Immune Gene Signature Delineates a Subclass of Papillary Thyroid Cancer with Unfavorable Clinical Outcomes
Abstract
1. Introduction
2. Results
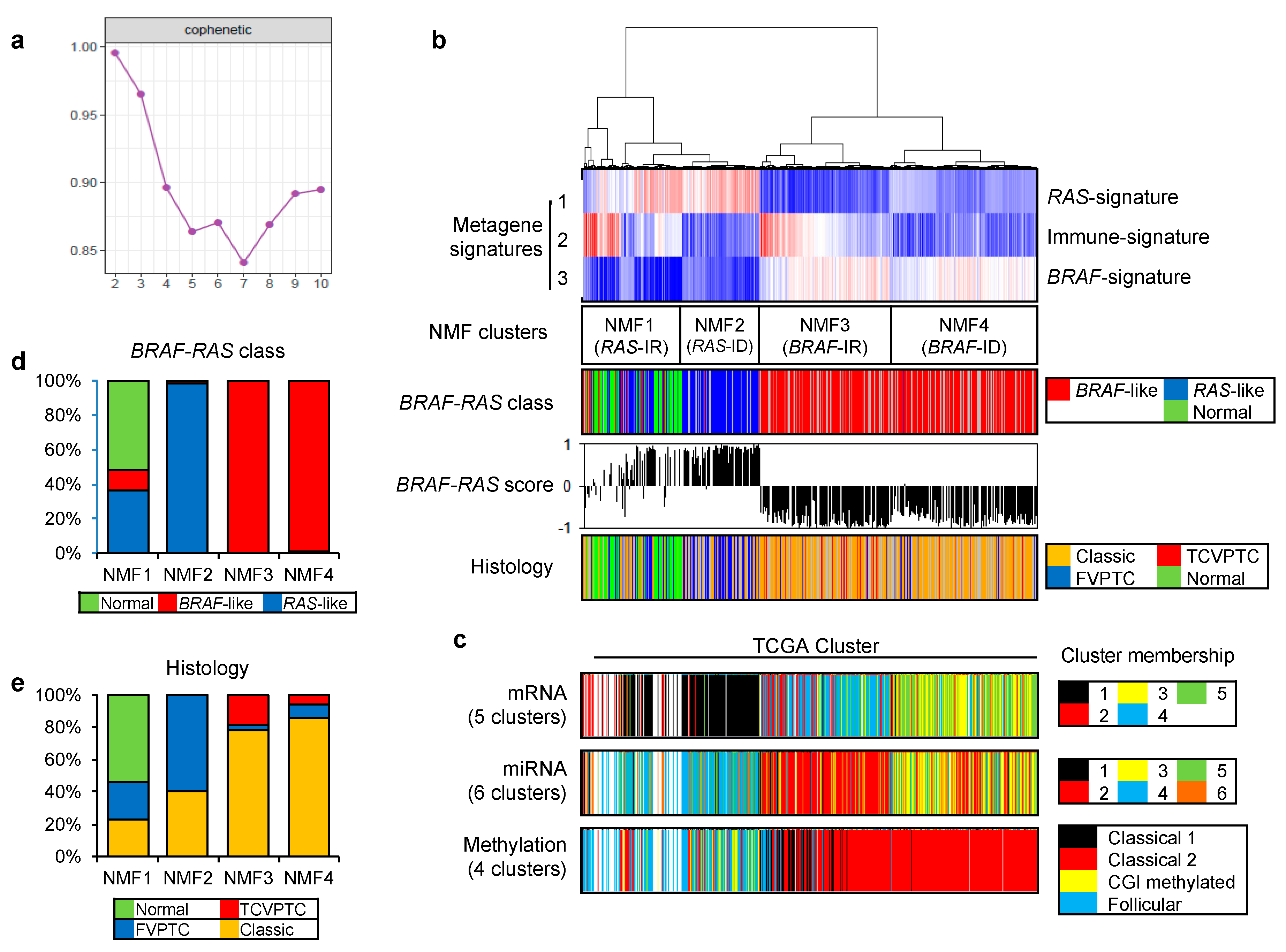
2.1. Deconvolution of PTC Expression Profiles into Key Metagene Signatures
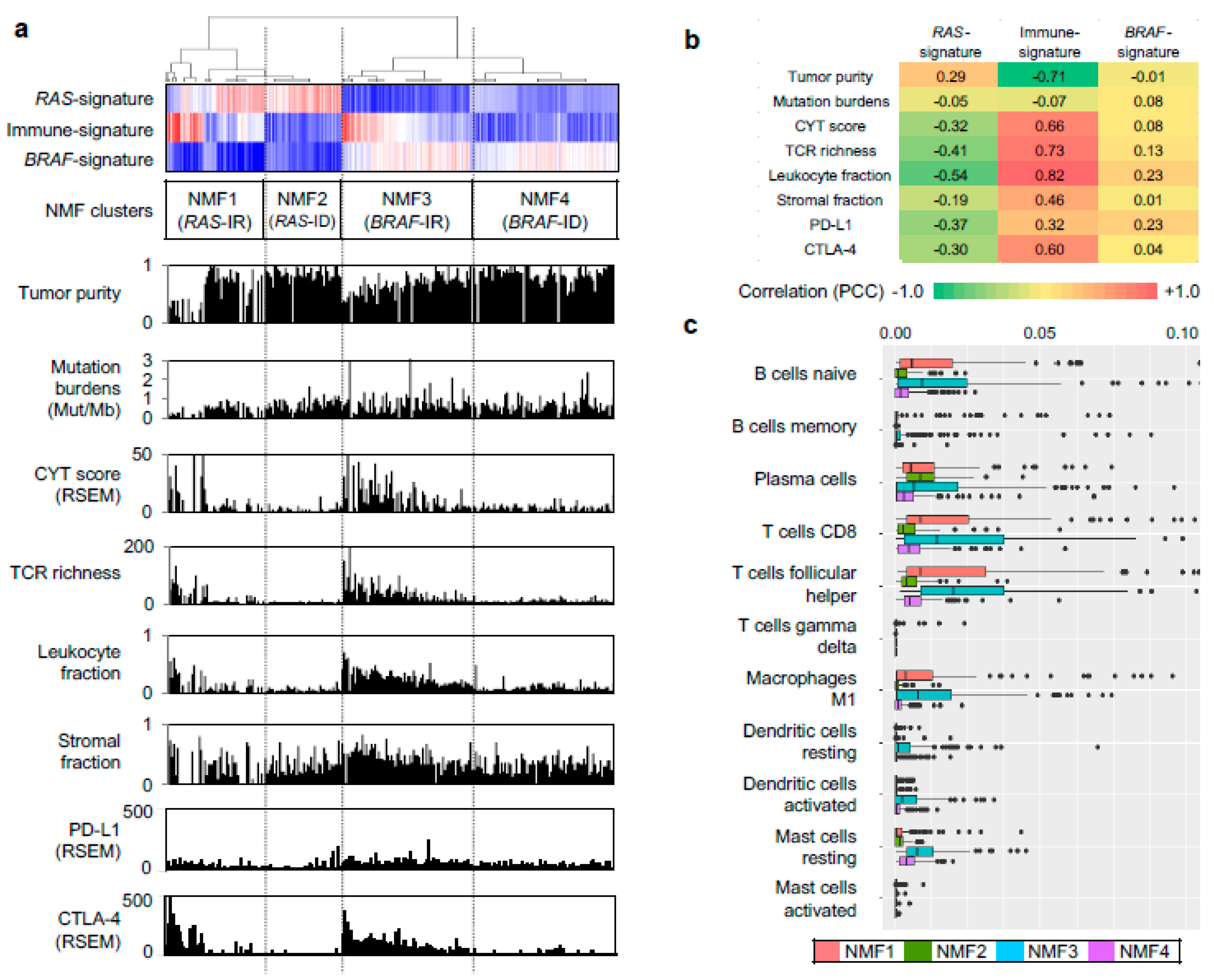
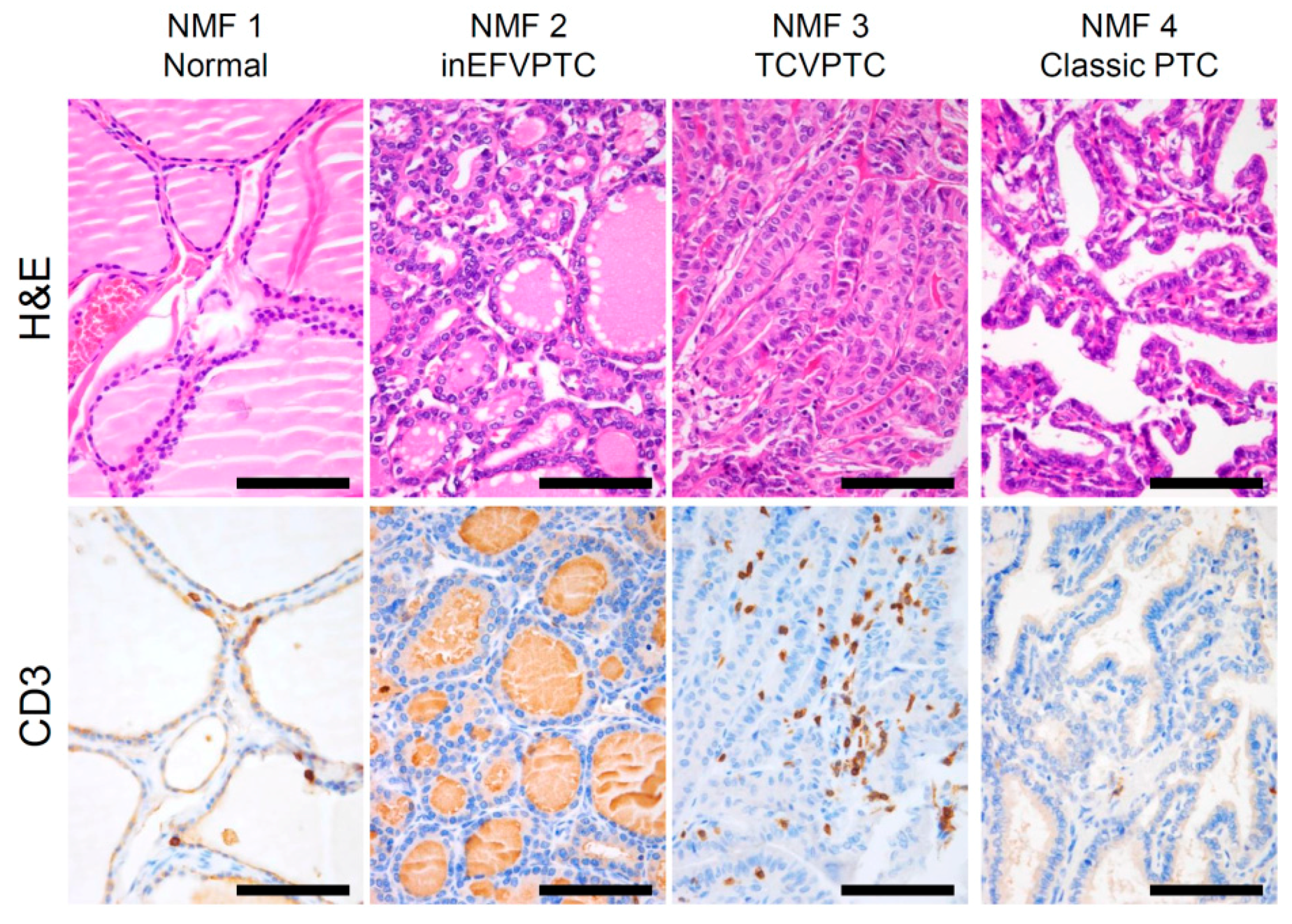
2.2. Functional Annotation of Immune-Related Metagene Signature
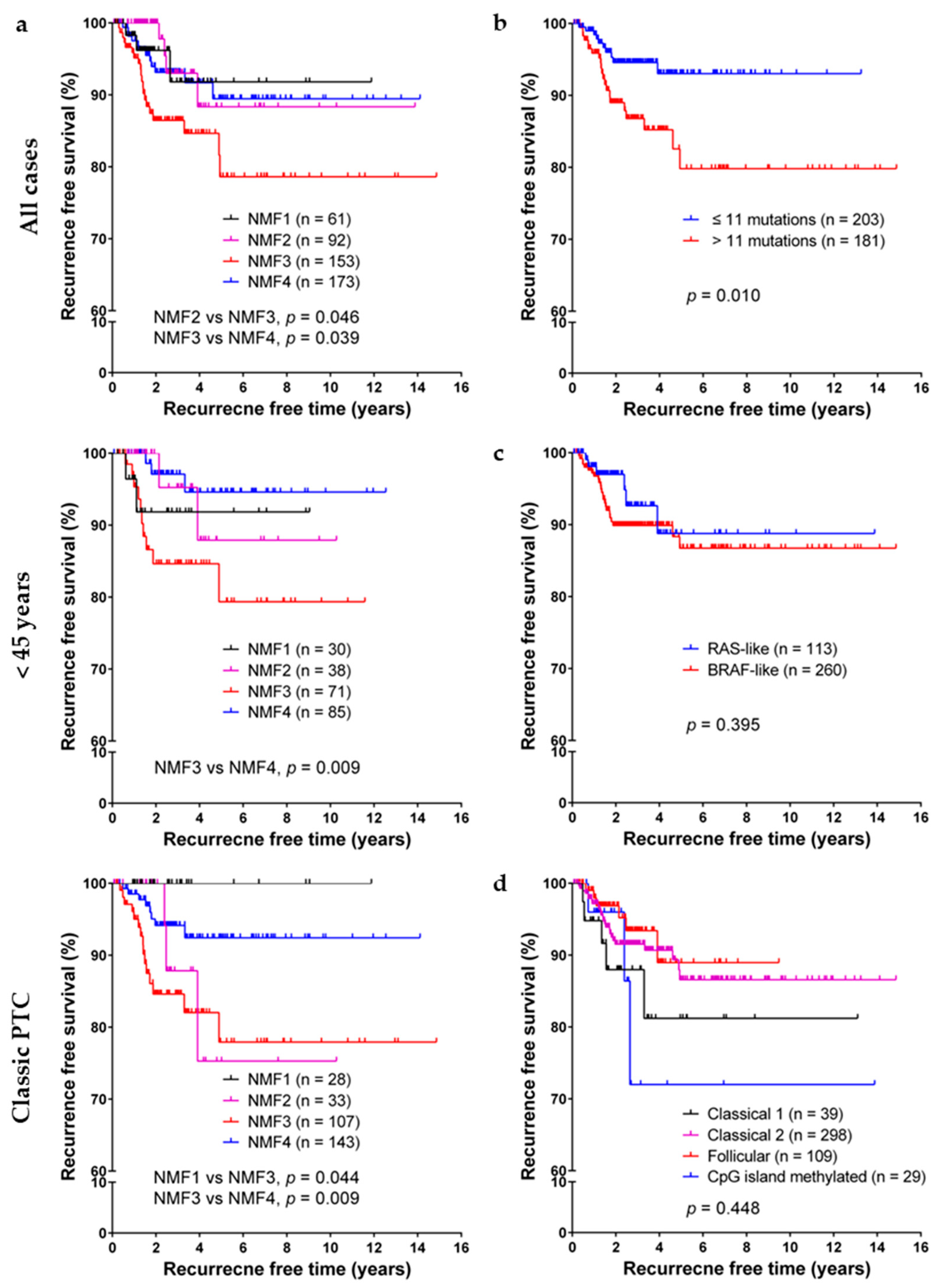
2.3. Prognostic Impact of NMF Clustering-Based PTC Classification in the TCGA Dataset
2.4. Validation of NMF Clustering-Based Classification in Two Different Cohorts
3. Discussion
4. Materials and Methods
4.1. Public PTC Transcriptome Data
4.2. Transcriptome Sequencing and Data Processing
4.3. NMF and Metagene Signatures of PTCs
4.4. Immunoprofiling
4.5. Immunohistochemistry
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ahn, H.S.; Kim, H.J.; Welch, H.G. Korea’s thyroid-cancer “epidemic”—Screening and overdiagnosis. N. Engl. J. Med. 2014, 371, 1765–1767. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Devesa, S.S.; Sosa, J.A.; Check, D.; Kitahara, C.M. Trends in Thyroid Cancer Incidence and Mortality in the United States, 1974–2013. JAMA 2017, 317, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Grogan, R.H.; Kaplan, S.P.; Cao, H.; Weiss, R.E.; Degroot, L.J.; Simon, C.A.; Embia, O.M.; Angelos, P.; Kaplan, E.L.; Schechter, R.B. A study of recurrence and death from papillary thyroid cancer with 27 years of median follow-up. Surgery 2013, 154, 1436–1446. [Google Scholar] [CrossRef] [PubMed]
- Nikiforov, Y.E.; Baloch, Z.W.; Belge, G.; Chan, J.K.C.; Derwahl, K.M.; Evans, H.L.; Fagin, J.A.; Ghossein, R.A.; Lloyd, R.V.; Oriola, J.; et al. Tumors of the thyroid gland. In WHO Classification of Tumours of Endocrine Organs, 4th ed.; Lloyd, R.V., Osamura, R.Y., Klöppel, G., Rosai, J., Eds.; WHO Press: Geneva, Switzerland, 2017; Volume 10, pp. 66–103. [Google Scholar]
- Haugen, B.R.; Alexander, E.K.; Bible, K.C.; Doherty, G.M.; Mandel, S.J.; Nikiforov, Y.E.; Pacini, F.; Randolph, G.W.; Sawka, A.M.; Schlumberger, M.; et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid 2016, 26, 1–133. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Hay, I.D.; Bergstralh, E.J.; Goellner, J.R.; Ebersold, J.R.; Grant, C.S. Predicting outcome in papillary thyroid carcinoma: Development of a reliable prognostic scoring system in a cohort of 1779 patients surgically treated at one institution during 1940 through 1989. Surgery 1993, 114, 1050–1057. [Google Scholar] [PubMed]
- Smallridge, R.C.; Chindris, A.M.; Asmann, Y.W.; Casler, J.D.; Serie, D.J.; Reddi, H.V.; Cradic, K.W.; Rivera, M.; Grebe, S.K.; Necela, B.M.; et al. RNA sequencing identifies multiple fusion transcripts, differentially expressed genes, and reduced expression of immune function genes in BRAF (V600E) mutant vs. BRAF wild-type papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2014, 99, E338–E347. [Google Scholar] [CrossRef] [PubMed]
- Na, K.J.; Choi, H. Immune landscape of papillary thyroid cancer and immunotherapeutic implications. Endocr. Relat. Cancer 2018, 25, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Kim, Y.E.; Ahn, S.; Kim, J.Y.; Ki, C.S.; Oh, Y.L.; Kim, K.; Yun, J.W.; Park, W.Y.; Choe, J.H.; et al. TERT promoter mutations and long-term survival in patients with thyroid cancer. Endocr. Relat. Cancer 2016, 23, 813–823. [Google Scholar] [CrossRef]
- Liu, R.; Xing, M. TERT promoter mutations in thyroid cancer. Endocr. Relat. Cancer 2016, 23, R143–155. [Google Scholar] [CrossRef]
- Xing, M.; Liu, R.; Liu, X.; Murugan, A.K.; Zhu, G.; Zeiger, M.A.; Pai, S.; Bishop, J. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J. Clin. Oncol. 2014, 32, 2718–2726. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Brunet, J.P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA 2004, 101, 4164–4169. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Sia, D.; Jiao, Y.; Martinez-Quetglas, I.; Kuchuk, O.; Villacorta-Martin, C.; Castro de Moura, M.; Putra, J.; Camprecios, G.; Bassaganyas, L.; Akers, N.; et al. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology 2017, 153, 812–826. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef]
- Yoo, S.K.; Lee, S.; Kim, S.J.; Jee, H.G.; Kim, B.A.; Cho, H.; Song, Y.S.; Cho, S.W.; Won, J.K.; Shin, J.Y.; et al. Comprehensive Analysis of the Transcriptional and Mutational Landscape of Follicular and Papillary Thyroid Cancers. PLoS Genet. 2016, 12, e1006239. [Google Scholar] [CrossRef]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef]
- Rhee, J.K.; Jung, Y.C.; Kim, K.R.; Yoo, J.; Kim, J.; Lee, Y.J.; Ko, Y.H.; Lee, H.H.; Cho, B.C.; Kim, T.M. Impact of Tumor Purity on Immune Gene Expression and Clustering Analyses across Multiple Cancer Types. Cancer Immunol. Res. 2018, 6, 87–97. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martinez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Kimura, E.T.; Nikiforova, M.N.; Zhu, Z.; Knauf, J.A.; Nikiforov, Y.E.; Fagin, J.A. High prevalence of BRAF mutations in thyroid cancer: Genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003, 63, 1454–1457. [Google Scholar] [PubMed]
- Soares, P.; Trovisco, V.; Rocha, A.S.; Lima, J.; Castro, P.; Preto, A.; Maximo, V.; Botelho, T.; Seruca, R.; Sobrinho-Simoes, M. BRAF mutations and RET/PTC rearrangements are alternative events in the etiopathogenesis of PTC. Oncogene 2003, 22, 4578–4580. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015, 372, 2521–2532. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef] [PubMed]
- French, J.D.; Bible, K.; Spitzweg, C.; Haugen, B.R.; Ryder, M. Leveraging the immune system to treat advanced thyroid cancers. Lancet Diabetes Endocrinol. 2017, 5, 469–481. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Lipson, E.J.; Forde, P.M.; Hammers, H.J.; Emens, L.A.; Taube, J.M.; Topalian, S.L. Antagonists of PD-1 and PD-L1 in Cancer Treatment. Semin. Oncol. 2015, 42, 587–600. [Google Scholar] [CrossRef]
- Kuo, C.-Y.; Liu, T.-P.; Yang, P.-S.; Cheng, S.-P. Characteristics of lymphocyte-infiltrating papillary thyroid cancer. J. Cancer Res. Pract. 2017, 4, 95–99. [Google Scholar] [CrossRef]
- Cunha, L.L.; Marcello, M.A.; Nonogaki, S.; Morari, E.C.; Soares, F.A.; Vassallo, J.; Ward, L.S. CD8+ tumour-infiltrating lymphocytes and COX2 expression may predict relapse in differentiated thyroid cancer. Clin. Endocrinol. 2015, 83, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Halvorsen, E.C.; Mahmoud, S.M.; Bennewith, K.L. Emerging roles of regulatory T cells in tumour progression and metastasis. Cancer Metastasis Rev. 2014, 33, 1025–1041. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.Y.; Cho, S.W.; Kim, Y.A.; Kim, D.; Oh, B.C.; Park, D.J.; Park, Y.J. Cancers with Higher Density of Tumor-Associated Macrophages Were Associated with Poor Survival Rates. J. Pathol. Transl. Med. 2015, 49, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Qing, W.; Fang, W.Y.; Ye, L.; Shen, L.Y.; Zhang, X.F.; Fei, X.C.; Chen, X.; Wang, W.Q.; Li, X.Y.; Xiao, J.C.; et al. Density of Tumor-Associated Macrophages Correlates with Lymph Node Metastasis in Papillary Thyroid Carcinoma. Thyroid 2012, 22, 905–910. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Tamayo, P.; Scanfeld, D.; Ebert, B.L.; Gillette, M.A.; Roberts, C.W.; Mesirov, J.P. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. Proc. Natl. Acad. Sci. USA 2007, 104, 5959–5964. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef]
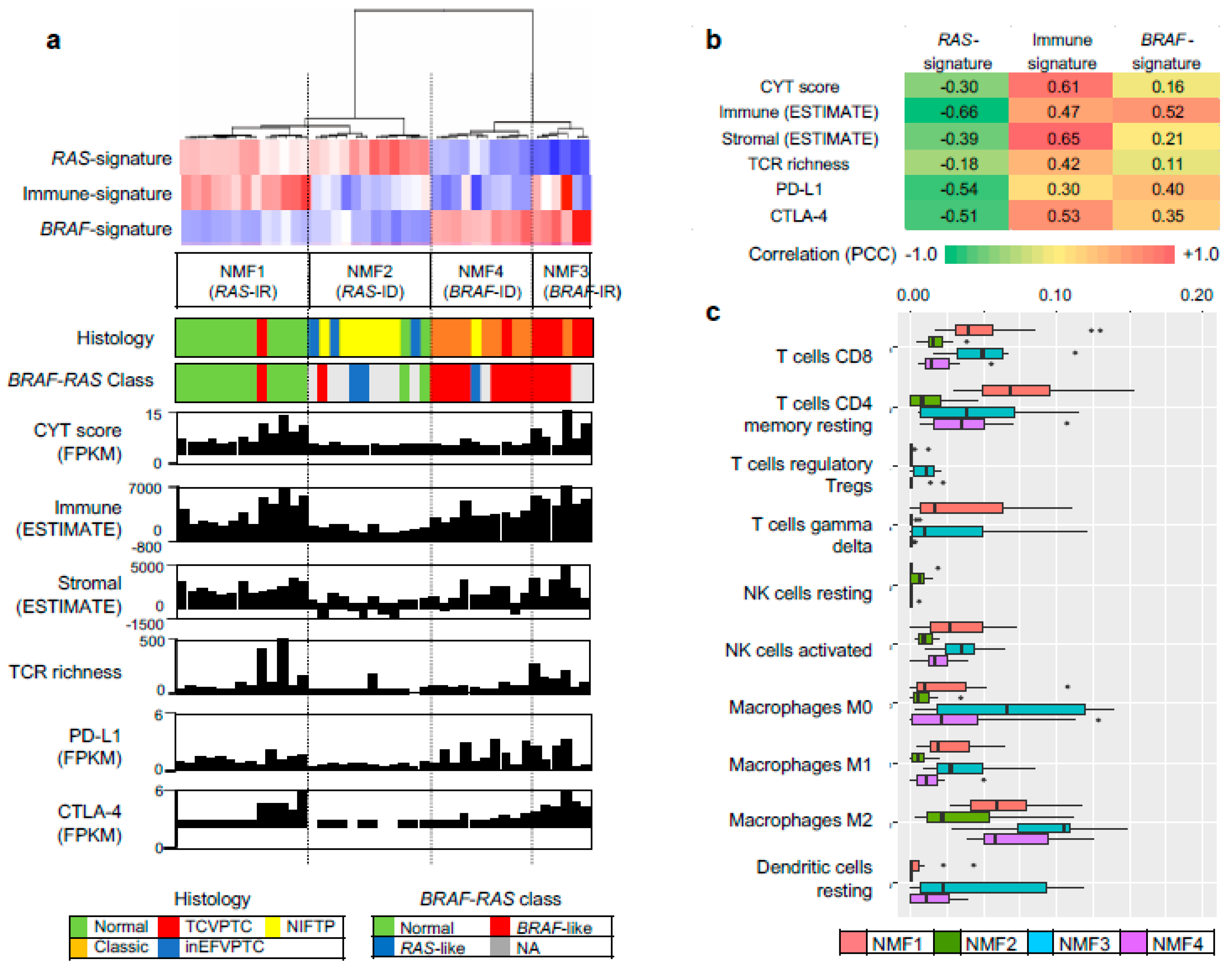
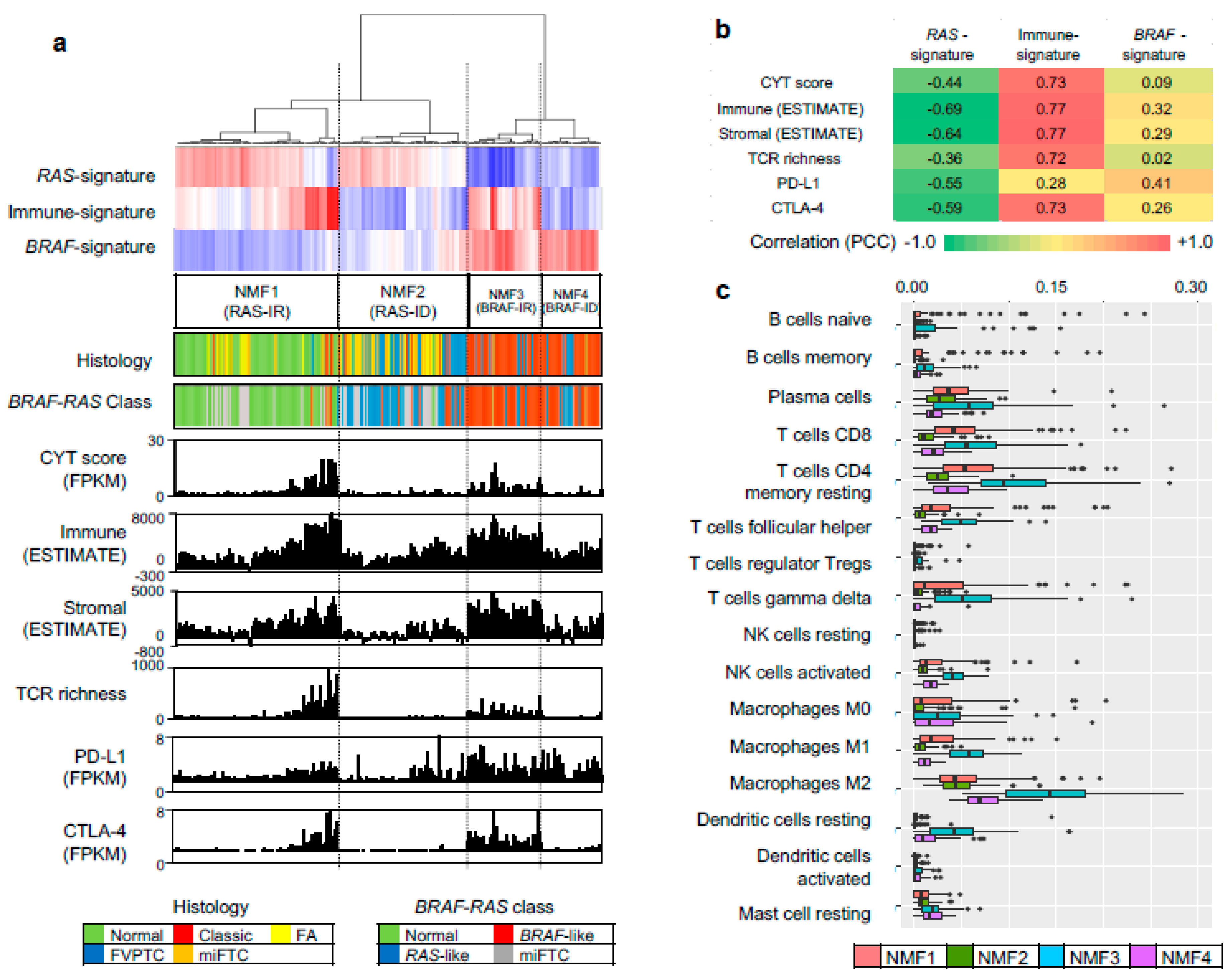
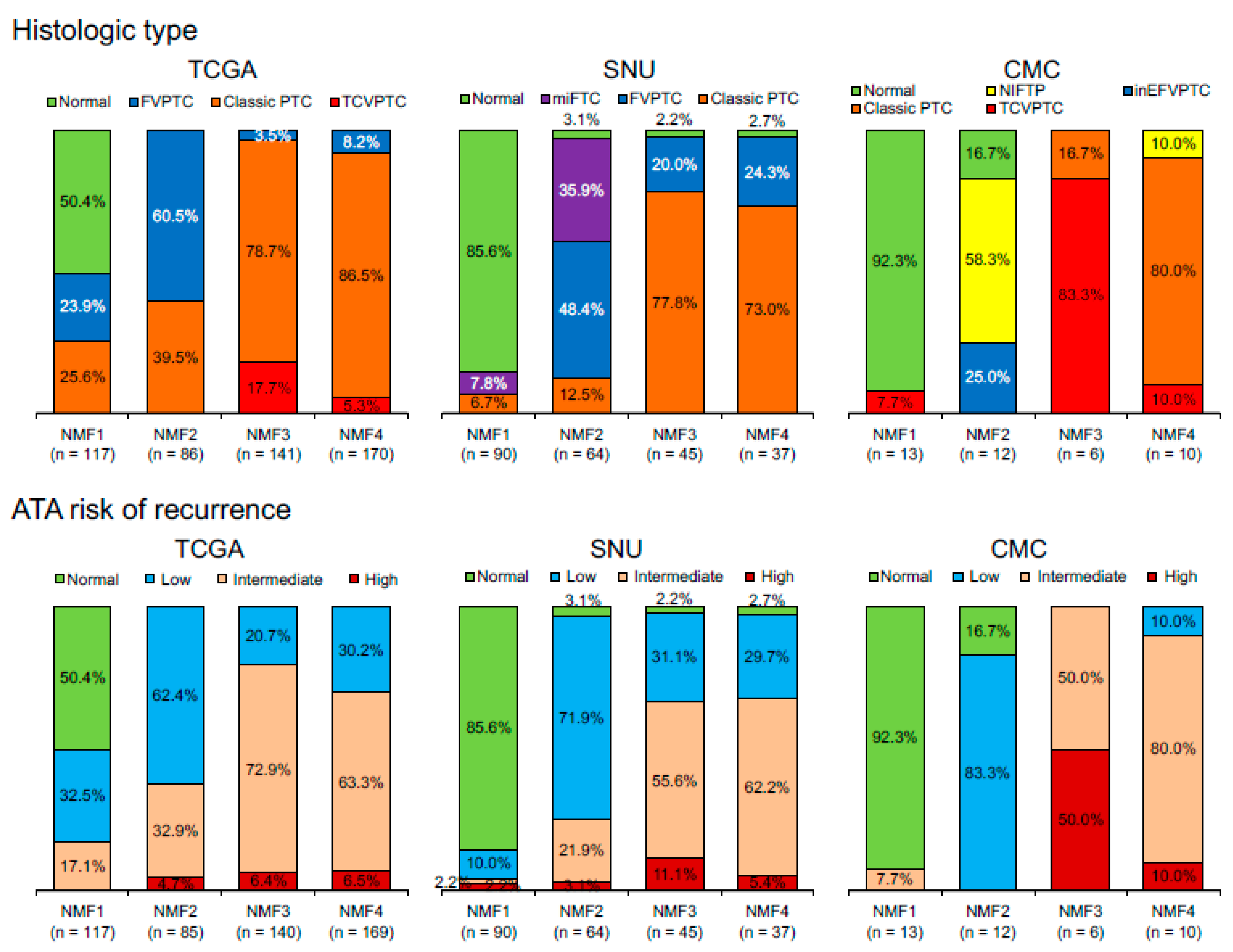
| Characteristic | n | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | p-Value | p-Value * |
|---|---|---|---|---|---|---|---|
| Age (years) | 500 | ||||||
| <45 | 228 | 30 (13.2%) | 40 (17.5%) | 72 (31.6%) | 86 (37.7%) | 0.788 | 0.720 |
| ≥45 | 272 | 34 (12.5%) | 57 (21.0%) | 86 (31.6%) | 95 (34.9%) | ||
| <55 | 335 | 44 (13.1%) | 57 (17.0%) | 102 (30.4%) | 132 (39.4%) | 0.095 | 0.096 |
| ≥55 | 165 | 20 (12.1%) | 40 (24.2%) | 56 (33.9%) | 49 (29.7%) | ||
| Sex | 0.965 | 0.828 | |||||
| Female | 340 | 42 (12.4%) | 63 (18.5%) | 108 (31.8%) | 127 (37.4%) | ||
| Male | 124 | 17 (13.7%) | 24 (19.4%) | 37 (29.8%) | 46 (37.1%) | ||
| Histologic variant | 464 | <0.001 | 0.008 | ||||
| Classic | 322 | 30 (9.3%) | 34 (10.6%) | 111 (34.5%) | 147 (45.7%) | ||
| Follicular variant | 99 | 28 (28.3%) | 52 (52.5%) | 5 (5.1%) | 14 (14.1%) | ||
| Tall cell variant | 34 | 0 (0.0%) | 0 (0.0%) | 25 (73.5%) | 9 (26.5%) | ||
| Other | 9 | 1 (11.1%) | 1 (11.1%) | 4 (44.4%) | 3 (33.3%) | ||
| Extrathyroidal extension | 500 | <0.001 | 0.009 | ||||
| Absent | 367 | 58 (15.8%) | 87 (23.7%) | 92 (25.1%) | 130 (35.4%) | ||
| Present | 133 | 6 (4.5%) | 10 (7.5%) | 66 (49.6%) | 51 (38.3%) | ||
| Pathologic T (pT) stage | 462 | <0.001 | 0.024 | ||||
| pT1 | 134 | 26 (19.4%) | 28 (20.9%) | 38 (28.4%) | 42 (31.3%) | ||
| pT2 | 157 | 18 (11.5%) | 38 (24.2%) | 34 (21.7%) | 67 (42.7%) | ||
| pT3 | 153 | 15 (9.8%) | 20 (13.1%) | 64 (41.8%) | 54 (35.3%) | ||
| pT4 | 18 | 0 (0.0%) | 1 (5.6%) | 8 (44.4%) | 9 (50.0%) | ||
| Pathologic N (pN) stage | 500 | <0.001 | 0.720 | ||||
| pN0/NX | 294 | 52 (17.7%) | 84 (28.6%) | 72 (24.5%) | 86 (29.3%) | ||
| pN1 | 206 | 12 (5.8%) | 13 (6.3%) | 86 (41.7%) | 95 (46.1%) | ||
| Initial distant metastasis | 500 | 0.470 | 1.000 | ||||
| Absent | 492 | 64 (13.0%) | 94 (19.1%) | 156 (31.7%) | 178 (36.2%) | ||
| Present | 8 | 0 | 3 (37.5%) | 2 (25.0%) | 3 (37.5%) | ||
| Tumor stage | 498 | <0.001 | 0.020 | ||||
| stage 1 | 283 | 44 (15.5%) | 52 (18.4%) | 86 (30.4%) | 101 (35.7%) | ||
| stage 2 | 51 | 10 (19.6%) | 19 (37.3%) | 4 (7.8%) | 18 (35.3%) | ||
| stage 3 | 109 | 10 (9.2%) | 18 (16.5%) | 45 (41.3%) | 36 (33.0%) | ||
| stage 4 | 55 | 0 (0.0%) | 7 (12.7%) | 23 (41.8%) | 25 (45.5%) | ||
| Thyroiditis | 0.004 | 0.002 | |||||
| Absent | 372 | 45 (12.1%) | 72 (19.4%) | 106 (28.5%) | 149 (40.1%) | ||
| Present | 70 | 14 (20.0%) | 9 (12.9%) | 31 (44.3%) | 16 (22.9%) | ||
| RAS and BRAF signature | <0.001 | 1.000 | |||||
| RAS-like | 118 | 40 (33.9%) | 77 (65.3%) | 0 | 1 (0.8%) | ||
| BRAF-like | 272 | 14 (5.1%) | 1 (0.4%) | 122 (44.9%) | 135 (49.5%) | ||
| MACIS | 442 | 0.318 | 0.050 | ||||
| <6 | 315 | 41 (13.0%) | 58 (18.4%) | 87 (27.6%) | 129 (41.0%) | ||
| 6–7 | 63 | 9 (14.3%) | 13 (20.6%) | 25 (39.7%) | 16 (25.4%) | ||
| 7–8 | 39 | 4 (10.3%) | 7 (17.9%) | 15 (38.5%) | 13 (33.3%) | ||
| >8 | 25 | 1 (4.0%) | 4 (16.0%) | 11 (44.0%) | 9 (36.0%) | ||
| ATA5 recurrence risk | 452 | <0.001 | 0.159 | ||||
| Low | 171 | 38 (22.2%) | 53 (31.0%) | 29 (17.0%) | 51 (29.8%) | ||
| Intermediate | 257 | 20 (7.8%) | 28 (10.9%) | 102 (39.7%) | 107 (41.6%) | ||
| High | 24 | 0 (0.0%) | 4 (16.7%) | 9 (37.5%) | 11 (45.8%) | ||
| Recurrence free status | 479 | 0.048 | 0.063 | ||||
| Disease free | 440 | 58 (13.2%) | 88 (20.0%) | 133 (30.2%) | 161 (36.6%) | ||
| Recurrent | 39 | 3 (7.7%) | 4 (10.3%) | 20 (51.3%) | 12 (30.8%) |
| Characteristic | No. of Recurrences | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| Hazard Ratio (95% CI) | p-Value | Hazard Ratio (95% CI) | p-Value | ||
| Age (years) | 0.032 | 0.180 | |||
| <55 | 22/334 (6.6%) | 1 (reference) | 1 (reference) | ||
| ≥55 | 19/152 (12.5%) | 2.03 (1.06–3.87) | 1.91 (0.74–4.89) | ||
| Sex | 0.214 | 0.476 | |||
| Female | 24/331 (7.3%) | 1 (reference) | 1 (reference) | ||
| Male | 13/119 (10.9%) | 1.57 (0.77–3.19) | 1.41 (0.55–3.58) | ||
| Histologic variant | 0.062 | 0.752 | |||
| Non-aggressive variant | 31/414 (7.5%) | 1 (reference) | 1 (reference) | ||
| Aggressive variant | 6/36 (16.7%) | 2.47 (0.96–6.39) | 1.22 (0.36–4.08) | ||
| Extrathyroidal extension | 0.008 | 0.374 | |||
| Absent | 23/359 (6.4%) | 1 (reference) | 1 (reference) | ||
| Present | 18/127 (14.2%) | 2.41 (1.26–4.64) | 1.49 (0.62–3.57) | ||
| Pathologic T (pT) stage | 0.007 | 0.501 | |||
| pT1-2 | 16/287 (5.6%) | 1 (reference) | 1 (reference) | ||
| pT3-4 | 21/161 (13.0%) | 2.54 (1.29–5.02) | 0.62 (0.15–2.49) | ||
| Pathologic N (pN) stage | 0.003 | ||||
| pN0/NX | 15/287 (5.2%) | 1 (reference) | 1 (reference) | 0.001 | |
| pN1 | 26/199 (13.1%) | 2.73 (1.40–5.29) | 5.27 (2.02–13.74) | ||
| RAS and BRAF signature | 0.206 | 0.548 | |||
| RAS-like | 6/113 (5.3%) | 1 (reference) | 1 (reference) | ||
| BRAF-like | 24/260 (9.2%) | 1.81 (0.72–4.57) | 0.65 (0.16–2.61) | ||
| Burden of nonsynonymous mutations | 0.020 | 0.021 | |||
| ≤11 mutations | 10/203 (4.9%) | 1 (reference) | 1 (reference) | ||
| >11 mutations | 21/181 (11.6%) | 2.53 (1.16–5.54) | 2.78 (1.17–6.62) | ||
| NMF cluster | 0.008 | 0.010 | |||
| Non-NMF cluster 3 | 19/326 (5.8%) | 1 (reference) | 1 (reference) | ||
| NMF cluster 3 | 20/153 (13.1%) | 2.43 (1.26–4.70) | 3.01 (1.31–6.95) | ||
| Variables | Univariate Analysis | Multivariate Analysis | |
|---|---|---|---|
| Hazard Ratio (95% CI) | p-Value | ||
| Age ≥ 55 years | 0.014 | 1.81 (0.75–4.38) | 0.187 |
| Male | 0.129 | 1.33 (0.56–3.14) | 0.521 |
| Aggressive variant | 0.028 | 1.39 (0.49–3.95) | 0.531 |
| Extrathyroidal extension | 0.017 | 1.28 (0.56–2.91) | 0.560 |
| Lymph node metastasis | 0.002 | 4.98 (2.00–12.38) | 0.001 |
| BRAF-like tumor | 0.395 | 1.63 (0.45–5.99) | 0.459 |
| Nonsynonymous mutations > 11 mutations | 0.010 | 2.68 (1.20–5.99) | 0.016 |
| NMF cluster 3 | 0.007 | 1.42 (1.10–1.84) | 0.007 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Jeon, S.; Kim, T.-M.; Jung, C.K. Immune Gene Signature Delineates a Subclass of Papillary Thyroid Cancer with Unfavorable Clinical Outcomes. Cancers 2018, 10, 494. https://doi.org/10.3390/cancers10120494
Kim K, Jeon S, Kim T-M, Jung CK. Immune Gene Signature Delineates a Subclass of Papillary Thyroid Cancer with Unfavorable Clinical Outcomes. Cancers. 2018; 10(12):494. https://doi.org/10.3390/cancers10120494
Chicago/Turabian StyleKim, Kyuryung, Sora Jeon, Tae-Min Kim, and Chan Kwon Jung. 2018. "Immune Gene Signature Delineates a Subclass of Papillary Thyroid Cancer with Unfavorable Clinical Outcomes" Cancers 10, no. 12: 494. https://doi.org/10.3390/cancers10120494
APA StyleKim, K., Jeon, S., Kim, T.-M., & Jung, C. K. (2018). Immune Gene Signature Delineates a Subclass of Papillary Thyroid Cancer with Unfavorable Clinical Outcomes. Cancers, 10(12), 494. https://doi.org/10.3390/cancers10120494







