On-Chip Isoniazid Exposure of Mycobacterium smegmatis Penicillin-Binding Protein (PBP) Mutant Using Time-Lapse Fluorescent Microscopy
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth of the Cells
2.2. Antibiotic Responses
2.3. Environmental Stresses
2.4. Minimum Inhibitory Concentration (MIC)
2.5. Transposon (Tn) Mutagenesis and Identification of the msm0031::Tn Mutant
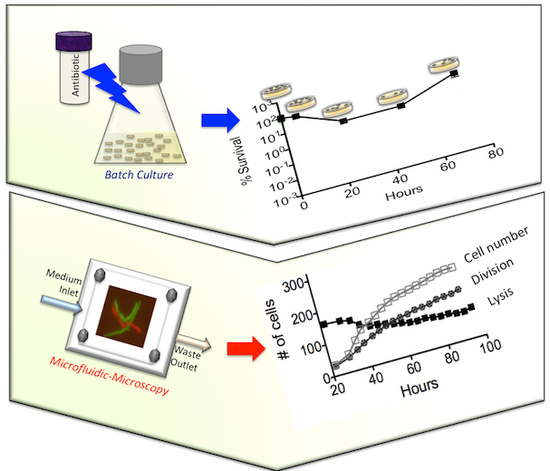
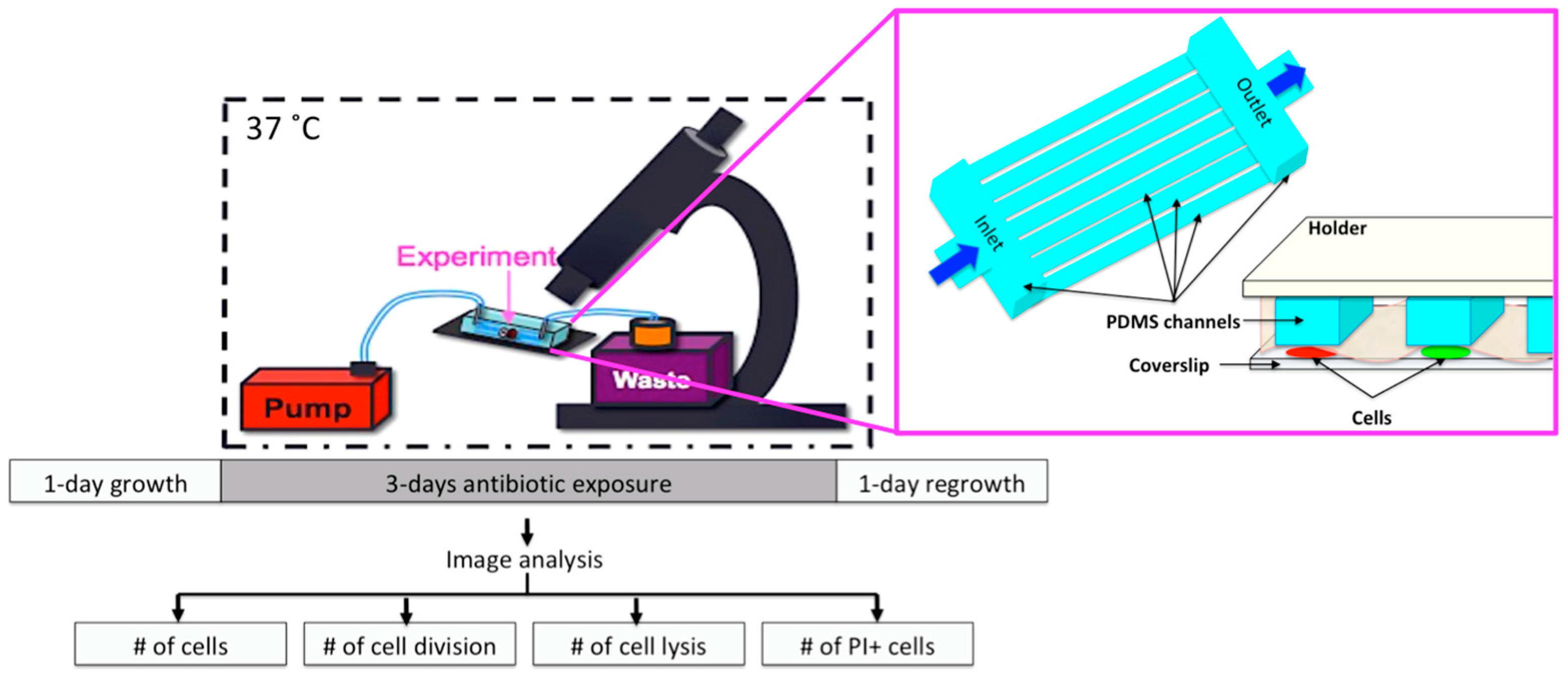
2.6. Microfluidic Chip and Live-Cell Imaging
2.7. Image Analysis
2.8. Statistical Analysis
3. Results
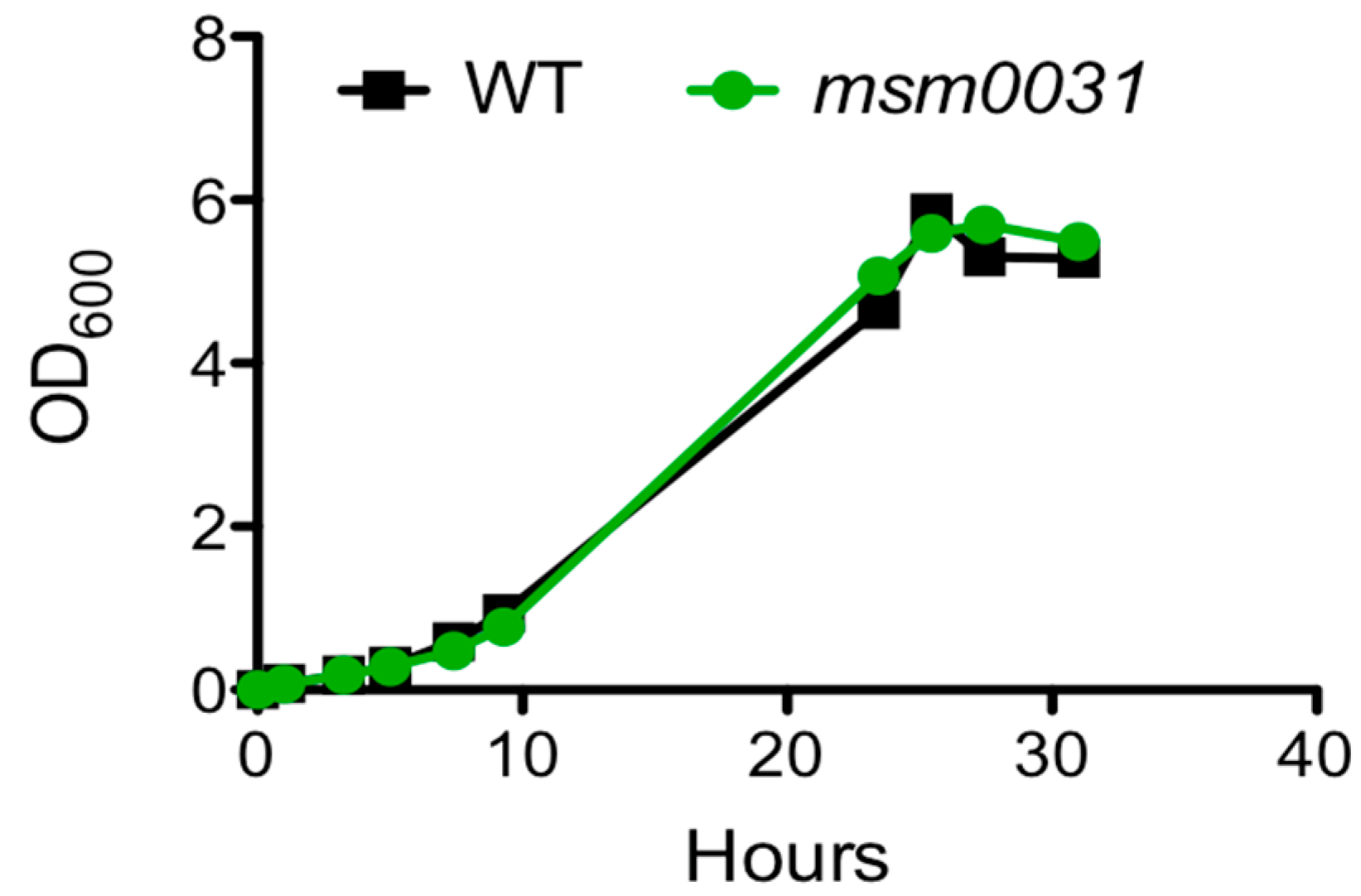
3.1. Batch-Cultue Behavior of the msm0031 Transposon Mutant and Wild-Type M. smegmatis Cells
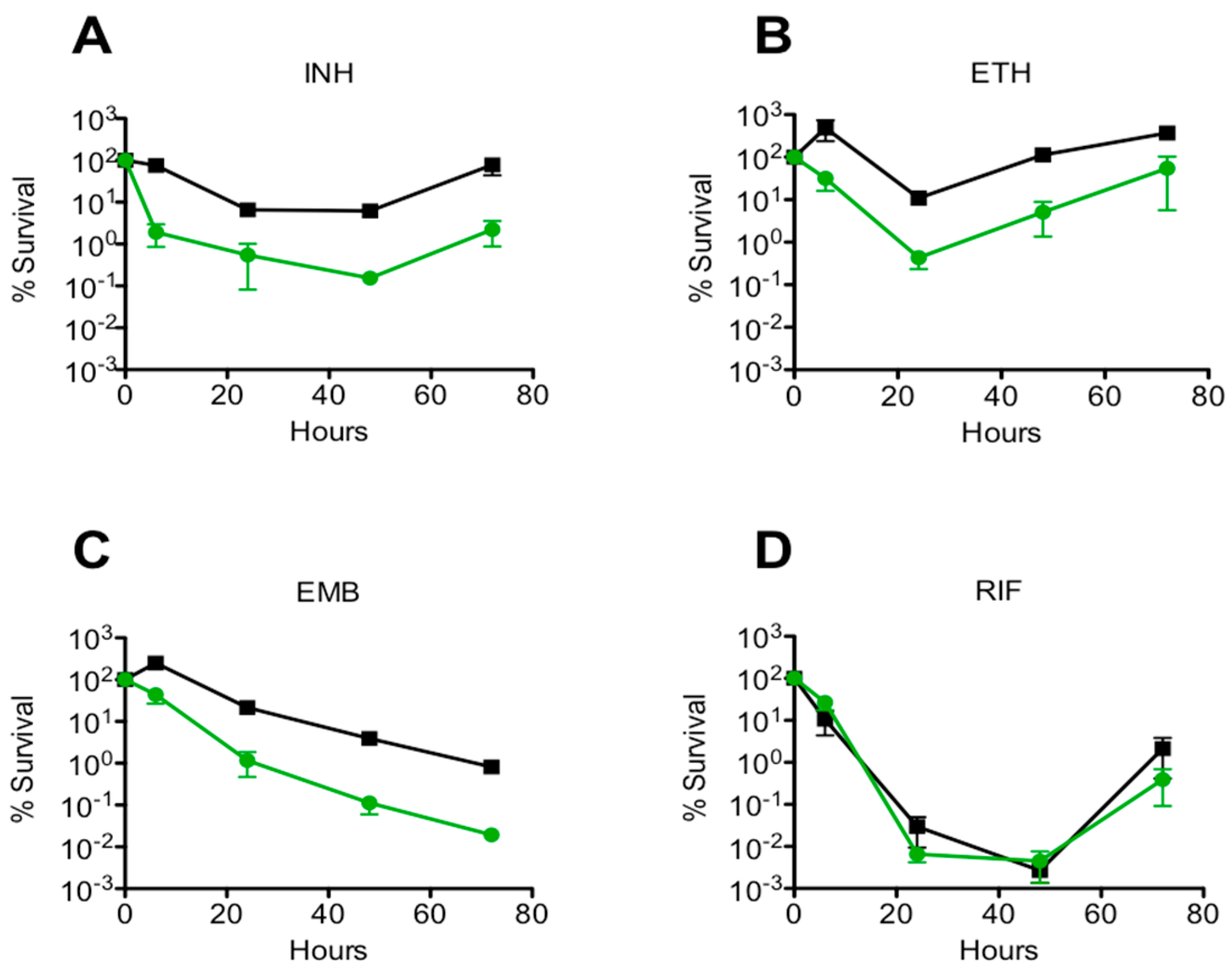
3.2. Drug Specificity
3.3. Stress Responses
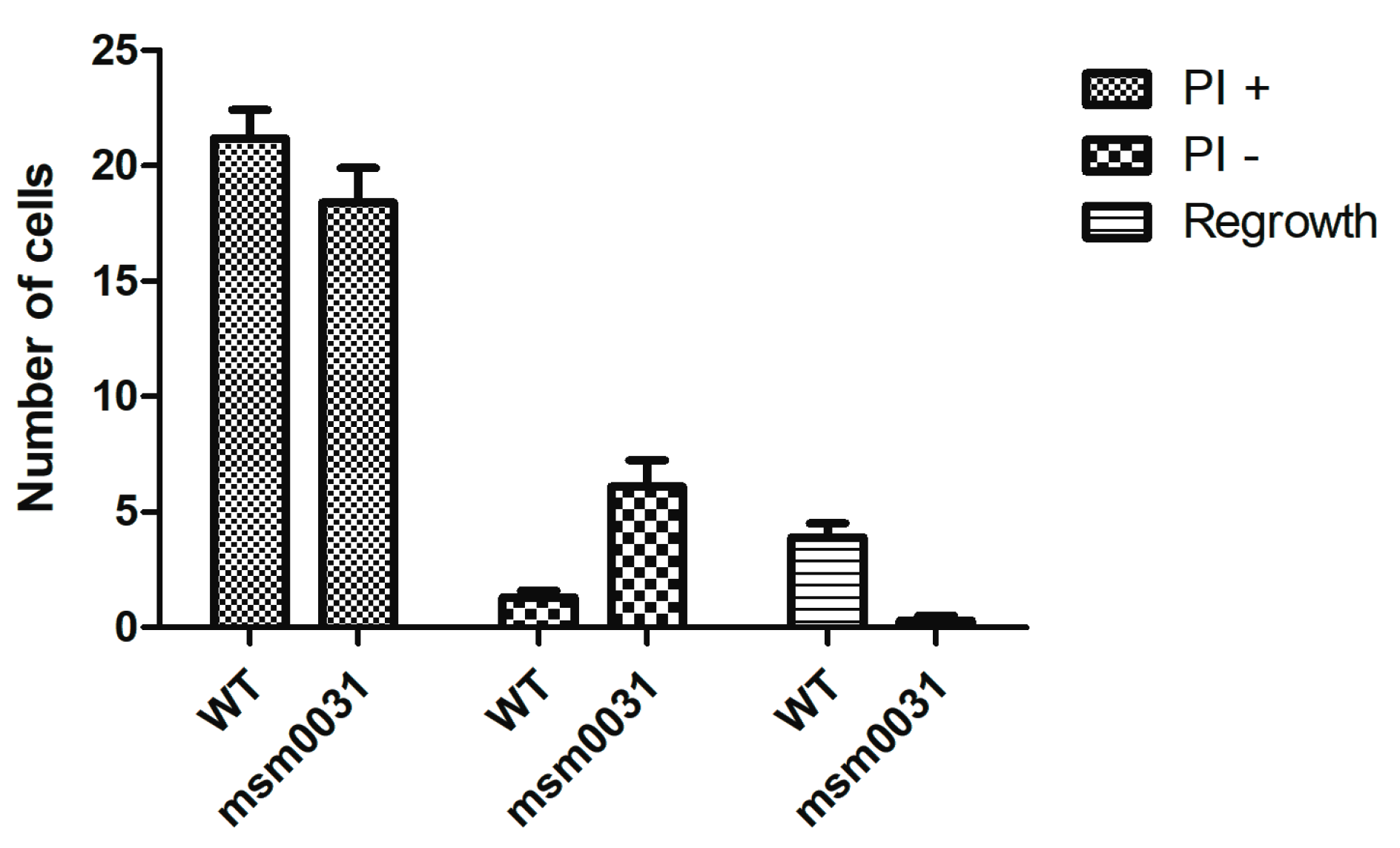
3.4. On-Chip Behavior of the msm0031 Transposon Mutant and Wild-Type M. smegmatis Cells
4. Discussion
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Schneider, E.K.; Reyes-Ortega, F.; Velkov, T.; Li, J. Antibiotic-non-antibiotic combinations for combating extremely drug-resistant Gram-negative ‘superbugs’. Essays Biochem. 2017, 61, 115–125. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO GAP AMR Newsletter; The World Health Organization (WHO): Geneve, Switzerland, 2017; Volume 27. [Google Scholar]
- Maltezu, H.C.; Theodoriduo, M.; Daikos, G.L. Antimicrobial resistance and the current refugee crisis. J. Glob. Antimicrob. Resist. 2017, 10, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Dhar, N.; McKinney, J.D. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 2007, 10, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Weibel, D.B.; DiLuzio, W.L.; Whitesides, G.M. Microfabrication meets microbiology. Nat. Rev. Microbiol. 2007, 5, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Rusconi, R.; Garren, M.; Stocker, R. Microfluidics expanding the frontiers of microbial ecology. Annu. Rev. Biophys. 2014, 43, 65–91. [Google Scholar] [CrossRef] [PubMed]
- Yawata, Y.; Nguyen, J.; Stocker, R.; Rusconi, R. Microfluidic Studies of Biofilm Formation in Dynamic Environments. J. Bacteriol. 2016, 198, 2589–2595. [Google Scholar] [CrossRef] [PubMed]
- Roggo, C.; van der Meer, J.R. Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr. Opin. Biotechnol. 2017, 45, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Brehm-Stecher, B.F.; Johnson, E.A. Single-cell microbiology: Tools, technologies, and applications. Microbiol. Mol. Biol. Rev. 2004, 268, 538–599. [Google Scholar] [CrossRef] [PubMed]
- Wakamato, Y.; Dhar, N.; Chait, R.; Schnieder, K.; Signorino-Gelo, F.; Liebler, S.; McKinney, J.D. Dynamic persistence of antibiotic-stressed mycobacteria. Science 2013, 339, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.J. Growing Unculturable Bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Hou, S.; Yongyat, C.; De Tore, S.; Ren, D. Patterned Biofilm Formation Reveals a Mechanism for Structural Heterogeneity in Bacterial Biofilms. Langmuir 2013, 29, 11145–11153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.P.; Be’er, A.; Florin, E.-L.; Swinney, H.L. Collective motion and density fluctuations in bacterial colonies. Proc. Natl. Acad. Sci. USA 2010, 107, 13626–13630. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.; García-Coca, M. Mycobacterium Biofilms. Front. Microbiol. 2017, 8, 2651. [Google Scholar] [CrossRef] [PubMed]
- Totani, T.; Nishiuchi, Y.; Tateishi, Y.; Yoshida, Y.; Kitanaka, H.; Niki, M.; Kaneko, Y.; Matsumoto, S. Effects of nutritional and ambient oxygen condition on biofilm formation in Mycobacterium avium subsp. hominissuis via altered glycolipid expression. Sci. Rep. 2017, 7, 41775. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Chauhan, A.; Rendueles, O.; Beloin, C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2013, 2, 288–356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lambert, G.; Liao, D.; Kim, H.; Robin, K.; Tung, C.-K.; Pourmand, N.; Austin, R.H. Acceleration of Emergence of Bacterial Antibiotic Resistance in Connected Microenvironments. Science 2011, 333, 1764–1767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bos, J.; Tarnopolskiy, G.; Sturm, J.G.; Kim, H.; Pourmand, N.; Austin, R.H. You cannot tell a book by looking at the cover: Cryptic complexity in bacterial evolution. Biomicrofluidics 2014, 9, 052004. [Google Scholar] [CrossRef] [PubMed]
- Lambert, G.; Vyawahare, S.; Austin, R.H. Bacteria and game theory: The rise and fall of cooperation in spatially heterogeneous environments. Interface Focus 2014, 4, 20140029. [Google Scholar] [CrossRef] [PubMed]
- Mathis, R.; Ackermann, M. Response of single bacterial cells to stress gives rise to complex history dependence at the population level. Proc. Natl. Acad. Sci. USA 2016, 113, 4224–4229. [Google Scholar] [CrossRef] [PubMed]
- Mathis, R.; Ackermann, M. Asymmetric cellular memory in bacteria exposed to antibiotics. BMC Evolut. Biol. 2017, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, F.; Littmann, S.; Lavik, G.; Escrig, S.; Meiborn, A.; Kuypers, M.M.; Ackermann, M. Phenotypic heterogeneity driven by nutrient limitation promotes growth in fluctuating environments. Nat. Microbiol. 2016, 1, 16055. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.R.; Angst, D.C.; Schiessl, K.T.; Ackermann, M. Costs of antibiotic resistance—Separating trait effects and selective effects. Evol. Appl. 2015, 8, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Benoît, Z.; Chami, M.; Houssin, C.; Dubochet, J.; Griffiths, G.; Daffé, M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 2008, 90, 5672–5680. [Google Scholar] [CrossRef]
- Hoffmann, C.; Leis, A.; Niederweis, M.; Plitzko, J.M.; Engelhardt, H. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA 2008, 105, 3963–3967. [Google Scholar] [CrossRef] [PubMed]
- Reyrat, J.M.; Kahn, D. Mycobacterium smegmatis: An absurd model for tuberculosis? Trends Microbiol. 2001, 9, 472–474. [Google Scholar] [CrossRef]
- Tyai, J.S.; Sharma, D. Mycobacterium smegmatis and tuberculosis. Tends Microbiol. 2002, 10, 68–69. [Google Scholar] [CrossRef]
- Haenni, M.; Moreillon, P. Fitness Cost and Impaired Survival in Penicillin-Resistant Streptococcus gordonii Isolates Selected in the Laboratory. Antimicrob. Agents Chemother. 2008, 52, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Elitas, M. Isoniazid Killing of Mycobacterium smegmatis NADH Pyrophosphatase Mutant at Single-Cell Level using Microfluidics and Time-Lapse Microscopy. Sci. Rep. 2017, 7, 10770. [Google Scholar] [CrossRef] [PubMed]
- Patru, M.M.; Pavelka, M.S., Jr. A Role for the class a penicillin-binding protein PonA2 in the survival of Mycobacterium smegmatis under conditions of nonreplication. J. Bacteriol. 2010, 192, 3043–3054. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Kar, D.; Murugan, R.A.; Mallick, S.; Dutta, M.; Pandey, S.D.; Chowdhury, C.; Gosh, A. A putative low-molecular mass (LMM) penicillin-binding protein (PBP) of Mycobacterium smegmatis exhibits prominent physiological characters of DD-Carboxypeptidase and beta-lactamase. Microbiology 2015, 161, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Enany, S.; Yoshida, Y.; Tateishi, Y.; Ozeki, Y.; Nishiyama, A.; Savitskaya, A.; Yamaguchi, T.; Ohara, Y.; Yamamoto, T.; Ato, M.; et al. Mycobacterial DNA-binding protein 1 is critical for long term survival of Mycobacterium smegmatis and simultaneously coordinates cellular functions. Sci. Rep. 2017, 7, 6810. [Google Scholar] [CrossRef] [PubMed]
- Kieser, K.J.; Baranowski, C.; Chao, M.C.; Long, J.E.; Sassetti, C.M.; Waldor, M.K.; Sacchettini, J.C.; Ioerger, T.R.; Rubin, E.J. Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proc. Natl. Acad. Sci. USA 2015, 112, 13087–13092. [Google Scholar] [CrossRef] [PubMed]
- Flores, A.R.; Parson, L.M.; Pavelka, M.S., Jr. Characterization of novel Mycobacterium tuberculosis and Mycobacterium smegmatis mutants hypersusceptible to beta-lactam antibiotics. J. Bacteriol. 2005, 187, 1892–1900. [Google Scholar] [CrossRef] [PubMed]
- Wendel, S.O.; Perera, A.S.; Pfromm, P.H.; Czermak, P.; Bossmann, S.H. Adaptation of Mycobacterium smegmatis to an industrial scale medium and isolation of the Mycobaterial PorinMspA. Open Microbiol. J. 2013, 7, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Belisle, J.T.; Sonnenberg, M.G. Isolation of genomic DNA from mycobacteria. Methods Mol. Biol. 1998, 101, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, S.; Humpel, A.; McLellan, A.D.; Cook, G.M. The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology 2008, 154, 2786–2795. [Google Scholar] [CrossRef] [PubMed]
- Marrakchi, H.; Lanéelle, G.; Quémard, A. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology 2000, 146, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Panchagnula, R. In vitro analysis of rifampicin and its effect on quality control tests of rifampicin containing dosage forms. Pharmazie 2004, 59, 775–781. [Google Scholar] [PubMed]
- Singh, S.; Mariappan, T.T.; Sharda, N.; Singh, B. Degradation of rifampicin, isoniazid and pyrazinamide from prepared mixtures and marketed single and combinational products under acid conditions. Pharm. Pharmacol. Commun. 2000, 6, 491–494. [Google Scholar] [CrossRef]
- Quan, S.; Venter, H.; Dabbs, E.R. Ribosylative inactivation of rifampin by Mycobacterium Smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob. Agents Chemother. 1997, 41, 2456–2460. [Google Scholar] [CrossRef] [PubMed]


| Antibiotics | FSR ± SE WT | FSR ± SE msm0031 |
|---|---|---|
| INH (50 µg/mL) | 1 ± 0.160 | 0.024 ± 0.040 *** |
| EMB (5 µg/mL) | 1 ± 0.630 | 0.028 ± 0.050 ** |
| RIF (200 µg/mL) | 1 ± 0.001 | 1.670 ± 0.003 |
| ETH (200 µg/mL) | 1 ± 3.000 | 0.040 ± 3.700 * |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elitas, M. On-Chip Isoniazid Exposure of Mycobacterium smegmatis Penicillin-Binding Protein (PBP) Mutant Using Time-Lapse Fluorescent Microscopy. Micromachines 2018, 9, 561. https://doi.org/10.3390/mi9110561
Elitas M. On-Chip Isoniazid Exposure of Mycobacterium smegmatis Penicillin-Binding Protein (PBP) Mutant Using Time-Lapse Fluorescent Microscopy. Micromachines. 2018; 9(11):561. https://doi.org/10.3390/mi9110561
Chicago/Turabian StyleElitas, Meltem. 2018. "On-Chip Isoniazid Exposure of Mycobacterium smegmatis Penicillin-Binding Protein (PBP) Mutant Using Time-Lapse Fluorescent Microscopy" Micromachines 9, no. 11: 561. https://doi.org/10.3390/mi9110561
APA StyleElitas, M. (2018). On-Chip Isoniazid Exposure of Mycobacterium smegmatis Penicillin-Binding Protein (PBP) Mutant Using Time-Lapse Fluorescent Microscopy. Micromachines, 9(11), 561. https://doi.org/10.3390/mi9110561