Epidemiology of Moderate Alcohol Consumption and Breast Cancer: Association or Causation?
Abstract
:1. Introduction
1.1. Molecular Characteristics of Breast Cancer Subtypes
1.2. Risk Factors for Breast Cancer
1.2.1. Genetic Factors
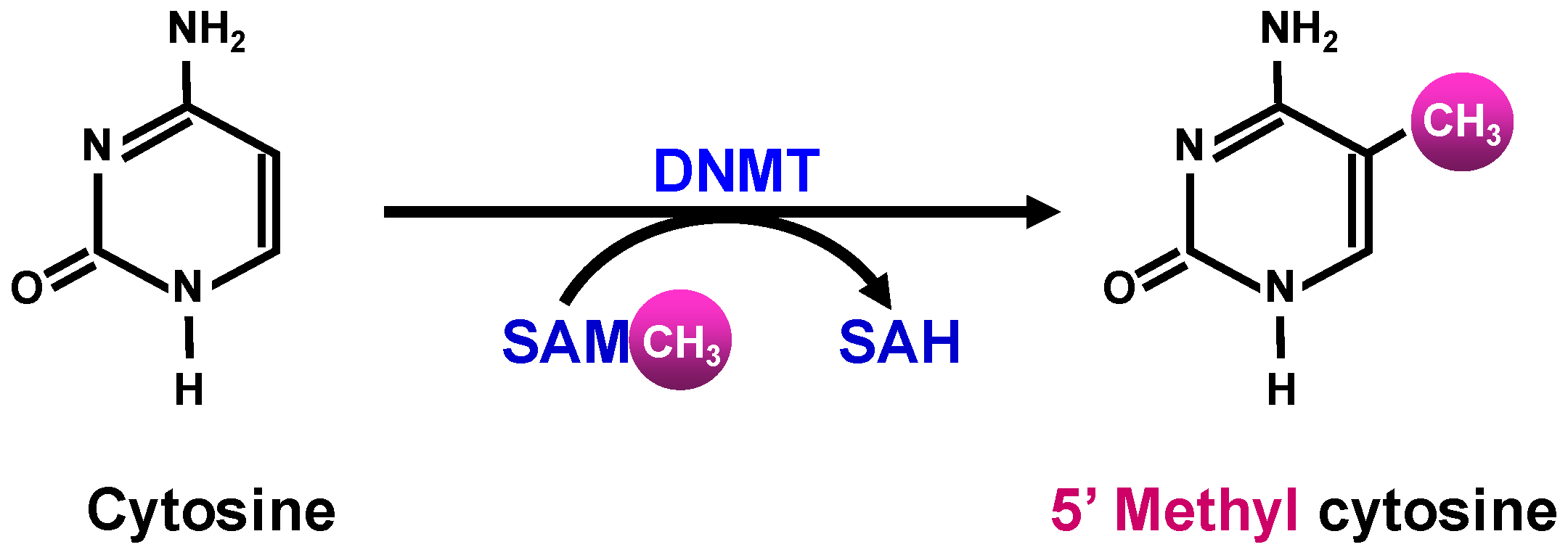
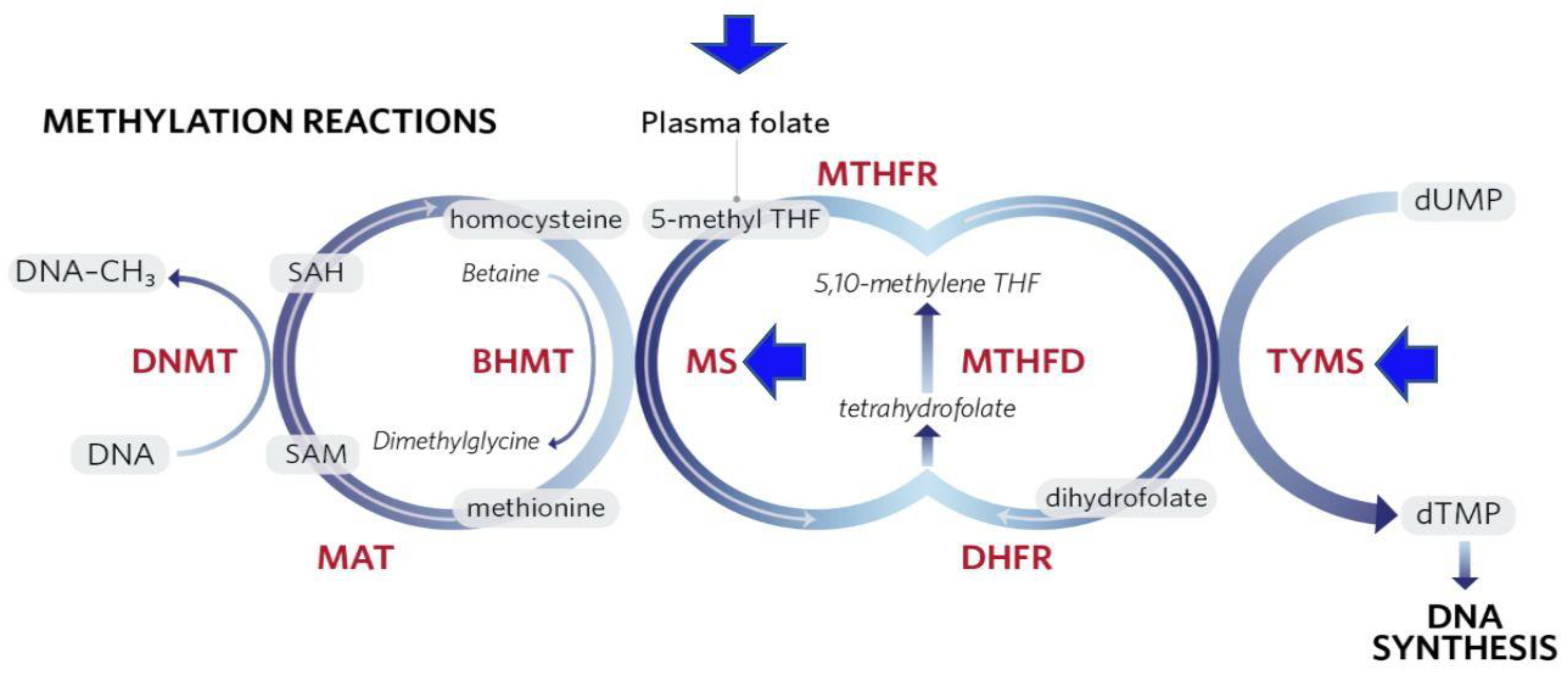
1.2.2. Epigenetic Modifications
2. Alcohol Consumption and Breast Cancer: Epidemiological Studies
3. Alcohol Consumption and Breast Cancer: Meta-Analyses
4. Association or Causation
4.1. Strengthening Causal Inference by Genetic Epidemiology through Mendelian Randomization
4.2. Marginal Structural Modeling and Agent-Based Modeling
4.3. Molecular Pathological Epidemiology
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Cancer Stat Facts: Female Breast Cancer. Available online: https://seer.cancer.gov/statfacts/html/breast.html (accessed on 22 January 2018).
- Benson, J.R.; Jatol, I. The global breast cancer burden. Future Oncol. 2012, 8, 697–702. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. World Health Organization Classification of Tumors of the Breast, 4th ed.; IARC Press: Lyon, France, 2012. [Google Scholar]
- Vinay, K.; Abul, K.A.; Jon, C.A.; Nelson, F. Robbins and Cotran Pathologic Basis of Disease, 8th ed.; Elsevier: Lyon, France, 2010. [Google Scholar]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Suprvised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Turkoz, F.P.; Solak, M.; Petekkaya, I.; Keskin, O.; Kertmen, N.; Sarici, F.; Arik, Z.; Babacan, T.; Ozisik, Y.; Altundag, K.; et al. Association between common risk factors and molecular subtypes in breast cancer patients. Breast 2013, 22, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millikan, R.C.; Newman, B.; Tse, C.K.; Moorman, P.G.; Conway, K.; Smith, L.V.; Labbok, M.H.; Geradts, J.; Bensen, J.T.; Jackson, S.; et al. Epidemiology of basal-like breast cancer. Breast Cancer Res. Treat. 2008, 109, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Kwan, M.L.; Kushi, L.H.; Weltzien, E.; Maring, B.; Kutner, S.E.; Fulton, R.S.; Lee, M.M.; Ambrosone, C.B.; Caan, B.J.; et al. Epidemiology of breast cancer subtypes in two prospective cohort studies of breast cancer survivors. Breast Cancer Res. 2009, 11, R31. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K. Breast cancer: Origins and evolution. J. Clin. Invest. 2007, 117, 3155–3163. [Google Scholar] [CrossRef] [PubMed]
- Zakhari, S.; Hoek, J. Alcohol and Breast Cancer: Reconciling Epidemiological and Molecular Data. Adv. Exp. Med. Biol. 2015, 815, 7–39. [Google Scholar] [PubMed]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, B.B.E.; Steindorf, K.; Hein, R.; Flesch-Janys, D.; Chang-Claude, J. Population attributable risk of invasive postmenopausal breast cancer and breast cancer subtypes for modifiable and non-modifiable risk factors. Cancer Epidemiol. 2011, 35, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Collaborative Group on Hormonal Factors in Breast Cancer. Familial breast cancer: Collaborative analysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet 2001, 358, 1389–1399. [Google Scholar] [CrossRef]
- Apostolou, P.; Fostira, F. Hereditary Breast Cancer: The Era of New Susceptibility Genes. Biomed. Res. Int. 2013, 2013, 747318. [Google Scholar] [CrossRef] [PubMed]
- Skol, A.D.; Sasaki, M.M.; Onel, K. The genetics of breast cancer risk in the post-genome era: Thoughts on study design to move past BRCA and towards clinical relevance. Breast Cancer Res. 2016, 18, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wooster, R.; Bignell, G.; Lancaster, J.; Swift, S.; Seal, S.; Mangion, J.; Collins, N.; Gregory, S.; Gumbs, C.; Micklem, G.; et al. Identification of the breast cancer susceptibility gene BRCA2. Nature 1995, 378, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Lasa, A.; Cajal, T.R.; Llort, G.; Suela, J.; Cigudosa, J.C.; Cornet, M.; Alonso, C.; Barnadas, A.; Baiget, M. Copy number variations are not modifiers of phenotypic expression in a pair of identical twins carrying a BRCA1 mutation. Breast Cancer Res. Treat. 2010, 123, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Fostira, F.; Tsitlaidou, M.; Papadimitriou, C.; Pertesi, M.; Timotheadou, E.; Stavropoulou, A.V.; Glentis, S.; Bournakis, E.; Bobos, M.; Pectasides, D.; et al. Prevalence of BRCA1 mutations among 403 women with triple-negative breast cancer: Implications for genetic screening selection criteria: A Hellenic Cooperative Oncology Group Study. Breast Cancer Res. Treat. 2012, 134, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Hirshfield, K.M.; Ganesan, S. Triple-negative breast cancer: Molecular subtypes and targeted therapy. Curr. Opin. Obstet. Gynecol. 2014, 26, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Shuen, A.W.; Foulkers, W.D. Inherited mutations in breast cancer genes—Risk and response. J. Mammary Gland. Biol. Neoplasia 2011, 16, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Dennis, J.; Ghadirian, P.; Little, J.; Lubinski, J.; Gronwald, J.; Kim-Sing, C.; Foulkes, W.; Moller, P.; Lynch, H.T.; Neuhausen, S.L.; et al. Alcohol consumption and the risk of breast cancer among BRCA1 and BRCA2 mutation carriers. Breast 2010, 19, 479–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGuire, V.; John, E.M.; Felberg, A.; Haile, R.W.; Boyd, N.F.; Thomas, D.C.; Jenkins, M.A.; Milne, R.L.; Daly, M.B.; Ward, J.; et al. No increased risk of breast cancer associated with alcohol consumption among carriers of BRCA1 and BRCA2 mutations ages <50 years. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1565–1567. [Google Scholar] [CrossRef] [PubMed]
- Shiovitz, S.; Korde, L.A. Genetics of breast cancer: A topic in evolution. Ann. Oncol. 2015, 26, 1291–1299. [Google Scholar] [CrossRef] [PubMed]
- Erkko, H.; Xia, B.; Nikkilä, J.; Schleutker, J.; Syrjäkoski, K.; Mannermaa, A.; Kallioniemi, A.; Pylkäs, K.; Karppinen, S.M.; Rapakko, K.; et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature 2007, 446, 316–319. [Google Scholar] [CrossRef] [PubMed]
- The CHEK2 breast cancer case-control consortium. CHECK2*1100delC and susceptibility to breast cancer: A collaborative analysis involving 10,860 breast cancer cases and 9065 controls from ten studies. Am. J. Hum. Genet. 2004, 74, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Renwick, A.; Thompson, D.; Seal, S.; Kelly, P.; Chagtai, T.; Ahmed, M.; North, B.; Jayatilake, H.; Barfoot, R.; Spanova, K.; et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat. Genet. 2006, 38, 837–875. [Google Scholar] [CrossRef] [PubMed]
- Seal, S.; Thompson, D.; Renwick, A.; Elliott, A.; Kelly, P.; Barfoot, R.; Chagtai, T.; Jayatilake, H.; Ahmed, M.; Spanova, K.; et al. Truncating mutations in the Fanconi anemia gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat. Genet. 2006, 38, 1239–1241. [Google Scholar] [CrossRef] [PubMed]
- Afghahi, A.; Telli, M.L.; Kurian, A.W. Genetics of triple-negative breast cancer: Implications for patient care. Curr. Probl. Cancer 2016, 40, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Easton, D.F.; Pooley, K.A.; Dunning, A.M.; Pharoah, P.D.; Thompson, D.; Ballinger, D.G.; Struewing, J.P.; Morrison, J.; Field, H.; Luben, R.; et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007, 447, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Pharoah, P.D.; Antoniou, A.; Bobrow, M.; Zimmern, R.L.; Easton, D.F.; Ponder, B.A. Polygenic susceptibility to breast cancer and implications for prevention. Nat. Genet. 2002, 31, 33–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haiman, C.A.; Chen, G.K.; Vachon, C.M.; Canzian, F.; Dunning, A.; Millikan, R.C.; Wang, X.; Ademuyiwa, F.; Ahmed, S.; Ambrosone, C.B.; et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat. Genet. 2011, 43, 1210–1215. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Galán, J.; Torres, B.; del Moral, R.; Muñoz-Gámez, J.A.; Martín-Oliva, D.; Villalobos, M.; Núñez, M.I.; Luna, J.D.D.; Oliver, F.J.; Almodóvar, J.M.R.D. Quantitative detection of methylated ESR1 and 14-3-3-sigma gene promoters in serum as candidate biomarkers for diagnosis of breast cancer and evaluation of treatment efficacy. Cancer Biol. Ther. 2008, 7, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Cen, Y.L.; Qi, M.L.; Li, H.G.; Su, Y.; Chen, L.J.; Lin, Y.; Chen, W.Q.; Xie, X.M.; Tang, L.Y.; Ren, Z.F. Associations of polymorphisms in the genes of FGFR2, FGF1, and RBFOX2 with breast cancer risk by estrogen/ progesterone receptor status. Mol. Carcinog. 2013, 52, E52–E59. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.W.; Trentham-Dietz, A.; Gangnon, R.E.; Hampton, J.M.; Figueroa, J.D.; Skinner, H.G.; Engelman, C.D.; Klein, B.E.; Titus, L.J.; Egan, K.M.; et al. Reproductive windows, genetic loci and breast cancer risk. Ann. Epidemiol. 2014, 24, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Purrington, K.S.; Slager, S.; Eccles, D.; Yannoukakos, D.; Fasching, P.A.; Miron, P.; Carpenter, J.; Chang-Claude, J.; Martin, N.G.; Montgomery, G.W.; et al. Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis 2014, 35, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, K.; Hall, P.; Gonzalez-Neira, A.; Ghoussaini, M.; Dennis, J.; Milne, R.L.; Schmidt, M.K.; Chang-Claude, J.; Bojesen, S.E.; Bolla, M.K.; et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013, 45, 353–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michailidou, K.; Beesley, J.; Lindstrom, S.; Canisius, S.; Dennis, J.; Lush, M.J.; Maranian, M.J.; Bolla, M.K.; Wang, Q.; Shah, M.; et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015, 47, 373–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoeps, A.; Rudolph, A.; Seibold, P.; Dunning, A.M.; Milne, R.L.; Bojesen, S.E.; Swerdlow, A.; Andrulis, I.; Brenner, H.; Behrens, S.; et al. Identification of New Genetic Susceptibility Loci for Breast Cancer Through Consideration of Gene-Environment Interactions. Genet. Epidemiol. 2014, 38, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Ruark, E.; Snape, K.; Humburg, P.; Loveday, C.; Bajrami, I.; Brough, R.; Rodrigues, D.N.; Renwick, A.; Seal, S.; Ramsay, E.; et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature 2013, 493, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, J. Association of MHTFR Ala222Val (rs1801133) polymorphism and breast cancer susceptibility: An update meta-analysis based on 51 research studies. Diagn. Pathol. 2012, 7, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.B.; Lackey, L.; Carpenter, M.A.; Rathore, A.; Land, A.M.; Leonard, B.; Refsland, E.W.; Kotandeniya, D.; Tretyakova, N.; Nikas, J.B.; et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013, 494, 366–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacPherson, G.; Healey, C.S.; Teare, M.D.; Balasubramanian, S.P.; Reed, M.W.; Pharoah, P.D.; Ponder, B.A.; Meuth, M.; Bhattacharyya, N.P.; Cox, A. Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J. Natl. Cancer Inst. 2004, 96, 1866–1869. [Google Scholar] [CrossRef] [PubMed]
- Frank, B.; Bermejo, J.L.; Hemminki, K.; Klaes, R.; Bugert, P.; Wappenschmidt, B.; Schmutzler, R.K.; Burwinkel, B. Re: Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J. Natl. Cancer Inst. 2005, 97, 1012. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.; Dunning, A.M.; Garcia-Closas, M.; Balasubramanian, S.; Reed, M.W.; Pooley, K.A.; Scollen, S.; Baynes, C.; Ponder, B.A.; Chanock, S.; et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 2007, 39, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Mullen, A.R.; Wheaton, W.W.; Jin, E.S.; Chen, P.H.; Sullivan, L.B.; Cheng, T.; Yang, Y.; Linehan, W.M.; Chandel, N.S.; DeBerardinis, R.J. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 2012, 481, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.L.; Kornbluth, S. The tangled circuitry of metabolism and apoptosis. Mol. Cell 2013, 49, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Ayyasamy, V.; Owens, K.M.; Koul, M.S.; Vujcic, M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J. Hum. Genet. 2009, 54, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Platek, M.; Mahasneh, A.; Ambrosone, C.B.; Zhao, H. Mitochondrial copy number and risk of breast cancer: A pilot study. Mitochondrion 2010, 10, 62–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alhomidi, M.A.; Vedicherla, B.; Movva, S.; Rao, P.K.; Ahuja, Y.R.; Hasan, Q. Mitochondrial D310 instability in Asian Indian breast cancer patients. Tumour. Biol. 2013, 34, 2427–2432. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, H.; Hattori, K.; Wada, R.; Ishikawa, K.; Fukuda, S.; Takenaga, K.; Nakada, K.; Hayashi, J.I. Mitochondrial DNA mutations regulate metastasis of human breast cancer cells. PLoS ONE 2011, 6, e23401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, N.; Chandra, D. Mitochondrial DNA mutations and breast tumorigenesis. Biochem. Biophys. Acta 2013, 1836, 336–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czarnecka, A.M.; Krawczyk, T.; Plak, K.; Klemba, A.; Zdrozny, M.; Arnold, R.S.; Kofler, B.; Golik, P.; Szybinska, A.; Lubinski, J.; et al. Mitochondrial genotype and breast cancer predisposition. Oncol. Rep. 2010, 24, 1521–1534. [Google Scholar] [PubMed]
- Bai, R.K.; Leal, S.M.; Covarrubias, D.; Liu, A.; Wong, L.J.C. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007, 67, 4687–4694. [Google Scholar] [CrossRef] [PubMed]
- Anderson, W.F.; Pfeiffer, R.M.; Dores, G.M.; Sherman, M.E. Comparison of age-distribution patterns of different histopathologic types of breast cancer. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1899–1905. [Google Scholar] [CrossRef] [PubMed]
- Garaycoechea, J.I.; Crossan, G.P.; Langevin, F.; Mulderrig, L.; Louzada, S.; Yang, F.; Guilbaud, G.; Park, N.; Roerink, S.; Nik-Zainal, S.; et al. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature 2018, 171, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.K.; Mueller, S. Alcohol and cancer: An overview with special emphasis on the role of acetaldehyde and cytochrome P450 2E1. Adv. Exp. Med. Biol. 2015, 815, 59–70. [Google Scholar] [PubMed]
- Hoek, J.B.; Cahill, A.; Pastorino, J.G. Alcohol and mitochondria: A dysfunctional relationship. Gastroenterology 2002, 122, 2049–2063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Z.; Lemasters, J.J. A Unifying Hypothesis Linking Hepatic Adaptations for Ethanol Metabolism to the Proinflammatory and Profibrotic Events of Alcoholic Liver Disease. Alcohol. Clin. Exp. Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Brooks, P.J.; Enoch, M.A.; Goldman, D.; Li, T.K.; Yokoyama, A. The Alcohol Flushing Response: An Unrecognized Risk Factor for Esophageal Cancer from Alcohol Consumption. PLoS Med. 2009, 6, e1000050. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Zakhari, S. Alcohol metabolism and epigenetics changes. Alcohol Res. Curr. Rev. 2013, 35, 6–16. [Google Scholar]
- Ndlovu, M.N.; Denis, H.; Fuke, F. Exposing the DNA methylome iceberg. Trends Biochem. Sci. 2011, 36, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Lo, P.K.; Sukumar, S. Epigenomics and breast cancer. Pharmacogenomics 2008, 9, 1879–1902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klarmann, G.; Decker, A.; Farrar, W.L. Epigenetic gene silencing in the Wnt pathway in breast cancer. Epigenetics 2008, 3, 59–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowsheen, S.; Aziz, K.; Tran, P.T.; Gorgoulis, V.G.; Yang, E.S.; Georgakilas, A.G. Epigenetic inactivation of DNA repair in breast cancer. Cancer Lett. 2014, 342, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Lapidus, R.G.; Nass, S.J.; Butash, K.A.; Parl, F.F.; Weitzman, S.A.; Graff, J.G.; Herman, J.G.; Davidson, N.E. Mapping of ER gene CpG island methylation-specific polymerase chain reaction. Cancer Res. 1998, 58, 2515–2519. [Google Scholar] [PubMed]
- Ferguson, A.T.; Lapidus, R.G.; Baylin, S.B.; Davidson, N.E. Demethylation of the estrogen receptor gene in estrogen receptor-negative breast cancer cells can reactivate estrogen receptor gene expression. Cancer Res. 1995, 55, 2279–2283. [Google Scholar] [PubMed]
- Locke, W.J.; Clark, S.J. Epigenome remodelling in breast cancer: Insights from an early in vitro model of carcinogenesis. Breast Cancer Res. 2012, 14, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, M. Cancer-linked DNA hypomethylation and its relationship to hpermethylation. Curr. Top. Microbiol. Immunol. 2006, 310, 251–274. [Google Scholar] [PubMed]
- Soares, J.; Pinto, A.E.; Cunha, C.V.; André, S.; Barao, I.; Sousa, J.M.; Cravo, M. Global DNA hypomethylation in breast carcinoma: Correlation with prognostic factors and tumor progression. Cancer 1999, 85, 112–118. [Google Scholar] [CrossRef]
- Fan, M.; Yan, P.S.; Hartman-Frey, C.; Chen, L.; Paik, H.; Oyer, S.L.; Salisbury, J.D.; Cheng, A.S.; Li, L.; Abbosh, P.H.; et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogen tamoxifen and fulvestrant. Cancer Res. 2006, 66, 11954–11966. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C.; Huang, Z.Z.; Yang, H.; Mato, J.M.; Avila, M.A.; Tsukamoto, H. Changes in methionine adenosyltransferase and S-adenosylmethionine homeostasis in the alcoholic rat liver. Am. J. Physiol. Gatrointest. Liver Physiol. 2000, 279, G178–G185. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.C.; Kelsey, K.T.; Zheng, S.; Houseman, E.A.; Marsit, C.J.; Wrensch, M.R.; Wiemels, J.L.; Nelson, H.H.; Karagas, M.R.; Kushi, L.H.; et al. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010, 6, e1001043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, K.; Davidson, N.E.; Hunter, S.; Yang, X.; Payne-Wilks, K.; Roland, C.L.; Phillips, D.; Bentley, C.; Dai, M.; Williams, S.M. Methyl-group dietary intake and risk of breast cancer among African-American women: A case-control study by methylation status of the estrogen receptor alpha genes. Cancer Causes Control 2003, 14, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Tao, M.H.; Marian, C.; Shields, P.G.; Nie, J.; McCann, S.E.; Millen, A.; Ambrosone, C.; Hutson, A.; Edge, S.B.; Krishnan, S.S.; et al. Alcohol consumption in relation to aberrant DNA methylation in breast tumors. Alcohol 2011, 45, 689–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, I.; Dnistrian, A.M.; Schwartz, M.; Toniolo, P.; Koenig, K.; Shore, R.E.; Akhmedkhanov, A.; Zeleniuch-Jacquotte, A.; Riboli, E. Serum folate, homocysteine and colorectal cancer risk in women: A nested case-study. Br. J. Cancer 1999, 79, 1917–1922. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.I. Folate and DNA methylation: A mechanistic link between folate deficiency and colorectal cancer? Cancer Epidemiol. Biomark. Prev. 2004, 13, 511–519. [Google Scholar]
- Nazki, F.H.; Sameer, A.S.; Ganaie, B.A. Folate: Metabolism, genes, polymorphisms and the associated diseases. Gene 2014, 533, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Ma, E.; Iwasaki, M.; Junko, I.; Hamada, G.S.; Nishimoto, I.N.; Carvalho, S.M.T.; Motola, J.; Laginha, F.M.; Tsugane, S. Dietary intake of folate, vitamin B6, and vitamin B12, genetic polymorphism of related enzymes, and risk of breast cancer; a case-study in Brazilian women. BMC Cancer 2009, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Blount, B.C.; Mack, M.M.; Wehr, C.M.; MacGregor, J.T.; Hiatt, R.A.; Wang, G.; Wickramasinghe, S.N.; Everson, R.B.; Ames, B.N. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: Implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. USA 1997, 94, 3290–3295. [Google Scholar] [CrossRef] [Green Version]
- Platek, M.E.; Shields, P.G.; Marian, C.; McCann, S.E.; Bonner, M.R.; Nie, J.; Ambrosone, C.B.; Millen, A.E.; Ochs-Balcom, H.M.; Quick, S.K.; et al. Alcohol consumption and genetic variation in methylenetetrahydro-folate reductase and 5-methyltetrahydrofolate-homocysteine methyltransferase in relation to breast cancer risk. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2453–2459. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Cui, L.H.; Ma, A.G.; Li, N.; Piao, J.M. Lack of effects of dietary folate intake on risk of breast cancer: An updated meta-analysis of prospective studies. Asian Pac. J. Cancer Prev. 2014, 15, 2323–2328. [Google Scholar] [CrossRef] [PubMed]
- Larsson, S.C.; Giovannucci, E.; Wolk, A. Folate and risk of breast cancer: A meta-analysis. J. Natl. Cancer Inst. 2007, 99, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Wade, P.A. Transcriptional control at regulatory checkpoints by histone deacetylases: Molecular connections between cancer and chromatin. Hum. Mol. Genet. 2001, 10, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Tryndyak, V.P.; Kovalchuk, O.; Pogribny, I.P. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferases 1, Suv4-20h2 histone methyltransferases and methyl-binding proteins. Cancer Biol. Ther. 2006, 5, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.Y.; Bang, Y.J.; Robertson, K.D. Histone deacetylase inhibitors for cancer therapy. Epigenetics 2006, 1, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Vadigepalli, R.; Hoek, J.B. Introduction to the Virtual Issue Alcohol and Epigenetic Regulation: Do the Products of Alcohol Metabolism Drive Epigenetic Control of Gene Expression in Alcohol-Related Disorders? ACER 2018, 42, 845. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.S.; Culhane, A.C.; Chan, M.W.; Venkataramu, C.R.; Ehrich, M.; Nasir, A.; Rodriguez, B.A.; Liu, J.; Yan, P.S.; Quackenbush, J.; et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008, 68, 1786–1796. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Deatherage, D.E.; Rodriguez, B.A.; Liyanarachchi, S.; Weng, Y.I.; Zuo, T.; Liu, J.; Cheng, A.S.; Huang, T.H. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res. 2009, 69, 5936–5945. [Google Scholar] [CrossRef] [PubMed]
- Hervouet, E.; Cartron, P.F.; Jouvenot, M.; Delage-Mourroux, R. Epigenetic regulation of estrogen signaling in breast cancer. Epigenetics 2013, 8, 237–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, B.N.; Hata, A. MicroRNA in cancer – the involvement of aberrant microRNA biogenesis regulatory pathways. Genes Cancer 2010, 1, 1100–1114. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Jothi, R.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Zhao, K. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009, 19, 1742–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guil, S.; Esteller, M. DNA methylomes, histone codes and miRNAs: Tying it all together. Int. J. Biochem. Cell Biol. 2009, 41, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, I.; Erlich, Y.; Muthuswamy, S.K.; Sachidanandam, R.; Hannon, G.J. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007, 21, 3238–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Haneda, S.; Imakawa, K.; Sakai, S.; Nagaoka, K. A microRNA, miR-101a, controls mammary gland development by regulating cyclooxygenase-2 expression. Differentiation 2009, 77, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Greene, S.B.; Gunaratne, P.H.; Hammond, S.M.; Rosen, J.M. A putative role for microRNA-205 in progenitors of mammary epithelial cells. J. Cell Sci. 2010, 123, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Place, R.F.; Li, L.C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequence. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Simonini, P.D.S.R.; Breiling, A.; Gupta, N.; Malekpour, M.; Youns, M.; Omranipour, R.; Malekpour, F.; Volinia, S.; Croce, C.M.; Najmabadi, H.; et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010, 70, 9175–9184. [Google Scholar] [CrossRef] [PubMed]
- Parrella, P. Epigenetic signature of breast cancer: Clinical perspective. Breast Care 2010, 5, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyle, P.; Boffetta, P. Alcohol Consumption and Breast Cancer Risk. 2009. Available online: http://breast-cancer-research.com/supplements/11/S3/S3 (accessed on 5 February 2018).
- Seitz, H.K.; Pelucchi, C.; Bagnard, V.; La Vecchia, C. Epidemiology and pathophysiology of alcohol and breast cancer: Update 2012. Alcohol Alcohol. 2012, 47, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Falk, R.T.; Maas, P.; Schairer, C.; Chatterjee, N.; Mabie, J.E.; Cunningham, C.; Buys, S.S.; Isaacs, C.; Ziegler, R.G. Alcohol and Risk of Breast Cancer in Postmenopausal Women: An Analysis of Etiological Heterogeneity by Multiple Tumor Characteristics. Am. J. Epidemiol. 2014, 180, 705–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullooly, M.; Khodr, Z.G.; Dallal, C.M.; Nyante, S.J.; Sherman, M.E.; Falk, R.; Liao, L.M.; Love, J.; Brinton, L.A.; Gierach, G.L. Epidemiologic Risk Factors for In Situ and Invasive Breast Cancers Among Postmenopausal Women in the National Institutes of Health-AARP Diet and Health Study. Am. J. Epidemiol. 2017, 186, 1329–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Jung, S.; Eliassen, A.H.; Chen, W.Y.; Willett, W.C.; Cho, E. Alcohol Consumption and Breast Cancer Risk in Younger Women According to Family History of Breast Cancer and Folate Intake. Am. J. Epidemiol. 2017, 186, 524–531. [Google Scholar] [CrossRef] [PubMed]
- White, A.J.; DeRoo, L.A.; Weinberg, C.R.; Sandler, D.P. Lifetime Alcohol Intake, Binge Drinking Behaviors, and Breast Cancer Risk. Am. J. Epidemiol. 2017, 186, 541–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirko, K.A.; Chen, W.Y.; Willett, W.C.; Rosner, B.A.; Hankinson, S.E.; Beck, A.H.; Tamimi, R.M.; Eliassen, A.H. Alcohol consumption and risk of breast cancer by molecular subtype: Prospective analysis of the nurses’ health study after 26 years of follow-up. Int. J. Cancer 2016, 138, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.A.; Olshan, A.F.; Tse, C.K.; Bell, M.E.; Troester, M.A. Alcohol intake and invasive breast cancer risk by molecular subtype and race in the Carolina Breast Cancer Study. Cancer Causes Control 2016, 27, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Dartois, L.; Fagherazzi, G.; Baglietto, L.; Boutron-Ruault, M.C.; Delaloge, S.; Mesrine, S.; Clavel-Chapelon, F. Proportion of premenopausal and postmenopausal breast cancers attributable to known risk factors: Estimates from the E3N-EPIC cohort. Int. J. Cancer 2016, 138, 2415–2427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romieu, I.; Scoccianti, C.; Chajes, V.; de Batlle, J.; Biessy, C.; Dossus, L.; Baglietto, L.; Clavel-Chapelon, F.; Overvad, K.; Olsen, A.; et al. Alcohol intake and breast cancer in the European prospective investigation into cancer and nutrition. Int. J. Cancer 2015, 137, 1921–1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strumylaite, L.; Sharp, S.J.; Kregzdyte, R.; Poskiene, L.; Bogusevicius, A.; Pranys, D. The Association of Low-To-Moderate Alcohol Consumption with Breast Cancer Subtypes Defined by Hormone Receptor Status. PLoS ONE 2015, 10, e0144680. [Google Scholar] [CrossRef] [PubMed]
- Shin, A.; Sandin, S.; Lof, M.; Margolis, K.L.; Kim, K.; Couto, E.; Adami, H.O.; Weiderpass, E. Alcohol consumption, body mass index and breast cancer risk by hormone receptor status: Women’ Lifestyle and Health Study. BMC Cancer 2015, 15, 881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhang, X.; Beck, A.H.; Collins, L.C.; Chen, W.Y.; Tamimi, R.M.; Hazra, A.; Brown, M.; Rosner, B.; Hankinson, S.E. Alcohol Consumption and Risk of Breast Cancer by Tumor Receptor Expression. Horm. Cancer 2015, 6, 237–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hvidtfeldt, U.A.; Tjønneland, A.; Keiding, N.; Lange, T.; Andersen, I.; Sørensen, T.I.; Prescott, E.; Hansen, Å.M.; Grønbæk, M.; Bojesen, S.E. Risk of Breast Cancer in Relation to Combined Effects of Hormone Therapy, Body Mass Index, and Alcohol Use, by Hormone-receptor Status. Epidemiology 2015, 26, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Ito, H.; Sueta, A.; Hosono, S.; Hirose, K.; Watanabe, M.; Iwata, H.; Tajima, K.; Tanaka, H.; Matsuo, K. Alcohol and dietary folate intake and the risk of breast cancer: A case-control study in Japan. Eur. J. Cancer Prev. 2013, 4, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Rosner, B.; Hankinson, S.E.; Colditz, G.A.; Willett, W.C. Moderate alcohol consumption during adult life, drinking patterns, and breast cancer risk. JAMA 2011, 30617, 1884–1890. [Google Scholar] [CrossRef] [PubMed]
- Kawai, M.; Minami, Y.; Kakizaki, M.; Kakugawa, Y.; Nishino, Y.; Fukao, A.; Tsuji, I.; Ohuchi, N. Alcohol consumption and breast cancer risk in Japanese women: The Miyagi Cohort study. Breast Cancer Res. Treat. 2011, 1283, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Iwasaki, M.; Inoue, M.; Sasazuki, S.; Sawada, N.; Yamaji, T.; Shimazu, T.; Tsugane, S. Alcohol consumption-associated breast cancer incidence and potential effect modifiers: The Japan Public Health Center-based Prospective Study. Int. J. Cancer 2010, 1273, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Bessaoud, F.; Daures, J.P. Patterns of alcohol (especially wine) consumption and breast cancer risk; a case-control study among a population in Southern France. Ann. Epidemiol. 2008, 6, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Dumitrescu, R.G.; Shields, P.G. The etiology of alcohol-induced breast cancer. Alcohol 2005, 353, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Mahabir, S.; Pfeiffer, R.; Xu, X.; Baer, D.J.; Taylor, P.R. Effects of low-to-moderate alcohol supplementation on urinary estrogen metabolites in postmenopausal women in a controlled feeding study. Cancer Med. 2017, 6, 2419–2423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sangrajrang, S.; Sato, Y.; Sakamoto, H.; Ohnami, S.; Khuhaprema, T.; Yoshida, T. Genetic polymorphisms in folate and alcohol metabolism and breast cancer risk: A case-control study in Thai women. Breast Cancer Res. Treat. 2010, 123, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Kawase, T.; Matsuo, K.; Hiraki, A.; Suzuki, T.; Watanabe, M.; Iwata, H.; Tanaka, H.; Tajima, K. Interaction of the effects of alcohol drinking and polymorphisms in alcohol-metabolizing enzymes on the risk of female breast cancer in Japan. J. Epidemiol. 2009, 19, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Abel, J.; Neuhaus, T.; Ko, Y.; Harth, V.; Hamajima, N.; Tajima, K.; Yoo, K.Y.; Park, S.K.; Noh, D.Y.; et al. Role of alcohol and genetic polymorphisms of CYP2E1 and ALDH2 in breast cancer development. Pharmacogenetics 2003, 13, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Playdon, M.C.; Ziegler, R.G.; Sampson, J.N.; Stolzenberg-Solomon, R.; Thompson, H.J.; Irwin, M.L.; Mayne, S.T.; Hoover, R.N.; Moore, S.C. Nutritional metabolomics and breast cancer risk in a prospective study. Am. J. Clin. Nutr. 2017, 106, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, M.; Ke, Z.; Luo, J. Cellular and Molecular Mechanism Underlying Alcohol-induced Aggressiveness of Breast Cancer. Pharmacol. Res. 2017, 115, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.D.; Ebrahim, S. ‘Mendelian Randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Peto, R.; Doll, R.; Buckley, J.D.; Sporn, M.B. Can dietary beta-carotene materially reduce human cancer rates? Nature 1981, 290, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, R.G.; Colavito, E.A.; Hartge, P.; McAdams, M.J.; Schoenberg, J.B.; Mason, T.J.; Fraumeni, J.F., Jr. Importance of a-carotene, b-carotene, and other phytochemicals in the etiology of lung cancer. J. Natl. Cancer Inst. 1996, 88, 612–615. [Google Scholar] [CrossRef] [PubMed]
- Gallicchio, L.; Boyd, K.; Matanoski, G.; Tao, X.; Chen, L.; Lam, T.K.; Shiels, M.; Hammond, E.; Robinson, K.A.; Caulfield, L.E.; et al. Carotenoids and the risk of developing lung cancer: A systematic review. Am. J. Clin. Nutr. 2008, 88, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.M.; Cook, N.R.; Gaziano, J.M.; Gordon, D.; Ridker, P.M.; Manson, J.E.; Hennekens, C.H.; Buring, J.E. Vitamin E in the primary prevention of cardiovascular disease and cancer: The Women’s Health Study: A randomized controlled trial. JAMA 2005, 294, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Tatsioni, A.; Bonitsis, N.G.; Ioannidis, J.P. Persistence of contradicted claims in the literature. JAMA 2007, 298, 2517–2526. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.N.; Smith, G.D. How independent are ‘independent’ effects? Relative risk estimation when correlated exposures are measured imprecisely. J. Clin. Epidemiol. 1991, 44, 1223–1231. [Google Scholar] [CrossRef]
- Phillips, A.N.; Smith, G.D. The design of prospective epidemiological studies: More subjects or better measurements? J. Clin. Epidemiol. 1993, 46, 1203–1211. [Google Scholar] [CrossRef]
- Longnecker, M.P. Alcoholic beverage consumption in relation to risk of breast cancer: Meta-analysis and review. Cancer Causes Control 1994, 51, 73–82. [Google Scholar] [CrossRef]
- Corrao, G.; Bagnardi, V.; Zambon, A.; La Vecchia, C. A meta-analysis of alcohol consumption and the risk of 15 diseases. Prev. Med. 2004, 385, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Key, J.; Hodgson, S.; Omar, R.Z.; Jensen, T.K.; Thompson, S.G.; Boobis, A.R.; Davies, D.S.; Elliott, P. Meta-analysis of studies of alcohol and breast cancer with consideration of the methodological issues. Cancer Causes Control 2006, 6, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Orsini, N.; Mignone, L.; Saji, S.; Wolk, A. Alcohol intake and risk of breast cancer defined by estrogen and progesterone receptor status-a meta-analysis of epidemiological studies. Int. J. Cancer 2008, 1228, 1832–1841. [Google Scholar] [CrossRef] [PubMed]
- Bagnardi, V.; Rota, M.; Botteri, E.; Tramacere, I.; Islami, F.; Fedirko, V.; Scotti, L.; Jenab, M.; Turati, F.; Pasquali, E.; et al. Light alcohol drinking and cancer: A meta-analysis. Ann. Oncol. 2013, 24, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Bagnardi, V.; Rota, M.; Botteri, E.; Tramacere, I.; Islami, F.; Fedirko, V.; Scotti, L.; Jenab, M.; Turati, F.; Pasquali, E.; et al. Alcohol consumption and site-specific cancer risk: A comprehensive dose-response meta-analysis. Br. J. Cancer 2015, 112, 580–593. [Google Scholar] [CrossRef] [PubMed]
- Jayasekara, H.; MacInnis, R.J.; Room, R.; English, D.R. Long-Term Alcohol Consumption and Breast, Upper Aero-Digestive Tract and Colorectal Cancer Risk: A Systematic Review and Meta-Analysis. Alcohol Alcohol. 2016, 51, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Myung, S.K.; Lee, J.H. Light Alcohol Drinking and Risk of Cancer: A Meta-analysis of Cohort Studies. Cancer Res. Treat. 2018, 50, 474–487. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Wang, M.; Anderson, K.; Baglietto, L.; Bergkvist, L.; Bernstein, L.; van den Brandt, P.A.; Brinton, L.; Buring, J.E.; et al. Alcohol consumption and breast cancer risk by estrogen receptor status: In a pooled analysis of 20 studies. Int. J. Epidemiol. 2016, 45, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, J.; Ioannidis, P.A. Is everything we eat associated with cancer? A Systematic cookbook review. Am. J. Clin. Nutr. 2013, 97, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Parekh, S.; Hooper, L.; Loke, Y.K.; Ryder, J.; Sutton, A.J.; Hing, C.; Kwok, C.S.; Pang, C.; et al. Dissemination and publication of research findings: An updated review of related biases. Health Technol. Assess. 2010, 14, 1–193. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.; Schneider, M.; Smith, G.D. Spurious precision? Meta-analysis of observational studies. BMJ 1988, 316, 140–144. [Google Scholar] [CrossRef]
- Ioannidis, J.P.A. The challenge of reforming nutritional epidemiologic research. JAMA 2018, 320, 969–970. [Google Scholar] [CrossRef]
- Theodoratou, E.; Timofeeva, M.; Li, X.; Meng, X.; Ioannidis, J.P.A. Nature, Nurture, and Cancer Risks: Genetic and Nutritional Contributions to Cancer. Annu. Rev. Nutr. 2017, 37, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Dietary Guidelines for Americans 2015–2020. Available online: https://health.gov/dietaryguidelines/2015/resources/2015-2020_Dietary_Guidelines.pdf (accessed on 18 February 2018).
- Klatsky, A.L.; Udaltsova, N.; Li, Y.; Baer, D.; Tran, H.N.; Friedman, G.D. Moderate alcohol intake and cancer: The role of underreporting. Cancer Causes Control 2014, 25, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Schatzkin, A.; Kipnis, V. Could exposure assessment problems give us wrong answers to nutrition and cancer questions? J. Natl. Cancer Inst. 2004, 96, 1564–1565. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, D.A.; Smith, G.D.; Kundu, D.; Bruckdorfer, K.R.; Ebrahim, S. Those confounded vitamins: What can we learn from the differences between observational versus randomised trial evidence? Lancet 2004, 363, 1724–1727. [Google Scholar] [CrossRef]
- Goodman, S.N.; Samet, J.A. Causal inference in cancer epidemiology. In Cancer Epidemiology and Prevention; Thun, M.J., Linet, M.S., Cerhan, J.R., Haiman, C.A., Schottenfeld, D., Eds.; Oxford University Press: Oxford, UK, 2017; pp. 97–104. [Google Scholar]
- Hill, A.B. The Environment and Disease: Association or Causation? Proc. R. Soc. Med. 1965, 58, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Tabak, L.A. Policy: NIH plans to enhance reproducibility. Nature 2014, 505, 612–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begley, C.G.; Ioannidis, J.P. Reproducibility in science: Improving the standard for basic and preclinical research. Circ. Res. 2015, 116, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.D.; Lawlor, D.A.; Harbord, R.; Timpson, N.; Day, I.; Ebrahim, S. Clustered Environments and Randomized Genes: A Fundamental Distinction between Conventional and Genetic Epidemiology. PLoS Med. 2007, 4, e352. [Google Scholar] [CrossRef] [PubMed]
- Haycock, P.C.; Burgess, S.; Wade, K.H.; Bowden, J.; Relton, C.; Smith, G.D. Best (but oft-forgotten) practices: The design, analysis, and interpretation of Mendelian randomization studies. Am. J. Clin. Nutr. 2016, 103, 965–978. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.D.; Galea, S. Formalizing the role of agent-based modeling in causal inference and epidemiology. Am. J. Epidemiol. 2015, 181, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Vanderweele, T.J.; Vansteelandt, S.; Robins, J.M. Marginal Structural Models for Sufficient Cause Interactions. Am. J. Epidemiol. 2010, 171, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernán, M.A. Invited Commentary: Agent-Based Models for Causal Inference—Reweighting Data and Theory in Epidemiology. Am. J. Epidemiol. 2015, 181, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Nishi, A.; Milner, D.A., Jr.; Giovannucci, E.L.; Nishihara, R.; Tan, A.S.; Kawachi, I.; Ogino, S. Integration of Molecular Pathology, Epidemiology, and Social Science for Global Precision Medicine. Expert Rev. Mol. Diagn. 2016, 16, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Nishihara, R.; VanderWeele, T.J.; Wang, M.; Nishi, A.; Lochhead, P.; Qian, Z.R.; Zhang, X.; Wu, K.; Nan, H.; et al. The role of molecular pathological epidemiology in the study of neoplastic and non-neoplastic diseases in the era of precision medicine. Epidemiology 2016, 27, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Stampfer, M. Lifestyle Factors and Microsatellite Instability in Colorectal Cancer: The Evolving Field of Molecular Pathological Epidemiology. JNCI 2010, 102, 365–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogino, S.; Lochhead, P.; Chan, A.T.; Nishihara, R.; Cho, E.; Wolpin, B.M.; Meyerhardt, J.A.; Meissner, A.; Schernhammer, E.S.; Fuchs, C.S.; et al. Molecular pathological epidemiology of epigenetics: Emerging integrative science to analyze environment, host, and disease. Mod. Pathol. 2013, 26, 465–484. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Nowak, J.A.; Hamada, T.; Phipps, A.I.; Peters, U.; Milner, D.A., Jr.; Giovannucci, E.L.; Nishihara, R.; Giannakis, M.; Garrett, W.S.; et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 2018, 67, 1168–1180. [Google Scholar] [CrossRef] [PubMed]
- Babic, A.; Shah, S.M.; Song, M.; Wu, K.; Meyerhardt, J.A.; Ogino, S.; Yuan, C.; Giovannucci, E.L.; Chan, A.T.; Stampfer, M.J.; et al. Soluble tumour necrosis factor receptor type II and survival in colorectal cancer. Br. J. Cancer 2016, 114, 995–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babic, A.; Bao, Y.; Qian, Z.R.; Yuan, C.; Giovannucci, E.L.; Aschard, H.; Kraft, P.; Amundadottir, L.T.; Stolzenberg-Solomon, R.Z.; Morales-Oyarvide, V.; et al. Pancreatic cancer risk associated with prediagnostic plasma levels of leptin and leptin receptor genetic polymorphisms. Cancer Res. 2016, 76, 7160–7167. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Clish, C.B.; Wu, C.; Mayers, J.R.; Kraft, P.; Townsend, M.K.; Zhang, M.; Tworoger, S.S.; Bao, Y.; Qian, Z.R.; et al. Circulating Metabolites and Survival Among Patients with Pancreatic Cancer. JNCI J. Natl. Cancer Inst. 2016, 108, djv409. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Nishihara, R.; Wu, K.; Qian, Z.R.; Kim, S.A.; Sukawa, Y.; Mima, K.; Inamura, K.; Masuda, A.; Yang, J.; et al. Marine omega-3 Polyunsaturated Fatty Acids and Risk of Colorectal Cancer According to Microsatellite Instability. J. Natl. Cancer Inst. 2015, 107, djv007. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Chan, A.T.; Fuchs, C.S.; Giovannucci, E. Molecular pathological epidemiology of colorectal neoplasia: An emerging transdisciplinary and interdisciplinary field. Gut 2011, 60, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Begg, C.B. A Strategy for Distinguishing Optimal Cancer Sub-Types. Int. J. Cancer 2011, 129, 931–937. [Google Scholar] [CrossRef] [PubMed]

| Number | Study Design | Age (yearrs) at Baseline | Data Collection | Unit of Measurement | Outcomes | Ref |
|---|---|---|---|---|---|---|
| 2017 | ||||||
| 1 | Longitudinal cohort NIH-AARP Diet and Health Study 190,325 postmenopausal women | 55–70 | Self-report | Avg. alcohol consumption (g/day) in 12 mo. before questionnaire completion |
| [108] |
| 2 | Prospective cohort Nurses’ Health Study II 93,835 US women | 27–44 | Semi-quantitative food questionnaire | Calculated total daily alcohol consumption |
| [109] |
| 3 | Sister Study 50,884 women | 35–74 | Self-report | Lifetime alcohol intake |
| [110] |
| 2016 | ||||||
| 4 | Prospective cohort Nurse’s Health Study 105,972 women | 30–55 | Semi-quantitative food frequency questionnaire | Cumulative average alcohol intake |
| [111] |
| 5 | Case-control Carolina Breast Cancer Study; 781 Afr. Am. women; 1014 White women | 25–50 | Alcohol intake (self-report) most proximal to diagnosis | Drinks per week |
| [112] |
| 2015 | ||||||
| 6 | Cohort study French E3N-EPIC 66,481 women | 40–65 | Self-report diet-history questionnaire | Cumulative average drinks/day |
| [113] |
| 7 | Prospective EPIC Study 334,850 women | 35–70 | Dietary and lifestyle questionnaires | Average lifetime alcohol intake |
| [114] |
| 8 | Case-control 585 cases | 28–90 | Self-administered questionnaire | Total number of alcoholic drinks per week |
| [115] |
| 9 | Prospective 45,233 women | 30–49 | Self-report | Current number of drinks/week converted to g/day |
| [116] |
| 10 | Prospective NHS | 30–55 | Self-report | Cumulative average intake per day |
| [117] |
| 11 | Prospective cohorts (2) Danish | 50+ | Self-report | Avg. drinks per week |
| [118] |
| 2013 and previous | ||||||
| 12 | Case Control, 2013 Japanese cohort 1754 pre- and postmenopausal women | 20–79 | Self-reported alcohol drinking | Avg. consumption g/day |
| [119] |
| 13 | Prospective observational, 2011 Nurses’ Health Study 105,986 women | Avg. 60 | Semiquantitative food frequency questionnaire | Avg. daily consumption in g/day |
| [120] |
| 14 | Prospective control, 2011 Japanese cohort 19,227 patients | 40–64 | Food frequency questionnaire | Avg. consumption g/day |
| [121] |
| 15 | Prospective, 2010 50,757 pre- and postmenopausal Japanese women | 40–69 | Self-reported questionnaires | Average consumption g/week |
| [122] |
| 16 | Case control, 2008 437 women | 25–85 | Structured questionnaire administered by two interviewers | Average consumption g/day |
| [123] |
| Study | # of Studies Included | Definitions of Drinking (g/day) | Relative Risk | Confidence Interval (95%) | Comments | Ref. |
|---|---|---|---|---|---|---|
| A | 38 10 follow-up 28 case-control | 13 g alcohol (~1 drink)/day | 1.10 | 1.08–1.17 | “Modest size of the association and variation in results across studies leaves the causal role of alcohol in question” | [139] |
| B | 29 24 case-control 5 cohort | 25 g/d (~1.8 drinks) 50 g/d (~3.6 drinks) 100 g/d (~7.1 drinks) | 1.25 1.55 2.41 | 1.20–1.29 1.44–1.67 2.07–2.80 | All doses are higher than moderate drinking | [140] |
| C | 85 77 retrospective 8 prospective studies | Drinkers vs. non-drinkers Dose response | 1.11 | 1.06–1.17 increased risk by 12% for 10 g/day | No quantification of amount of alcohol consumed | [141] |
| D | 16 4 prospective cohort 12 case-control | Dose response for all ER+, ER−, PR+ & PR-tumors. Increased risk 10 g ethanol/day | 12% ER+ 07% ER− 11% ER+/PR+ 15% ER+/PR− ER−/PR− | 8%–15% 0%–14% 7%–14% 2%–30% No significant association |
| [142] |
| E | 110 39 cohort 71 case-control | 1.05 | 1.02–1.08 |
| [143] | |
| F | 118 43 cohort 75 case-control | Light (≤12.5 g or ~1 drink) Moderate (≤50 g or ~3.6 drinks) Heavy (>50 g or >3.6 drinks) | 1.04 1.23 1.61 | 1.01–1.07 1.19–1.28 1.33–1.94 | According to US dietary guidelines moderate drinking is no more than one drink/day | [144] |
| G | 16 13 case-control 3 cohort | Highest vs. lowest category of alcohol intake | 1.28 | 1.07–1.52 |
| [145] |
| H | 34 cohort studies | ≤0.5 drink/day ≤1.0 drink/day | 1.04 1.09 | 1.01–1.07 1.06–1.12 | A small number of cohort studies in Asian populations were included | [146] |
| I | 20 prospective cohort 1,089,273 women | ≥30 g/day 5 to <15 g/day | 1.35 ER+ 1.28 ER− BC even among women with high folate intake 1.12 ER+ 1.19 ER− | 1.23–1.48 1.10–1.49 1.07–1.18 1.08–1.31 |
| [147] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakhari, S.; Hoek, J.B. Epidemiology of Moderate Alcohol Consumption and Breast Cancer: Association or Causation? Cancers 2018, 10, 349. https://doi.org/10.3390/cancers10100349
Zakhari S, Hoek JB. Epidemiology of Moderate Alcohol Consumption and Breast Cancer: Association or Causation? Cancers. 2018; 10(10):349. https://doi.org/10.3390/cancers10100349
Chicago/Turabian StyleZakhari, Samir, and Jan B. Hoek. 2018. "Epidemiology of Moderate Alcohol Consumption and Breast Cancer: Association or Causation?" Cancers 10, no. 10: 349. https://doi.org/10.3390/cancers10100349
APA StyleZakhari, S., & Hoek, J. B. (2018). Epidemiology of Moderate Alcohol Consumption and Breast Cancer: Association or Causation? Cancers, 10(10), 349. https://doi.org/10.3390/cancers10100349




