Electrospun Polycaprolactone (PCL) Nanofibers Induce Elongation and Alignment of Co-Cultured Primary Cortical Astrocytes and Neurons
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Characterization of Nanofibers
2.3. Nanofiber Construct Characterization
2.4. Preparation of Fiber Constructs and Glass Coverslips for Cell Culture
2.5. Primary Mouse Neuronal–Glial Co-Culture
2.6. Seeding Cells onto Nanofiber Constructs
2.7. Fixation and Immunostaining of Cultures
2.8. Fiber Diameter and Directionality Analyses (SEM and Light Microscopy)
2.9. Fluorescence Microscopy and Image Analysis, Cultures
2.10. Directionality/Alignment Analysis
2.11. Statistical Analyses
3. Results
3.1. Neural Cell Responses to Nanofibers
3.1.1. Astrocyte and Neuron Proportions Were Similar on Both Coverslips and Fibers
3.1.2. Astrocyte and Neuron Morphologies Differed on Coverslips and Fibers
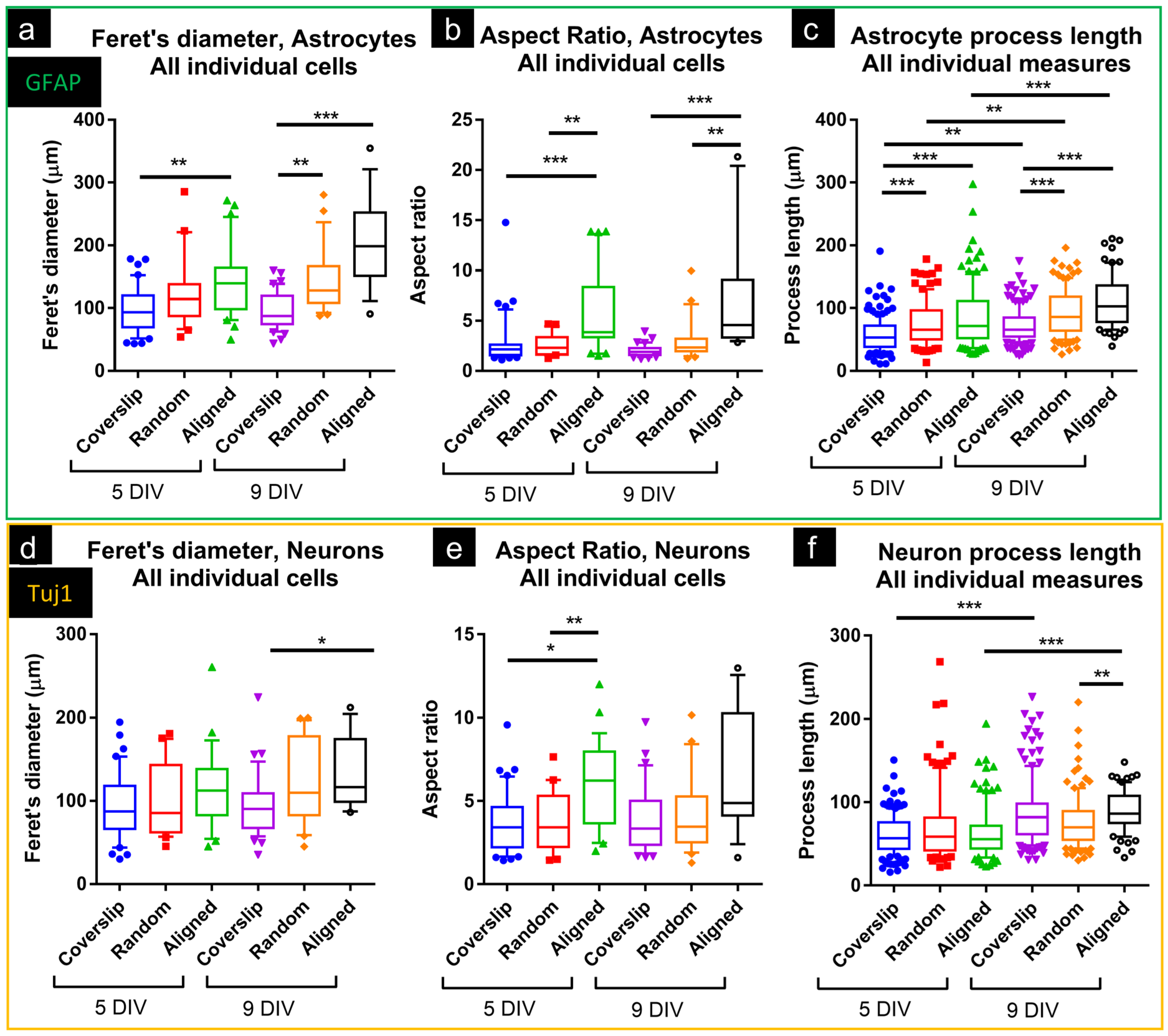
3.1.3. Astrocytes and Neurons Showed Elongated Morphologies on Nanofibers
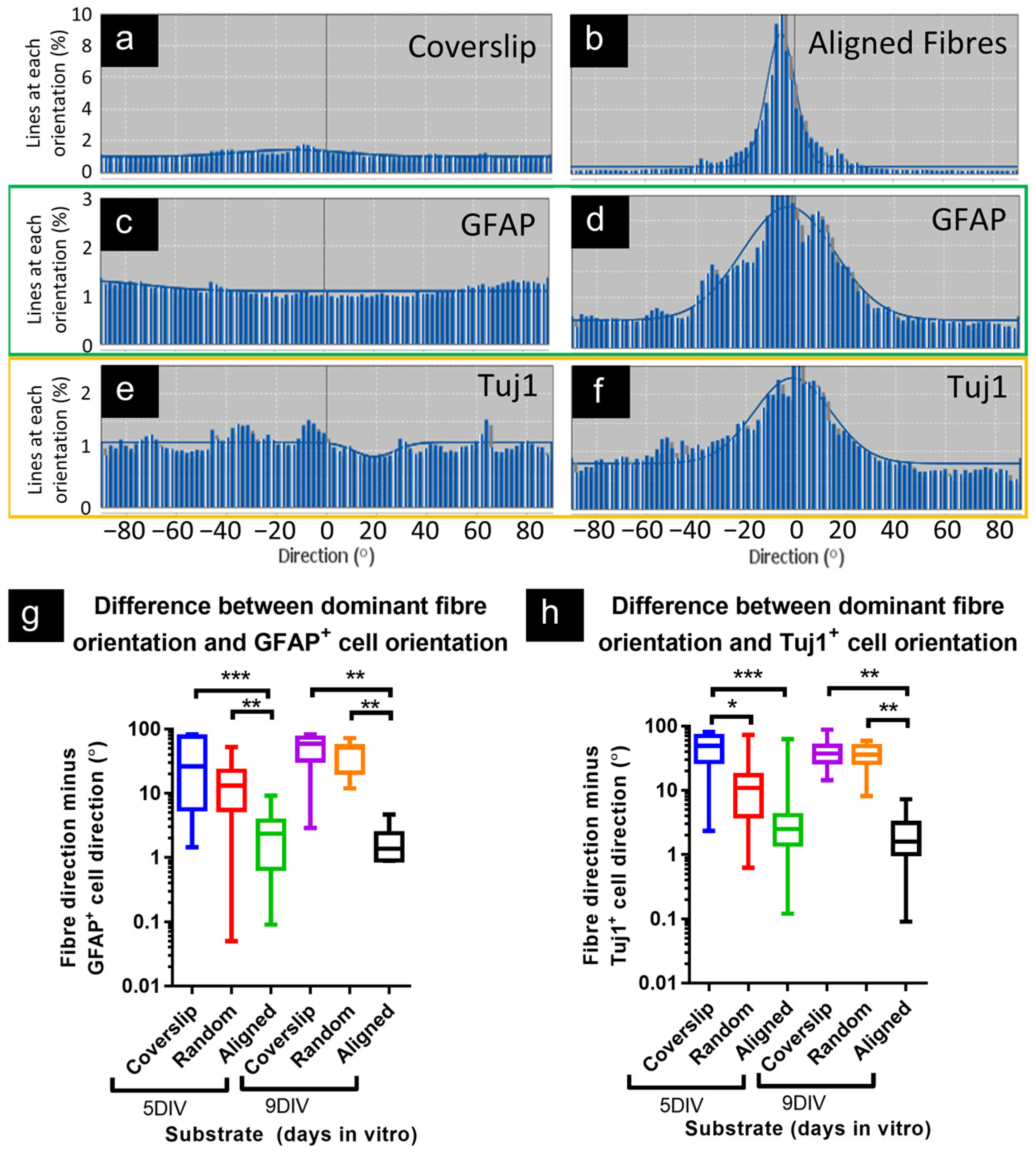
3.1.4. Astrocytes and Neurons Demonstrated Coincident Orientation with Fibers
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wiseman, J.; Basit, R.H.; Suto, A.; Middya, S.; Kabiri, B.; Evans, M.; George, V. A macro-transection model of brain trauma for neuromaterial testing with functional electrophysiological readouts. Neural Regen. Res. 2025, 20, 3539–3552. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.M.S.; Burch, R.L. The Principles of Humane Experimental Technique; Methuen & Co., Ltd.: London, UK, 1959; ISBN 0900767782. [Google Scholar]
- Balls, M. The origins and early days of the Three Rs concept. Altern. Lab. Anim. 2009, 37, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, Z.; Liu, G.; Li, X.; Yang, Z. Developmental Origins of Human Cortical Oligodendrocytes and Astrocytes. Neurosci. Bull. 2022, 38, 47–68. [Google Scholar] [CrossRef]
- Xue, J.; Wu, T.; Dai, Y.; Xia, Y. Electrospinning and electrospun nanofibers: Methods, materials, and applications. Chem. Rev. 2019, 119, 5298–5415. [Google Scholar] [CrossRef]
- Schüttler, K.F.; Bauhofer, M.W.; Ketter, V.; Giese, K.; Eschbach, D.A.; Yenigün, M.; Fuchs-Winkelmann, S.; Paletta, J.R.J. Direct incorporation of mesenchymal stem cells into a Nanofiber scaffold—In vitro and in vivo analysis. Sci. Rep. 2020, 10, 9557. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; MacEwan, M.R.; Schwartz, A.G.; Xia, Y. Electrospun nanofibers for neural tissue engineering. Nanoscale 2010, 2, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Rho, K.S.; Jeong, L.; Lee, G.; Seo, B.M.; Park, Y.J.; Hong, S.D.; Roh, S.; Cho, J.J.; Park, W.H.; Min, B.M. Electrospinning of collagen nanofibers: Effects on the behavior of normal human keratinocytes and early-stage wound healing. Biomaterials 2006, 27, 1452–1461. [Google Scholar] [CrossRef]
- Brennan, D.A.; Conte, A.A.; Kanski, G.; Turkula, S.; Hu, X.; Kleiner, M.T.; Beachley, V. Mechanical Considerations for Electrospun Nanofibers in Tendon and Ligament Repair. Adv. Healthc. Mater. 2018, 7, e1701277. [Google Scholar] [CrossRef]
- Stocco, T.D.; Silva, M.C.M.; Corat, M.A.F.; Lima, G.G.; Lobo, A.O. Towards Bioinspired Meniscus-Regenerative Scaffolds: Engineering a Novel 3D Bioprinted Patient-Specific Construct Reinforced by Biomimetically Aligned Nanofibers. Int. J. Nanomed. 2022, 17, 1111–1124. [Google Scholar] [CrossRef]
- Antonova, O.Y.; Kochetkova, O.Y.; Shlyapnikov, Y.M. ECM-Mimetic Nylon Nanofiber Scaffolds for Neurite Growth Guidance. Nanomaterials 2021, 11, 516. [Google Scholar] [CrossRef]
- Wu, J.; Xie, L.; Lin, W.Z.Y.; Chen, Q. Biomimetic nanofibrous scaffolds for neural tissue engineering and drug development. Drug Discov. Today 2017, 22, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Entekhabi, E.; Haghbin Nazarpak, M.; Moztarzadeh, F.; Sadeghi, A. Design and manufacture of neural tissue engineering scaffolds using hyaluronic acid and polycaprolactone nanofibers with controlled porosity. Mater. Sci. Eng. C 2016, 69, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.H.; Kim, J.R.; Kwon, G.B.; Namgung, U.; Song, K.S.; Lee, J.H. Effect of surface pore structure of nerve guide conduit on peripheral nerve regeneration. Tissue Eng. Part C Methods 2013, 19, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.M.; Silva, T.H.; Reis, R.L.; Mano, J.F. Nanocoatings containing sulfated polysaccharides prepared by layer-by-layer assembly as models to study cell–material interactions. J. Mater. Chem. B 2013, 1, 4406. [Google Scholar] [CrossRef] [PubMed]
- Schnell, E.; Klinkhammer, K.; Balzer, S.; Brook, G.; Klee, D.; Dalton, P.; Mey, J. Guidance of glial cell migration and axonal growth on electrospun nanofibers of poly-epsilon-caprolactone and a collagen/poly-epsilon-caprolactone blend. Biomaterials 2007, 28, 3012–3025. [Google Scholar] [CrossRef]
- Horne, M.K.; Nisbet, D.R.; Forsythe, J.S.; Parish, C.L. Three-dimensional nanofibrous scaffolds incorporating immobilized bdnf promote proliferation and differentiation of cortical neural stem cells. Stem Cells Dev. 2010, 19, 843–852. [Google Scholar] [CrossRef]
- Hamad, K.; Kaseem, M.; Yang, H.W.; Deri, F.; Ko, Y.G. Properties and medical applications of polylactic acid: A review. Express Polym. Lett. 2015, 9, 435–455. [Google Scholar] [CrossRef]
- Ramazani, S.; Karimi, M. Investigating the influence of temperature on electrospinning of polycaprolactone solutions. E-Polymers 2014, 14, 323–333. [Google Scholar] [CrossRef]
- Corey, J.M.; Gertz, C.C.; Wang, B.S.; Birrell, L.K.; Johnson, S.L.; Martin, D.C.; Feldman, E.L. The design of electrospun PLLA nanofiber scaffolds compatible with serum-free growth of primary motor and sensory neurons. Acta Biomater. 2008, 4, 863–875. [Google Scholar] [CrossRef]
- Luo, B.; Tian, L.; Chen, N.; Ramakrishna, S.; Thakor, N.; Yang, I.H. Electrospun nanofibers facilitate better alignment, differentiation, and long-term culture in an: In vitro model of the neuromuscular junction (NMJ). Biomater. Sci. 2018, 6, 3262–3272. [Google Scholar] [CrossRef]
- Xie, J.; MacEwan, M.R.; Li, X.; Sakiyama-Elbert, S.E.; Xia, Y. Neurite outgrowth on nanofiber scaffolds with different orders, structures, and surface properties. ACS Nano 2009, 3, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Mondrinos, M.J.; Chen, X.; Gandhi, M.R.; Ko, F.K.; Lelkes, P.I. Elastin Blends for Tissue Engineering Scaffolds. J. Biomed. Mater. Res. Part A 2006, 79, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Mogas Barcons, A.; Chowdhury, F.; Chari, D.M.; Adams, C. Systematic Alignment Analysis of Neural Transplant Cells in Electrospun Nanofibre Scaffolds. Materials 2022, 16, 124. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Jin, L.; Li, C.; Kuddannayai, S.; Zhang, Y. The effect of electrical stimulation on cortical cells in 3D nanofibrous scaffolds. RSC Adv. 2018, 8, 11027–11035. [Google Scholar] [CrossRef]
- Christopherson, G.T.; Song, H.; Mao, H.Q. The influence of fiber diameter of electrospun substrates on neural stem cell differentiation and proliferation. Biomaterials 2009, 30, 556–564. [Google Scholar] [CrossRef]
- Cerrone, F.; Pozner, T.; Siddiqui, A.; Ceppi, P.; Winner, B.; Rajendiran, M.; Babu, R.; Ibrahim, H.S.; Rodriguez, B.J.; Winkler, J.; et al. Polyhydroxyphenylvalerate/polycaprolactone nanofibers improve the life-span and mechanoresponse of human IPSC-derived cortical neuronal cells. Mater. Sci. Eng. C 2020, 111, 110832. [Google Scholar] [CrossRef]
- Bourke, J.L.; Coleman, H.A.; Pham, V.; Forsythe, J.S.; Parkington, H.C. Neuronal electrophysiological function and control of neurite outgrowth on electrospun polymer nanofibers are cell type dependent. Tissue Eng.—Part A 2014, 20, 1089–1095. [Google Scholar] [CrossRef]
- Weightman, A.P.; Jenkins, S.I.; Pickard, M.; Chari, D.M.; Yang, Y. Alignment of multiple glial cell populations in 3D nanofiber scaffolds: Toward the development of multicellular implantable scaffolds for repair of neural injury. Nanomed. NBM 2014, 10, 291–295. [Google Scholar] [CrossRef]
- Weightman, A.P. Enhancing the Complexity of Neural Tissue Engineering Platforms for Repair of Neurological Injury. Ph.D. Thesis, Keele University, Newcastle, UK, 2015. [Google Scholar]
- Weir, N.; Stevens, B.; Wagner, S.; Miles, A.; Ball, G.; Howard, C.; Chemmarappally, J.; McGinnity, M.; Hargreaves, A.J.; Tinsley, C. Aligned Poly-l-lactic Acid Nanofibers Induce Self-Assembly of Primary Cortical Neurons into 3D Cell Clusters. ACS Biomater. Sci. Eng. 2022, 8, 765–776. [Google Scholar] [CrossRef]
- Reyes-Ramos, A.M.; Álvarez-García, Y.R.; Solodin, N.; Almodovar, J.; Alarid, E.T.; Torres-Garcia, W.; Domenech, M. Collagen i Fibrous Substrates Modulate the Proliferation and Secretome of Estrogen Receptor-Positive Breast Tumor Cells in a Hormone-Restricted Microenvironment. ACS Biomater. Sci. Eng. 2021, 7, 2430–2443. [Google Scholar] [CrossRef]
- Schoenenberger, A.D.; Foolen, J.; Moor, P.; Silvan, U.; Snedeker, J.G. Substrate fiber alignment mediates tendon cell response to inflammatory signaling. Acta Biomater. 2018, 71, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, B.J.; Koh, S.; Cho, H.J.; Jin, X.; Kim, B.G.; Choi, J.Y. A primary culture method for the easy, efficient, and effective acquisition of oligodendrocyte lineage cells from neonatal rodent brains. Heliyon 2024, 10, e29359. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Tan, L.; Wu, P.; Yin, Y.; Liu, X.; Meng, H.; Cui, G.; Wu, N.; Lin, J.; Hu, R.; et al. Poly-L-ornithine promotes preferred differentiation of neural stem/progenitor cells via ERK signalling pathway. Sci. Rep. 2015, 5, 15535. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, S.I.; Pickard, M.R.; Khong, M.; Smith, H.L.; Mann, C.L.A.; Emes, R.D.; Chari, D.M. Identifying the cellular targets of drug action in the central nervous system following corticosteroid therapy. ACS Chem. Neurosci. 2014, 5, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Zanier, E.R.; Fumagalli, S.; Perego, C.; Pischiutta, F.; De Simoni, M.-G. Shape descriptors of the “never resting” microglia in three different acute brain injury models in mice. Intensive Care Med. Exp. 2015, 3, 7. [Google Scholar] [CrossRef]
- Brown, L.S.; King, N.E.; Courtney, J.M.; Gasperini, R.J.; Foa, L.; Howells, D.W.; Sutherland, B.A. Brain pericytes in culture display diverse morphological and functional phenotypes. Cell Biol. Toxicol. 2023, 39, 2999–3014. [Google Scholar] [CrossRef]
- Lau, C.L.; Kovacevic, M.; Tingleff, T.S.; Forsythe, J.S.; Cate, H.S.; Merlo, D.; Cederfur, C.; Maclean, F.L.; Parish, C.L.; Horne, M.K.; et al. 3D Electrospun scaffolds promote a cytotrophic phenotype of cultured primary astrocytes. J. Neurochem. 2014, 130, 215–226. [Google Scholar] [CrossRef]
- Xia, H.; Xia, Y. An in vitro study of non-aligned or aligned electrospun poly(methyl methacrylate) nanofibers as primary rat astrocytes-loading scaffold. Mater. Sci. Eng. C 2018, 91, 228–235. [Google Scholar] [CrossRef]
- Miranda-Negrón, Y.; García-Arrarás, J.E. Radial glia and radial glia-like cells: Their role in neurogenesis and regeneration. Front. Neurosci. 2022, 16, 1006037. [Google Scholar] [CrossRef]
- Yeh, C.; Wu, K.; Huang, G.; Verkhratsky, A. Radial stem astrocytes (aka neural stem cells): Identity, development, physio-pathology, and therapeutic potential. Acta Physiol. 2023, 238, e13967. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.P.; Warren, P.M.; Silver, J. New insights into glial scar formation after spinal cord injury. Cell Tissue Res. 2022, 387, 319–336. [Google Scholar] [CrossRef]
- Bradke, F. Mechanisms of Axon Growth and Regeneration: Moving between Development and Disease. J. Neurosci. 2022, 42, 8393–8405. [Google Scholar] [CrossRef] [PubMed]
- Maclean, F.L.; Horne, M.K.; Williams, R.J.; Nisbet, D.R. Review: Biomaterial systems to resolve brain inflammation after traumatic injury. APL Bioeng. 2018, 2, 021502. [Google Scholar] [CrossRef] [PubMed]
- Kwiecien, J.M. Barriers to axonal regeneration after spinal cord injury: A current perspective. Neural Regen. Res. 2022, 17, 85–86. [Google Scholar] [CrossRef]
- Xie, J.; Liu, W.; Macewan, M.R.; Bridgman, P.C.; Xia, Y. Neurite outgrowth on electrospun nanofibers with uniaxial alignment: The effects of fiber density, surface coating, and supporting substrate. ACS Nano 2014, 8, 1878–1885. [Google Scholar] [CrossRef]
- Yang, T.C.; Chuang, J.H.; Buddhakosai, W.; Wu, W.J.; Lee, C.J.; Chen, W.S.; Yang, Y.P.; Li, M.C.; Peng, C.H.; Chen, S.J. Elongation of Axon Extension for Human iPSC-Derived Retinal Ganglion Cells by a Nano-Imprinted Scaffold. Int. J. Mol. Sci. 2017, 18, 2013. [Google Scholar] [CrossRef]
- Kilinc, D.; Blasiak, A.; Lee, G.U. Microtechnologies for studying the role of mechanics in axon growth and guidance. Front. Cell. Neurosci. 2015, 9, 282. [Google Scholar] [CrossRef]
- Melrose, J.; Hayes, A.J.; Bix, G. The CNS/PNS extracellular matrix provides instructive guidance cues to neural cells and neuroregulatory proteins in neural development and repair. Int. J. Mol. Sci. 2021, 22, 5583. [Google Scholar] [CrossRef]
- Hoffman-Kim, D.; Mitchel, J.A.; Bellamkonda, R.V. Topography, Cell Response, and Nerve Regeneration. Annu. Rev. Biomed. Eng. 2010, 12, 203–231. [Google Scholar] [CrossRef]
- Hemati-Gourabi, M.; Cao, T.; Romprey, M.K.; Chen, M. Capacity of astrocytes to promote axon growth in the injured mammalian central nervous system. Front. Neurosci. 2022, 16, 955598. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nutt, K.; Dombros-Ryan, Z.; Birea, R.; Franks, E.V.; Eastham, S.; Godwin, M.; Adams, C.F.; Chari, D.M.; Jenkins, S.I. Electrospun Polycaprolactone (PCL) Nanofibers Induce Elongation and Alignment of Co-Cultured Primary Cortical Astrocytes and Neurons. Micromachines 2025, 16, 256. https://doi.org/10.3390/mi16030256
Nutt K, Dombros-Ryan Z, Birea R, Franks EV, Eastham S, Godwin M, Adams CF, Chari DM, Jenkins SI. Electrospun Polycaprolactone (PCL) Nanofibers Induce Elongation and Alignment of Co-Cultured Primary Cortical Astrocytes and Neurons. Micromachines. 2025; 16(3):256. https://doi.org/10.3390/mi16030256
Chicago/Turabian StyleNutt, Kayleigh, Zoe Dombros-Ryan, Ruxandra Birea, Emily Victoria Franks, Sarah Eastham, Morgan Godwin, Chris F. Adams, Divya Maitreyi Chari, and Stuart Iain Jenkins. 2025. "Electrospun Polycaprolactone (PCL) Nanofibers Induce Elongation and Alignment of Co-Cultured Primary Cortical Astrocytes and Neurons" Micromachines 16, no. 3: 256. https://doi.org/10.3390/mi16030256
APA StyleNutt, K., Dombros-Ryan, Z., Birea, R., Franks, E. V., Eastham, S., Godwin, M., Adams, C. F., Chari, D. M., & Jenkins, S. I. (2025). Electrospun Polycaprolactone (PCL) Nanofibers Induce Elongation and Alignment of Co-Cultured Primary Cortical Astrocytes and Neurons. Micromachines, 16(3), 256. https://doi.org/10.3390/mi16030256





