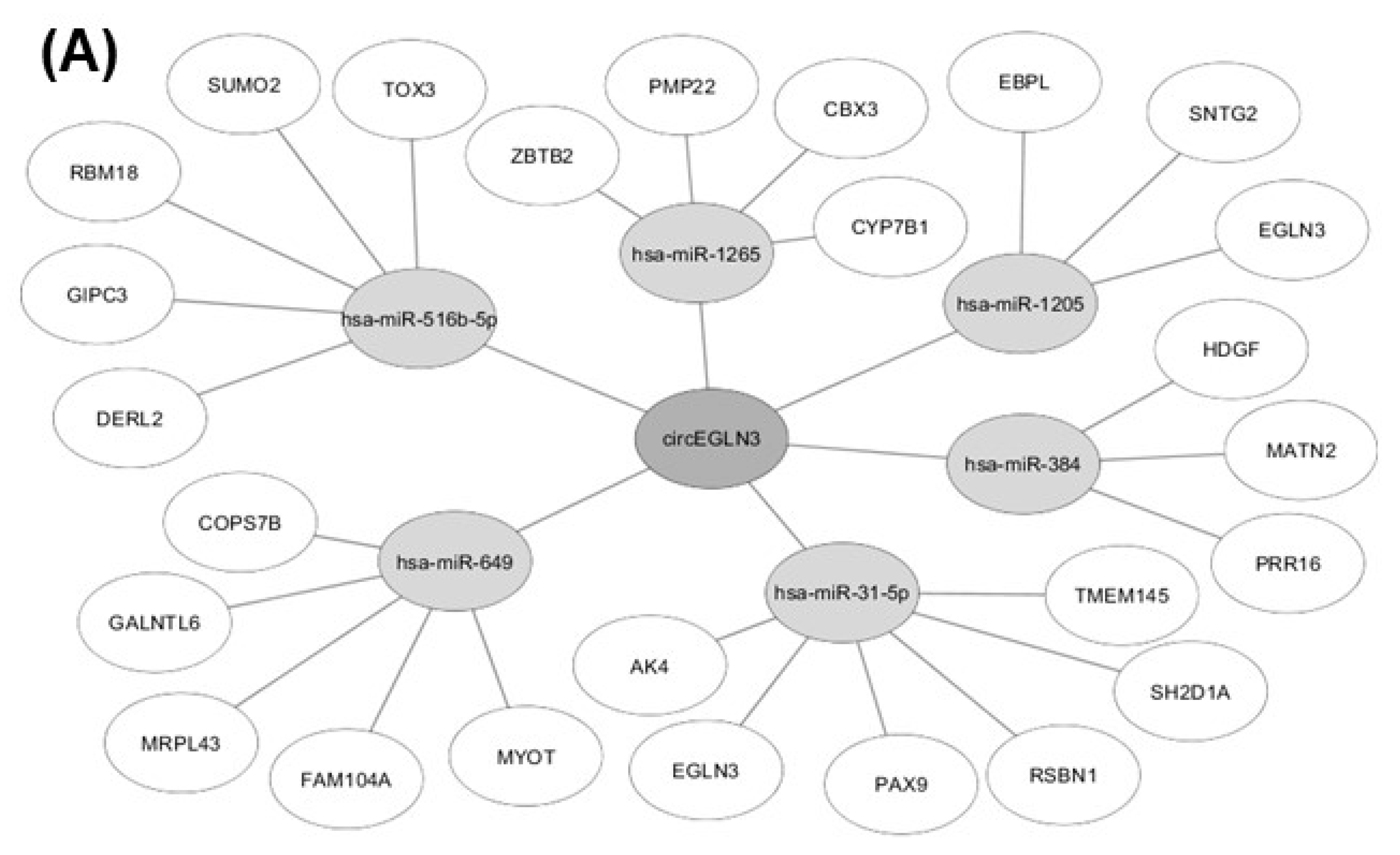
2.5. In-silico Analysis of circRNA-miRNA-Gene Interaction
3. Discussion
4. Materials and Methods
4.1. Patients and Tissue Samples
4.2. Analytical Methods
4.2.1. Total RNA Samples and Their Characteristics
4.2.2. Microarray Detection of circRNAs
4.2.3. RT-qPCR Methodology and circRNA Validation Methods
4.3. Statistics and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bensalah, K.; Dabestani, S.; Fernández-Pello, S.; Giles, R.H.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur. Urol. 2019, 75, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Dabestani, S.; Beisland, C.; Stewart, G.D.; Bensalah, K.; Gudmundsson, E.; Lam, T.B.; Gietzmann, W.; Zakikhani, P.; Marconi, L.; Fernández-Pello, S.; et al. Long-term Outcomes of Follow-up for Initially Localised Clear Cell Renal Cell Carcinoma: RECUR Database Analysis. Eur. Urol. Focus 2018. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Shariat, S.F.; Cheng, C.; Ficarra, V.; Murai, M.; Oudard, S.; Pantuck, A.J.; Zigeuner, R.; Karakiewicz, P.I. Prognostic Factors and Predictive Models in Renal Cell Carcinoma: A Contemporary Review. Eur. Urol. 2011, 60, 644–661. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.H.; Cheng, L.; Rioux-Leclercq, N.; Merino, M.J.; Netto, G.; Reuter, V.E.; Shen, S.S.; Grignon, D.J.; Montironi, R.; Egevad, L.; et al. Renal tumors: Diagnostic and prognostic biomarkers. Am. J. Surg. Pathol. 2013, 37, 1518–1531. [Google Scholar] [CrossRef] [PubMed]
- Klatte, T.; Seligson, D.B.; LaRochelle, J.; Shuch, B.; Said, J.W.; Riggs, S.B.; Zomorodian, N.; Kabbinavar, F.F.; Pantuck, A.J.; Belldegrun, A.S. Molecular Signatures of Localized Clear Cell Renal Cell Carcinoma to Predict Disease-Free Survival after Nephrectomy. Cancer Epidemiol. Prev. Biomark. 2009, 18, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Rini, B.; Goddard, A.; Knezevic, D.; Maddala, T.; Zhou, M.; Aydin, H.; Campbell, S.; Elson, P.; Koscielny, S.; Lopatin, M.; et al. A 16-gene assay to predict recurrence after surgery in localised renal cell carcinoma: Development and validation studies. Lancet Oncol. 2015, 16, 676–685. [Google Scholar] [CrossRef]
- Shi, D.; Qu, Q.; Chang, Q.; Wang, Y.; Gui, Y.; Dong, D. A five-long non-coding RNA signature to improve prognosis prediction of clear cell renal cell carcinoma. Oncotarget 2017, 8, 58699–58708. [Google Scholar] [CrossRef][Green Version]
- Haddad, A.Q.; Luo, J.H.; Krabbe, L.M.; Darwish, O.; Gayed, B.; Youssef, R.; Kapur, P.; Rakheja, D.; Lotan, Y.; Sagalowsky, A.; et al. Prognostic value of tissue-based biomarker signature in clear cell renal cell carcinoma. BJU Int. 2017, 119, 741–747. [Google Scholar] [CrossRef]
- Ghatalia, P.; Rathmell, W.K. Systematic Review: ClearCode 34—A Validated Prognostic Signature in Clear Cell Renal Cell Carcinoma (ccRCC). Kidney Cancer 2018, 2, 23–29. [Google Scholar] [CrossRef]
- Zuo, S.; Wang, L.; Wen, Y.; Dai, G. Identification of a universal 6-lncRNA prognostic signature for three pathologic subtypes of renal cell carcinoma. J. Cell. Biochem. 2019, 120, 7375–7385. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.; Rabien, A.; Stephan, C.; Ralla, B.; Fuchs, S.; Jung, K.; Fendler, A. Circular RNAs: A new class of biomarkers as a rising interest in laboratory medicine. Clin. Chem. Lab. Med. 2018, 56, 1992–2003. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Sun, Y.; Tao, W.; Fei, X.; Chang, C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017, 394, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Zhang, Y.; Sun, Y.; Chen, J.; Chang, C.; Wang, X.; Yeh, S. ERβ-mediated alteration of circATP2B1 and miR-204-3p signaling promotes invasion of clear cell renal cell carcinoma. Cancer Res. 2018, 78, 2550–2563. [Google Scholar] [CrossRef]
- Zhou, B.; Zheng, P.; Li, Z.; Li, H.; Wang, X.; Shi, Z.; Han, Q. CircPCNXL2 sponges miR-153 to promote the proliferation and invasion of renal cancer cells through upregulating ZEB2. Cell Cycle 2018, 17, 2644–2654. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, Y.; Jia, L.; Liu, C.; Xu, F. Circular RNA ABCB10 promotes tumor progression and correlates with pejorative prognosis in clear cell renal cell carcinoma. Int. J. Biol. Markers 2019, 34, 176–183. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-Type Specific Features of Circular RNA Expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Maass, P.G.; Glažar, P.; Memczak, S.; Dittmar, G.; Hollfinger, I.; Schreyer, L.; Sauer, A.V.; Toka, O.; Aiuti, A.; Luft, F.C.; et al. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 2017, 95, 1179–1189. [Google Scholar] [CrossRef]
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Glažar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Campisano, S.; Bertran, E.; Caballero-Díaz, D.; La Colla, A.; Fabregat, I.; Chisari, A.N. Paradoxical role of the NADPH oxidase NOX4 in early preneoplastic stages of hepatocytes induced by amino acid deprivation. Biochim. Biophys. Acta (BBA) Gen. Subj. 2019, 1863, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Crosas-Molist, E.; Bertran, E.; Rodriguez-Hernandez, I.; Herraiz, C.; Cantelli, G.; Fabra, A.; Sanz-Moreno, V.; Fabregat, I. The NADPH oxidase NOX4 represses epithelial to amoeboid transition and efficient tumour dissemination. Oncogene 2017, 36, 3002–3014. [Google Scholar] [CrossRef] [PubMed]
- Miikkulainen, P.; Högel, H.; Seyednasrollah, F.; Rantanen, K.; Elo, L.L.; Jaakkola, P.M. Hypoxia-inducible factor (HIF)-prolyl hydroxylase 3 (PHD3) maintains high HIF2A mRNA levels in clear cell renal cell carcinoma. J. Biol. Chem. 2019, 294, 3760–3771. [Google Scholar] [CrossRef]
- Tanaka, T.; Torigoe, T.; Hirohashi, Y.; Sato, E.; Honma, I.; Kitamura, H.; Masumori, N.; Tsukamoto, T.; Sato, N. Hypoxia-inducible factor (HIF)-independent expression mechanism and novel function of HIF prolyl hydroxylase-3 in renal cell carcinoma. J. Cancer Res. Clin. Oncol. 2014, 140, 503–513. [Google Scholar] [CrossRef]
- Zhang, C.S.; Liu, Q.; Li, M.; Lin, S.Y.; Peng, Y.; Wu, D.; Li, T.Y.; Fu, Q.; Jia, W.; Wang, X.; et al. RHOBTB3 promotes proteasomal degradation of HIFalpha through facilitating hydroxylation and suppresses the Warburg effect. Cell Res. 2015, 25, 1025–1042. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.H.; Bindereif, A. Exon Circularization Requires Canonical Splice Signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Ramankulov, A.; Roigas, J.; Johannsen, M.; Ringsdorf, M.; Kristiansen, G.; Jung, K. In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC Mol. Biol. 2007, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Camp, R.L. X-Tile: A New Bio-Informatics Tool for Biomarker Assessment and Outcome-Based Cut-Point Optimization. Clin. Cancer Res. 2004, 10, 7252–7259. [Google Scholar] [CrossRef]
- Steyerberg, E.W.; Pencina, M.J.; Lingsma, H.F.; Kattan, M.W.; Vickers, A.J.; Van, C.B. Assessing the incremental value of diagnostic and prognostic markers: A review and illustration. Eur. J. Clin. Investig. 2012, 42, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Wagenmakers, E.J.; Farrell, S. AIC model selection using Akaike weights. Psychon. Bull. Rev. 2004, 11, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
- Littman, B.H.; Di, M.L.; Plebani, M.; Marincola, F.M. What’s next in translational medicine? Clin. Sci. 2007, 112, 217–227. [Google Scholar] [CrossRef]
- Klatte, T.; Rossi, S.H.; Stewart, G.D. Prognostic factors and prognostic models for renal cell carcinoma: A literature review. World J. Urol. 2018, 36, 1943–1952. [Google Scholar] [CrossRef]
- Li, S.; Teng, S.; Xu, J.; Su, G.; Zhang, Y.; Zhao, J.; Zhang, S.; Wang, H.; Qin, W.; Lu, Z.J.; et al. Microarray is an efficient tool for circRNA profiling. Brief. Bioinform. 2018. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.L.; Lim, B.T.; Anene-Nzelu, C.G.; Ackers-Johnson, M.; Dashi, A.; See, K.; Tiang, Z.; Lee, D.P.; Chua, W.W.; Luu, T.D.; et al. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017, 113, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, V.; Xu, X.; Livingstone, J.; Soares, F.; Jeon, J.; Zeng, Y.; Hua, J.T.; Petricca, J.; Guo, H.; et al. Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell 2019, 176, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Siede, D.; Rapti, K.; Gorska, A.; Katus, H.; Altmüller, J.; Boeckel, J.; Meder, B.; Maack, C.; Völkers, M.; Müller, O.; et al. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J. Mol. Cell. Cardiol. 2017, 109, 48–56. [Google Scholar] [CrossRef]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881. [Google Scholar] [CrossRef]
- Cieślik, M.; Chugh, R.; Wu, Y.M.; Wu, M.; Brennan, C.; Lonigro, R.; Su, F.; Wang, R.; Siddiqui, J.; Mehra, R.; et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res. 2015, 25, 1372–1381. [Google Scholar] [CrossRef]
- Gerszten, R.E.; Wang, T.J. The search for new cardiovascular biomarkers. Nature 2008, 451, 949–952. [Google Scholar] [CrossRef]
- Ralla, B.; Busch, J.; Flörcken, A.; Westermann, J.; Zhao, Z.; Kilic, E.; Weickmann, S.; Jung, M.; Fendler, A.; Jung, K. miR-9-5p in Nephrectomy Specimens is a Potential Predictor of Primary Resistance to First-Line Treatment with Tyrosine Kinase Inhibitors in Patients with Metastatic Renal Cell Carcinoma. Cancers 2018, 10, 321. [Google Scholar] [CrossRef]
- Steyerberg, E.W.; E Harrell, F.; Borsboom, G.J.; Eijkemans, M.J.; Vergouwe, Y.; Habbema, J.D. Internal validation of predictive models: Efficiency of some procedures for logistic regression analysis. J. Clin. Epidemiol. 2001, 54, 774–781. [Google Scholar] [CrossRef]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Yu, C.; Cui, S.; Wang, H.; Jin, H.; Wang, C.; Li, B.; Qin, M.; Yang, C.; He, J.; et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat. Commun. 2019, 10, 3200. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Liu, T.; Feng, H.; Yang, R.; Zhao, X.; Chen, W.; Jiang, B.; Qin, H.; Guo, X.; Liu, M.; et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol. Cancer 2019, 18, 111. [Google Scholar] [CrossRef] [PubMed]
- Högel, H.; Miikkulainen, P.; Bino, L.; Jaakkola, P.M. Hypoxia inducible prolyl hydroxylase PHD3 maintains carcinoma cell growth by decreasing the stability of p27. Mol. Cancer 2015, 14, 143. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Quan, J.; Chen, F.; Pan, X.; Zhuang, C.; Xiong, T.; Zhuang, C.; Li, J.; Huang, X.; Ye, J.; et al. MiR-31-5p acts as a tumor suppressor in renal cell carcinoma by targeting cyclin-dependent kinase 1 (CDK1). Biomed. Pharmacother. 2019, 111, 517–526. [Google Scholar] [CrossRef]
- Lin, H.; Huang, Z.P.; Liu, J.; Qiu, Y.; Tao, Y.P.; Wang, M.C.; Yao, H.; Hou, K.Z.; Gu, F.M.; Xu, X.F. MiR-494-3p promotes PI3K/AKT pathway hyperactivation and human hepatocellular carcinoma progression by targeting PTEN. Sci. Rep. 2018, 8, 10461. [Google Scholar] [CrossRef]
- Wang, J.; Chen, H.; Liao, Y.; Chen, N.; Liu, T.; Zhang, H.; Zhang, H. Expression and clinical evidence of miR-494 and PTEN in non-small cell lung cancer. Tumor Biol. 2015, 36, 6965–6972. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; Vandesompele, J.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Altman, D.G.; McShane, L.M.; Sauerbrei, W.; Taube, S.E. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and elaboration. PLoS Med. 2012, 9, e1001216. [Google Scholar] [CrossRef]
- Bossuyt, P.M.; Reitsma, J.B.; Bruns, D.E.; Gatsonis, C.A.; Glasziou, P.P.; Irwig, L.; Lijmer, J.G.; Moher, D.; Rennie, D.; De Vet, H.C.; et al. STARD 2015: An Updated List of Essential Items for Reporting Diagnostic Accuracy Studies. Radiology 2015, 277, 826–832. [Google Scholar] [CrossRef]
- Jung, M.; Mollenkopf, H.J.; Grimm, C.; Wagner, I.; Albrecht, M.; Waller, T.; Pilarsky, C.; Johannsen, M.; Stephan, C.; Lehrach, H.; et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J. Cell. Mol. Med. 2009, 13, 3918–3928. [Google Scholar] [CrossRef] [PubMed]
- Wotschofsky, Z.; Meyer, H.A.; Jung, M.; Fendler, A.; Wagner, I.; Stephan, C.; Busch, J.; Erbersdobler, A.; Disch, A.C.; Mollenkopf, H.J.; et al. Reference genes for the relative quantification of microRNAs in renal cell carcinomas and their metastases. Anal. Biochem. 2011, 417, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Ralla, B.; Magheli, A.; Kempkensteffen, C.; Miller, K.; Jung, M.; Trujillo, E.; Wotschofsky, Z.; Kilic, E.; Fendler, A.; Jung, K.; et al. 773 Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. Eur. Urol. Suppl. 2015, 14, e773. [Google Scholar] [CrossRef][Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Vickers, A.J.; Elkin, E.B. Decision curve analysis: A novel method for evaluating prediction models. Med. Decis. Mak. 2006, 26, 565–574. [Google Scholar] [CrossRef]
- Stephan, C.; Jung, K.; Semjonow, A.; Schulze-Forster, K.; Cammann, H.; Hu, X.; Meyer, H.A.; Bogemann, M.; Miller, K.; Friedersdorff, F. Comparative assessment of urinary prostate cancer antigen 3 and TMPRSS2:ERG gene fusion with the serum [-2]proprostate-specific antigen-based prostate health index for detection of prostate cancer. Clin. Chem. 2013, 59, 280–288. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]


| Characteristics | Total | ccRCC, | ccRCC, | p-value b |
|---|---|---|---|---|
| Non-Metastatic a | Metastatic a | |||
| Patients, no. | 99 | 82 | 17 | |
| Sex, female/male; n (%) | 30/69 | 25/57 (30/70) | 5/12 (29/70) | 1 |
| Age, yrs, median (range) | 65 (36–87) | 65 (36–87) | 65 (37–78) | 0.982 |
| Pathological stage, no. (%) | ||||
| pT1 | 43 | 40 (49) | 3 (18) | 0.004 |
| pT2 | 8 | 8 (10) | 0 (0) | |
| pT3 | 47 | 34 (41) | 13 (76) | |
| pT4 | 1 | 0 (0) | 1 (6) | |
| TNM stage grouping, no. (%) c | ||||
| I | 40 | 40 | - | |
| II | 8 | 8 | - | |
| III | 34 | 34 | - | |
| IV | 17 | 0 | 17 | |
| Tumor size, median mm (range) | 57 (20–220) | 50 (20–220) | 75 (35–150) | 0.029 |
| Surgical margin, no. (%) | ||||
| R0 | 85 | 73 (89) | 12 (71) | 0.138 |
| R1/2 | 11 | 7 (9) | 4 (23) | |
| Unclassified | 3 | 2 (2) | 1 (6) | |
| Fuhrman grade, no. (%) | ||||
| G1 | 9 | 8 | 1 | 0.025 |
| G2 | 68 | 60 | 8 | |
| G3 | 20 | 12 | 8 | |
| G4 | 2 | 2 | 0 | |
| Events during follow-up, n, (%) | ||||
| Metastasis | - | 22 (27) | - | |
| Cancer-specific death | 26 (26) | 14 (17) | 12 (71) | <0.0001 |
| Overall death | 42 (42) | 30 (37) | 12 (71) | 0.014 |
| Survival, months, median/mean (95% CI) d | ||||
| Cancer-specific | 125 (111–139) | 140 (127–153) | 27.2 (14.1–40.4) | |
| Recurrence-free | - | 127 (113–141) | - | |
| Overall | 126 (90.4–166) | 160 (103–160) | 11.8 (8.2–39.9) | |
| circRNA in Manuscript | circRNA ID in ArrayStar a,b | circRNA ID in circBase a,c | Fold Change Expression in Tumor vs. Normal Tissue (p-value) | Best Transcript | Official Gene Symbol |
|---|---|---|---|---|---|
| Up-regulated circRNAs | |||||
| circEGLN3 | circRNA_405198 | circ_0101692 | 7.32 (0.0033) | NM_022073 | EGLN3 |
| - | circRNA_101202 | circ_0029340 | 5.68 (0.0006) | NM_005505 | SCARB1 |
| - | circRNA_101341 | circ_0031594 | 5.16 (0.0038) | NM_022073 | EGLN3 |
| - | circRNA_101803 | circ_0003520 | 4.39 (0.0011) | NM_018092 | NETO2 |
| - | circRNA_103980 | circ_0006528 | 4.01 (0.0024) | NM_138492 | PRELID2 |
| Down-regulated circRNAs | |||||
| - | circRNA_103093 | circ_0060937 | −12.3 (0.0049) | NM_000782 | CYP24A1 |
| - | circRNA_101120 | circ_0027821 | −6.73 (0.0342) | NR_024037 | RMST |
| circNOX4 | circRNA_100933 | circ_0023984 | −6.44 (0.0475) | NM_016931 | NOX4 |
| - | circRNA_100562 | circ_0006577 | −5.87 (0.0093) | NM_012425 | RSU1 |
| - | circRNA_031282 | circ_0031282 | −5.58 (0.0028) | NM_012244 | SLC7A8 |
| - | circRNA_103091 | circ_0060927 | −5.58 (0.0048) | NM_000782 | CYP24A1 |
| - | circRNA_023983 | circ_0008350 | −5.26 (0.0239) | NM_016931 | NOX4 |
| - | circRNA_1011001 | circ_0025135 | −4.89 (0.0132) | NM_001038 | SCNN1A |
| - | circRNA_035435 | circ_0035435 | −4.84 (0.0002) | NM_032866 | CGNL1 |
| circRHOBTB3 | circRNA_007444 | circ_0007444 | −4.45 (0.0013) | NM_014899 | RHOBTB3 |
| - | circRNA_101528 | circ_0035436 | −4.18 (0.0001) | NM_032866 | CGNL1 |
| RNA | Repeatability a | Reproducibility b | ||
|---|---|---|---|---|
| Cq Value Mean (%RSD) | Concentration (AU) Mean (%RSD) | Cq Value Mean ± SD (%RSD) | Concentration (AU) Mean ± SD (%RSD) | |
| circEGLN3 | 23.47 (0.284) | 1.185 (4.51) | 22.49 ± 0.138 (0.62) | 2.132 ± 0.199 (9.33) |
| circNOX4 | 24.16 (0.493) | 0.808 (9.35) | 22.77 ± 0.108 (0.47) | 1.118 ± 0.082 (7.30) |
| circRHOBTB3 | 25.64 (0.459) | 0.0268 (9.27) | 24.81 ± 0.190 (0.76) | 0.036 ± 0.005 (14.2) |
| linEGLN3 | 20.83 (0.521) | 32.72 (7.00) | 23.93 ± 0.042 (0.18) | 1.658 ± 0.046 (2.75) |
| linNOX4 | 27.65 (0.405) | 0.204 (7.67) | 25.50 ± 0.046 (0.18) | 0.483 ± 0.015 (3.10) |
| linRHOBTB3 | 24.90 (0.214) | 1.901 (3.43) | 23.43 ± 0.147 (0.63) | 3.791 ± 0.386 (10.2) |
| PPIA | 19.33 (0.329) | 32.01 (4.16) | 19.18 ± 0.081 (0.42) | 33.36 ± 1.745 (5.23) |
| TBP | 25.18 (0.331) | 2.330 (5.07) | 24.99 ± 0.104 (0.42) | 2.423 ± 0.156 (6.44) |
| RNAs | AUC (95% CI) | p-Value Different to AUC = 0.5 | Differentiating Ability at the Youden Index a | % Overall Correct Classification | |
|---|---|---|---|---|---|
| Sensitivity (95% CI) | Specificity (95% CI) | ||||
| circEGLN3 | 0.98 (0.95–0.99) | <0.0001 | 95 (89–99) | 95 (88–99) | 94.6 |
| circNOX4 | 0.81 (0.74–0.86) | <0.0001 | 91 (83–96) | 71 (60–80) | 80.4 |
| circRHOBTB3 | 0.82 (0.76–0.87) | <0.0001 | 72 (62–80) | 91 (82–96) | 69.0 |
| linEGLN3 | 0.98 (0.96–0.99) | <0.0001 | 96 (89–98) | 99 (94–100) | 95.7 |
| linNOX4 | 0.85 (0.79–0.90) | <0.0001 | 99 (95–100) | 78 (67–86) | 88.0 |
| linRHOBTB3 | 0.86 (081–0.91) | <0.0001 | 72 (62–80) | 94 (87–98) | 75.0 |
| circEGLN3 +linEGLN3 b | 0.99 (0.96–1.00) | <0.0001 | 95 (89–98) | 99 (94–100) | 95.7 |
| Variable b | Cancer-Specific Survival | Recurrence-Free Survival | Overall Survival | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| RNA signature. c,d,e | ||||||
| circEGLN3 | 0.24 (0.10–0.57) | 0.001 | 0.53 (0.18–1.03) | 0.074 | 0.49 (0.23–0.95) | 0.037 |
| circRHOBTB3 | 0.26 (0.09–0.73) | 0.010 | 0.14 (0.04–0.49) | 0.003 | 0.15 (0.05–0.49) | 0.002 |
| linRHOBTB3 | 2.57 (0.95–6.90) | 0.062 | 11.1 (2.79–43.8) | 0.001 | 4.46 (1.52–13.0) | 0.006 |
| Clinical model | ||||||
| Tumor stage grouping (III+IV/I+II) | 3.54 (1.16–10.8) | 0.027 | 0.79 (0.25–2.48) | 0.685 | 2.02 (0.95–4.27) | 0.064 |
| Fuhrman grading (3+4/1+2) | 2.68 (1.03–7.02) | 0.044 | 13.4 (4.06–44.3) | <0.0001 | 3.00 (1.32–6.82) | 0.009 |
| Surgical margin (R1/R0) | 2.82 (1.05–7.59) | 0.040 | 7.09 (1.75–28.7) | 0.006 | 2.26 (0.91–5.59) | 0.078 |
| Tumor size (≥7 cm<) | 1.10 (0.42–2.87) | 0.838 | 1.03 (0.35–2.97) | 0.963 | 0.99 (0.46–2.14) | 0.988 |
| Clinical model + RNA signature | ||||||
| Tumor stage grouping (III+IV/I+II) | 3.16 (0.93-10.8) | 0.066 | 0.67 (0.19–2.37) | 0.536 | 2.81 (1.23–6.42) | 0.014 |
| Fuhrman grading (3+4/1+2) | 2.04 (0.73–5.74) | 0.177 | 9.98 (2.99–33.4) | 0.0002 | 2.25 (0.99–5.12) | 0.053 |
| Surgical margin (R1 vs. R0) | 3.97 (1.28–12.3) | 0.017 | 3.48 (0.76–15.9) | 0.109 | 2.12 (0.87–5.18) | 0.099 |
| Tumor size (≥7 cm<) | 1.16 (0.44–3.11) | 0.762 | 1.22 (0.36–4.13) | 0.746 | 0.76 (0.36–1.64) | 0.490 |
| circEGLN3 | 0.33 (0.13–0.79) | 0.014 | 0.69 (0.22–2.19) | 0.532 | 0.51 (0.24–1.09) | 0.084 |
| circRHOBTB3 | 0.25 (0.08–0.80) | 0.019 | 0.21 (0.05–0.94) | 0.041 | 0.16 (0.05–0.53) | 0.003 |
| linRHOBTB3 | 3.84 (1.35–10.9) | 0.012 | 7.71 (1.55–38.3) | 0.013 | 5.26 (1.74–15.9) | 0.003 |
| Clinical model + RNA signature after backward elimination | ||||||
| Tumor stage grouping (III+IV/I+II) | 3.98 (1.32–12.0) | 0.014 | – | – | 2.89 (1.39–5.99) | 0.005 |
| Fuhrman grading (3+4/1+2) | – | – | 8.53 (3.08–23.6) | <0.0001 | 2.52 (1.15–5.53) | 0.021 |
| Surgical margin (R1 vs. R0) | 5.68 (2.09–15.4) | 0.0007 | 3.54 (1.02–12.3) | 0.047 | – | – |
| Tumor size (≥7 cm<) | – | – | – | – | – | – |
| circEGLN3 | 0.28 (0.12–0.68) | 0.005 | – | – | 0.50 (0.24–1.05) | 0.067 |
| circRHOBTB3 | 0.21 (0.07–0.65) | 0.007 | 0.18 (0.04–0.75) | 0.018 | 0.17 (0.05–0.56) | 0.004 |
| linRHOBTB3 | 3.59 (1.28–10.0) | 0.015 | 8.19 (1.81–37.1) | 0.006 | 5.37 (1.80–16.1) | 0.003 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franz, A.; Ralla, B.; Weickmann, S.; Jung, M.; Rochow, H.; Stephan, C.; Erbersdobler, A.; Kilic, E.; Fendler, A.; Jung, K. Circular RNAs in Clear Cell Renal Cell Carcinoma: Their Microarray-Based Identification, Analytical Validation, and Potential Use in a Clinico-Genomic Model to Improve Prognostic Accuracy. Cancers 2019, 11, 1473. https://doi.org/10.3390/cancers11101473
Franz A, Ralla B, Weickmann S, Jung M, Rochow H, Stephan C, Erbersdobler A, Kilic E, Fendler A, Jung K. Circular RNAs in Clear Cell Renal Cell Carcinoma: Their Microarray-Based Identification, Analytical Validation, and Potential Use in a Clinico-Genomic Model to Improve Prognostic Accuracy. Cancers. 2019; 11(10):1473. https://doi.org/10.3390/cancers11101473
Chicago/Turabian StyleFranz, Antonia, Bernhard Ralla, Sabine Weickmann, Monika Jung, Hannah Rochow, Carsten Stephan, Andreas Erbersdobler, Ergin Kilic, Annika Fendler, and Klaus Jung. 2019. "Circular RNAs in Clear Cell Renal Cell Carcinoma: Their Microarray-Based Identification, Analytical Validation, and Potential Use in a Clinico-Genomic Model to Improve Prognostic Accuracy" Cancers 11, no. 10: 1473. https://doi.org/10.3390/cancers11101473
APA StyleFranz, A., Ralla, B., Weickmann, S., Jung, M., Rochow, H., Stephan, C., Erbersdobler, A., Kilic, E., Fendler, A., & Jung, K. (2019). Circular RNAs in Clear Cell Renal Cell Carcinoma: Their Microarray-Based Identification, Analytical Validation, and Potential Use in a Clinico-Genomic Model to Improve Prognostic Accuracy. Cancers, 11(10), 1473. https://doi.org/10.3390/cancers11101473