Capture of Circulating Tumour Cell Clusters Using Straight Microfluidic Chips
Abstract
1. Introduction
2. Results
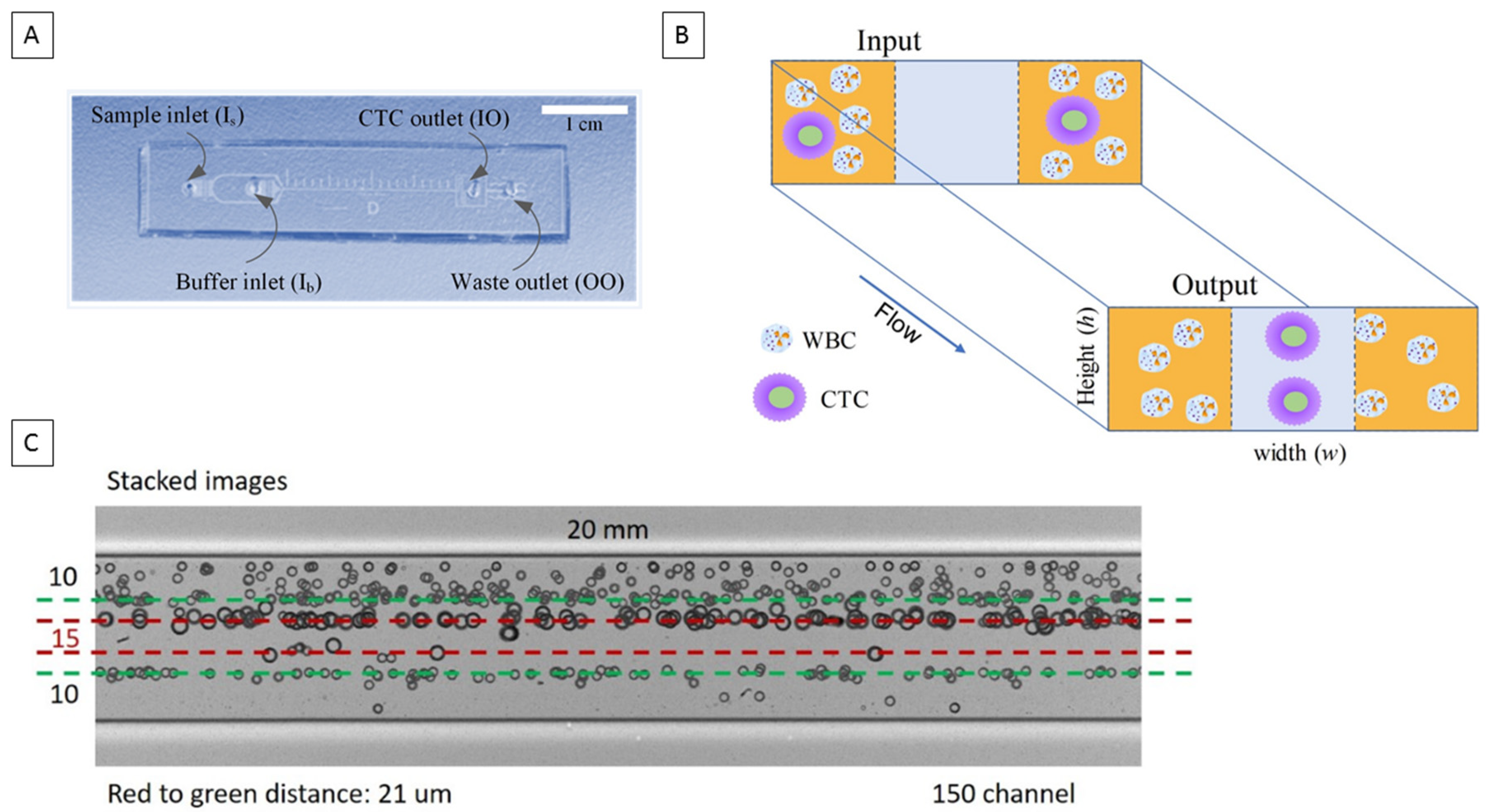
2.1. Microfluidic Chip Design
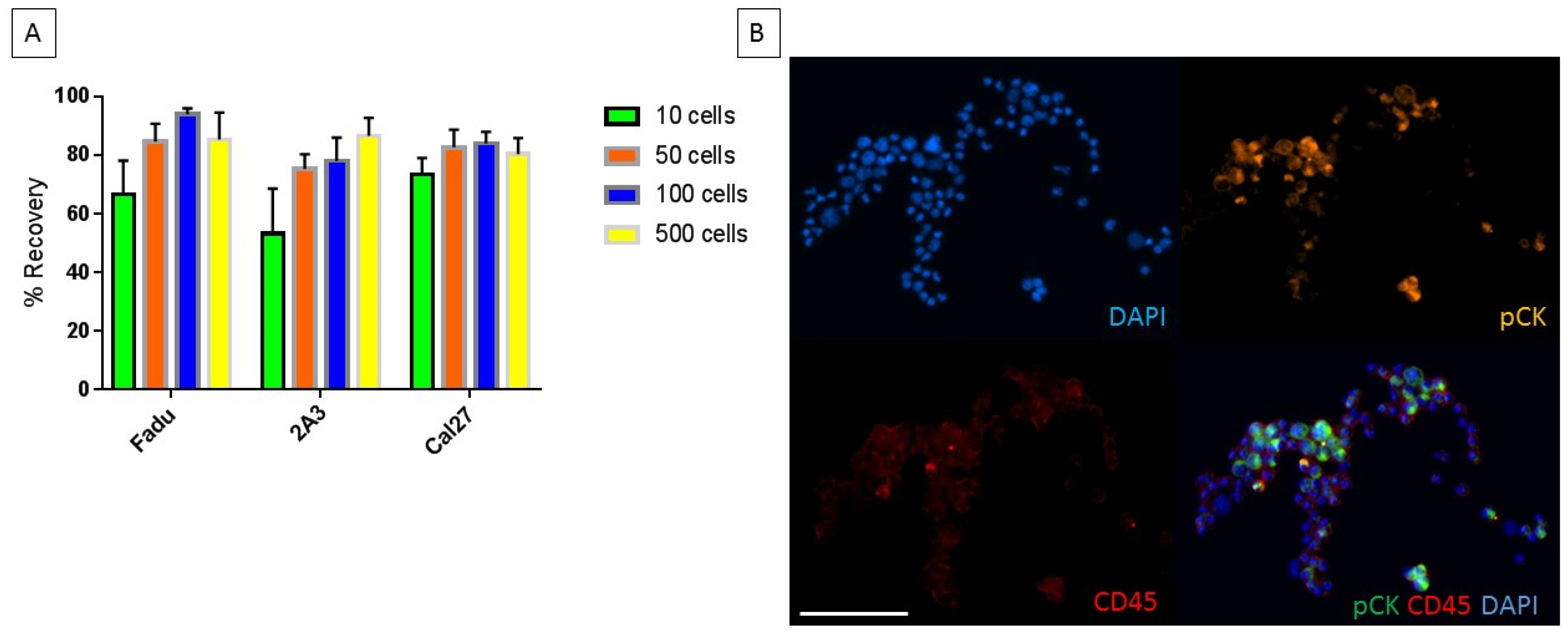
2.2. Characterization of Chip with Head and Neck Cancer Cell Lines
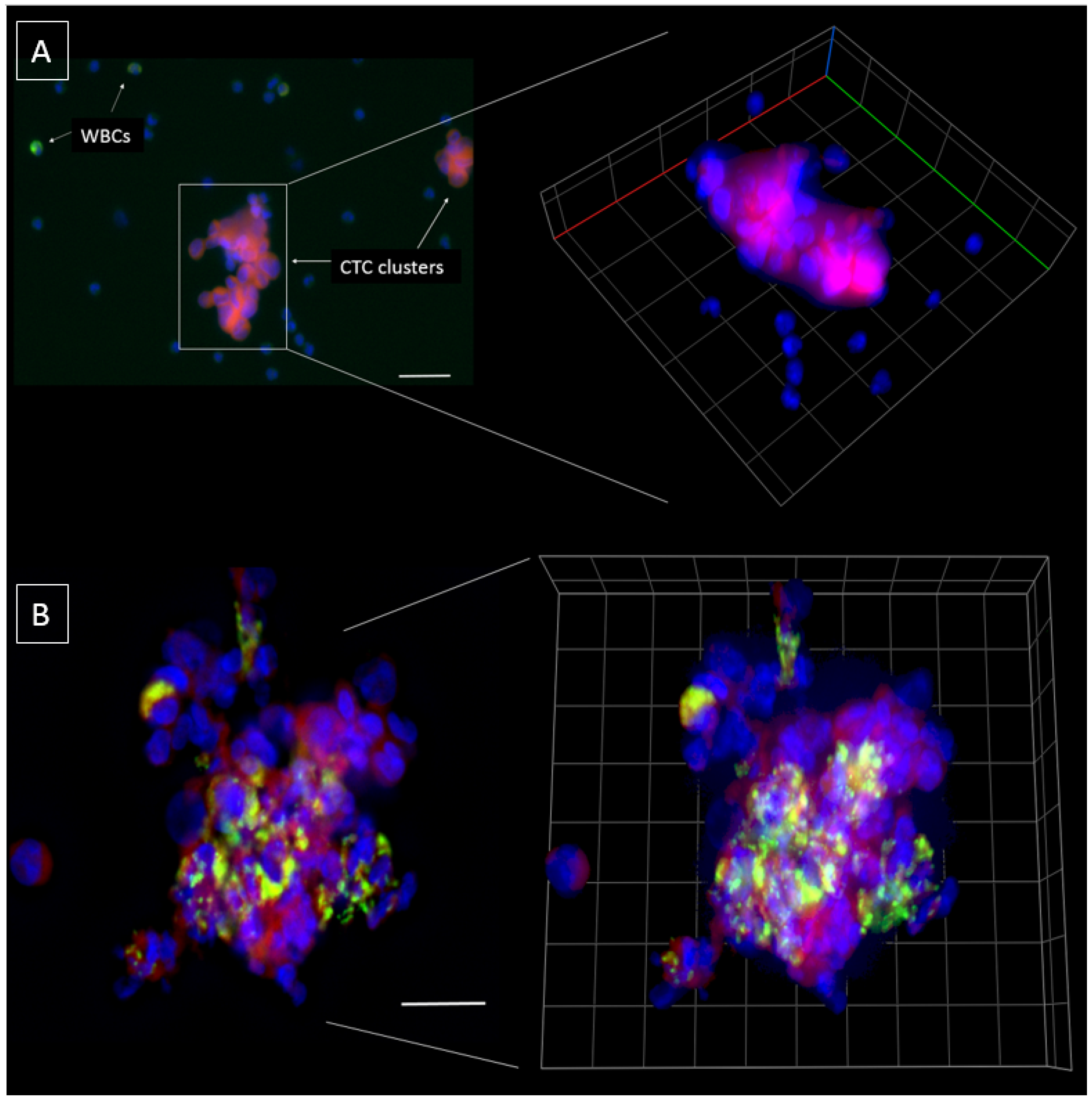
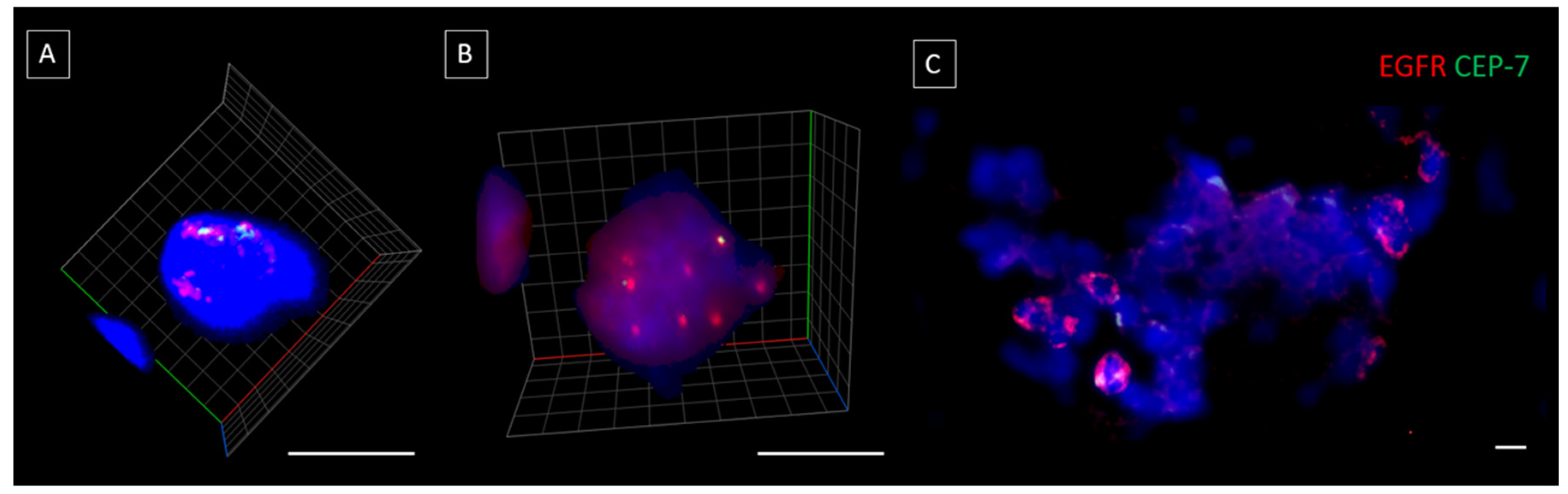
2.3. Characterization of the Chip Enriched Head and Neck Cancer Patient Blood Samples
3. Discussion
4. Materials and Methods
4.1. Chip Fabrication
4.2. Chip Operation and Workflow
4.3. Chip Characterization Using Cell Lines
4.4. Patient Recruitment
4.5. Enrichment and Phenotyping of CTCs/CTMs
4.6. Cell Imaging
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ashworth, T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust. Med. J. 1869, 14, 146–147. [Google Scholar]
- Watanabe, S. The metastasizability of tumor cells. Cancer 1954, 7, 215–223. [Google Scholar] [CrossRef]
- Kulasinghe, A.; Kenny, L.; Perry, C.; Thiery, J.P.; Jovanovic, L.; Vela, I.; Nelson, C.; Punyadeera, C. Impact of label-free technologies in head and neck cancer circulating tumour cells. Oncotarget 2016, 7, 71223–71234. [Google Scholar] [CrossRef] [PubMed]
- Kulasinghe, A.; Perry, C.; Jovanovic, L.; Nelson, C.; Punyadeera, C. Circulating tumour cells in metastatic head and neck cancers. Int. J. Cancer 2015, 136, 2515–2523. [Google Scholar] [CrossRef] [PubMed]
- Fachin, F.; Spuhler, P.; Martel-Foley, J.M.; Edd, J.F.; Barber, T.A.; Walsh, J.; Karabacak, M.; Pai, V.; Yu, M.; Smith, K.; et al. Monolithic Chip for High-throughput Blood Cell Depletion to Sort Rare Circulating Tumor Cells. Sci. Rep. 2017, 7, 10936. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science (N. Y.) 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.W.; Nagaraj, S.H. Transition states that allow cancer to spread. Nature 2018, 556, 442–444. [Google Scholar] [CrossRef]
- Klein, C.A. Selection and adaptation during metastatic cancer progression. Nature 2013, 501, 365–372. [Google Scholar] [CrossRef]
- Warkiani, M.E.; Khoo, B.L.; Wu, L.; Tay, A.K.; Bhagat, A.A.; Han, J.; Lim, C.T. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat. Protoc. 2016, 11, 134–148. [Google Scholar] [CrossRef]
- Khojah, R.; Stoutamore, R.; Di Carlo, D. Size-tunable microvortex capture of rare cells. Lab Chip 2017, 17, 2542–2549. [Google Scholar] [CrossRef]
- Kulasinghe, A.; Tran, T.H.; Blick, T.; O’Byrne, K.; Thompson, E.W.; Warkiani, M.E.; Nelson, C.; Kenny, L.; Punyadeera, C. Enrichment of circulating head and neck tumour cells using spiral microfluidic technology. Sci. Rep. 2017, 7, 42517. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Papautsky, I. Fundamentals of inertial focusing in microchannels. Lab Chip 2013, 13, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, D. Inertial microfluidics. Lab Chip 2009, 9, 3038–3046. [Google Scholar] [CrossRef] [PubMed]
- Long, E.; Ilie, M.; Bence, C.; Butori, C.; Selva, E.; Lalvee, S.; Bonnetaud, C.; Poissonnet, G.; Lacour, J.P.; Bahadoran, P.; et al. High expression of TRF2, SOX10, and CD10 in circulating tumor microemboli detected in metastatic melanoma patients. A potential impact for the assessment of disease aggressiveness. Cancer Med. 2016, 5, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Mascalchi, M.; Falchini, M.; Maddau, C.; Salvianti, F.; Nistri, M.; Bertelli, E.; Sali, L.; Zuccherelli, S.; Vella, A.; Matucci, M.; et al. Prevalence and number of circulating tumour cells and microemboli at diagnosis of advanced NSCLC. J. Cancer Res. Clin. Oncol. 2016, 142, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Kermanshah, L.; Poudineh, M.; Ahmed, S.; Nguyen, L.N.M.; Srikant, S.; Makonnen, R.; Pena Cantu, F.; Corrigan, M.; Kelley, S.O. Dynamic CTC phenotypes in metastatic prostate cancer models visualized using magnetic ranking cytometry. Lab Chip 2018, 18, 2055–2064. [Google Scholar] [CrossRef]
- Khoo, B.L.; Warkiani, M.E.; Tan, D.S.; Bhagat, A.A.; Irwin, D.; Lau, D.P.; Lim, A.S.; Lim, K.H.; Krisna, S.S.; Lim, W.T.; et al. Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells. PLoS ONE 2014, 9, e99409. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Grenci, G.; Jing, T.; Lim, Y.B.; Lee, S.C.; Thiery, J.P.; Han, J.; Lim, C.T. Liquid biopsy and therapeutic response: Circulating tumor cell cultures for evaluation of anticancer treatment. Sci. Adv. 2016, 2, e1600274. [Google Scholar] [CrossRef]
- Sheng, W.; Ogunwobi, O.O.; Chen, T.; Zhang, J.; George, T.J.; Liu, C.; Fan, Z.H. Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip 2014, 14, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Au, S.H.; Storey, B.D.; Moore, J.C.; Tang, Q.; Chen, Y.L.; Javaid, S.; Sarioglu, A.F.; Sullivan, R.; Madden, M.W.; O’Keefe, R.; et al. Clusters of circulating tumor cells traverse capillary-sized vessels. Proc. Natl. Acad. Sci. USA 2016, 113, 4947–4952. [Google Scholar] [CrossRef] [PubMed]
- Au, S.H.; Edd, J.; Stoddard, A.E.; Wong, K.H.K.; Fachin, F.; Maheswaran, S.; Haber, D.A.; Stott, S.L.; Kapur, R.; Toner, M. Microfluidic Isolation of Circulating Tumor Cell Clusters by Size and Asymmetry. Sci. Rep. 2017, 7, 2433. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Kulasinghe, A.; Bogseth, A.; O’Byrne, K.; Punyadeera, C.; Papautsky, I. Isolation of circulating tumor cells in non-small-cell-lung cancer patients using a multi-flow microfluidic channel. Nat. Microsyst. Nanotechnol. 2018, in press. [Google Scholar]
- Martel, J.M.; Toner, M. Inertial focusing in microfluidics. Annu. Rev. Biomed. Eng. 2014, 16, 371–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yan, S.; Yuan, D.; Alici, G.; Nguyen, N.T.; Ebrahimi Warkiani, M.; Li, W. Fundamentals and applications of inertial microfluidics: A review. Lab Chip 2016, 16, 10–34. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tu, C.; Liang, Y.; Huang, B.; Fang, Y.; Liang, X.; Papautsky, I.; Ye, X. Isolation of cells from whole blood using shear-induced diffusion. Sci. Rep. 2018, 8, 9411. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Zhou, J.; Liang, Y.; Huang, B.; Fang, Y.; Liang, X.; Ye, X. A flexible cell concentrator using inertial focusing. Biomed. Microdevices 2017, 19, e83. [Google Scholar] [CrossRef]
- Zhou, J.; Giridhar, P.V.; Kasper, S.; Papautsky, I. Modulation of aspect ratio for complete separation in an inertial microfluidic channel. Lab Chip 2013, 13, 1919–1929. [Google Scholar] [CrossRef]
- Che, J.; Yu, V.; Dhar, M.; Renier, C.; Matsumoto, M.; Heirich, K.; Garon, E.B.; Goldman, J.; Rao, J.; Sledge, G.W.; et al. Classification of large circulating tumor cells isolated with ultra-high throughput microfluidic Vortex technology. Oncotarget 2016, 7, 12748–12760. [Google Scholar] [CrossRef]
- Aya-Bonilla, C.A.; Marsavela, G.; Freeman, J.B.; Lomma, C.; Frank, M.H.; Khattak, M.A.; Meniawy, T.M.; Millward, M.; Warkiani, M.E.; Gray, E.S.; et al. Isolation and detection of circulating tumour cells from metastatic melanoma patients using a slanted spiral microfluidic device. Oncotarget 2017, 8, 67355–67368. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Gao, X.; Cheng, K.; Yu, L. Correlation between the presence of circulating tumor cells and the pathologic type and staging of non-small cell lung cancer during the early postoperative period. Oncol. Lett. 2017, 14, 5825–5830. [Google Scholar] [CrossRef] [PubMed]
- Umer, M.; Vaidyanathan, R.; Nguyen, N.T.; Shiddiky, M.J.A. Circulating tumor microemboli: Progress in molecular understanding and enrichment technologies. Biotechnol. Adv. 2018, 36, 1367–1389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gong, S.; Liu, Y.; Liang, L.; He, S.; Zhang, Q.; Si, M.; Yu, Z. Enumeration and molecular characterization of circulating tumor cell using an in vivo capture system in squamous cell carcinoma of head and neck. Chin. J. Cancer Res. Chung-Kuo Yen Cheng Yen Chiu 2017, 29, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Braunholz, D.; Saki, M.; Niehr, F.; Öztürk, M.; Borràs Puértolas, B.; Konschak, R.; Budach, V.; Tinhofer, I. Spheroid Culture of Head and Neck Cancer Cells Reveals an Important Role of EGFR Signalling in Anchorage Independent Survival. PLoS ONE 2016, 11, e0163149. [Google Scholar] [CrossRef] [PubMed]
- Kulasinghe, A.; Schmidt, H.; Perry, C.; Whitfield, B.; Kenny, L.; Nelson, C.; Warkiani, M.E.; Punyadeera, C. A Collective Route to Head and Neck Cancer Metastasis. Sci. Rep. 2018, 8, 746. [Google Scholar] [CrossRef] [PubMed]
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 2015, 12, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Krol, I.; Castro-Giner, F.; Maurer, M.; Gkountela, S.; Szczerba, B.M.; Scherrer, R.; Coleman, N.; Carreira, S.; Bachmann, F.; Anderson, S.; et al. Detection of circulating tumour cell clusters in human glioblastoma. Br. J. Cancer 2018, 119, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef]
- Cheung, K.J.; Padmanaban, V.; Silvestri, V.; Schipper, K.; Cohen, J.D.; Fairchild, A.N.; Gorin, M.A.; Verdone, J.E.; Pienta, K.J.; Bader, J.S.; et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. USA 2016, 113, E854–E863. [Google Scholar] [CrossRef]
- Choi, J.W.; Kim, J.K.; Yang, Y.J.; Kim, P.; Yoon, K.H.; Yun, S.H. Urokinase exerts antimetastatic effects by dissociating clusters of circulating tumor cells. Cancer Res. 2015, 75, 4474–4482. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Fang, F.; Zhang, Q. Circulating tumor cell clusters: What we know and what we expect (Review). Int. J. Oncol. 2016, 49, 2206–2216. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Mu, Z.; Chervoneva, I.; Austin, L.; Ye, Z.; Rossi, G.; Palazzo, J.P.; Sun, C.; Abu-Khalaf, M.; Myers, R.E.; et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res. Treat. 2017, 161, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Network, T.C.G.A. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Y.; Xu, J.; Zhang, A.; Wang, X.; Tang, R.; Zhang, X.; Yin, H.; Liu, M.; Wang, D.D.; et al. Quantified postsurgical small cell size CTCs and EpCAM+ circulating tumor stem cells with cytogenetic abnormalities in hepatocellular carcinoma patients determine cancer relapse. Cancer Lett. 2018, 412, 99–107. [Google Scholar] [CrossRef] [PubMed]

| Head and Neck Cancer | n |
|---|---|
| Total | 21 |
| Gender | |
| Male | 17 |
| Female | 4 |
| Age, y | |
| <60 | 12 |
| >60 | 9 |
| Tumour Type | |
| Oral Cavity | 12 |
| Oropharynx | 9 |
| Tumour Stage | |
| I | 4 |
| II | 4 |
| III | 8 |
| IV | 5 |
| HPV Status (p16 immunostainings) | |
| HPV-positive | 14 |
| HPV-negative | 7 |
| CTC findings/mL blood | |
| CTC+ (single cells) | 10 (Range 2–24) |
| CTC clusters | 9 (Range 1–6) |
| Single CTCs and clusters | 5 |
| CTC (single or cluster)+ | 14/21 (66.7%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulasinghe, A.; Zhou, J.; Kenny, L.; Papautsky, I.; Punyadeera, C. Capture of Circulating Tumour Cell Clusters Using Straight Microfluidic Chips. Cancers 2019, 11, 89. https://doi.org/10.3390/cancers11010089
Kulasinghe A, Zhou J, Kenny L, Papautsky I, Punyadeera C. Capture of Circulating Tumour Cell Clusters Using Straight Microfluidic Chips. Cancers. 2019; 11(1):89. https://doi.org/10.3390/cancers11010089
Chicago/Turabian StyleKulasinghe, Arutha, Jian Zhou, Liz Kenny, Ian Papautsky, and Chamindie Punyadeera. 2019. "Capture of Circulating Tumour Cell Clusters Using Straight Microfluidic Chips" Cancers 11, no. 1: 89. https://doi.org/10.3390/cancers11010089
APA StyleKulasinghe, A., Zhou, J., Kenny, L., Papautsky, I., & Punyadeera, C. (2019). Capture of Circulating Tumour Cell Clusters Using Straight Microfluidic Chips. Cancers, 11(1), 89. https://doi.org/10.3390/cancers11010089







