A Novel Microfluidic Platform for Circulating Tumor Cell Identification in Non-Small-Cell Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Specifications of Microfluidic Chips
2.2. Construction of Microfluidic Instruments
2.3. Experiment on Simulating Cells with Polystyrene Microspheres
2.4. Study on the Capture Efficiency of A549 Cells by Acoustic Microfluidic Technology
2.5. Labeling A549 Cells and Investigating Their Capture Utilizing an Acoustic Microfluidic Chip
2.6. Capture of CTCs from the Peripheral Blood of Patients with Lung Cancer
2.6.1. Inclusion and Exclusion Criteria for Patients with Lung Cancer
2.6.2. Collection of Peripheral Blood from Patients with Lung Cancer for CTC Capture by Acoustic Microfluidic Technique
2.6.3. Peripheral Blood Was Collected from the Same Patient, and CTCs Were Detected Using Microfluidics Combined with Immunomagnetic Bead Technology, and Comparison of the Differences Between the Two Methods
2.7. Detection of EGFR Genetic Mutations in Patients with Lung Cancer Using the ARMS-PCR Method
2.8. Data Analysis
3. Results
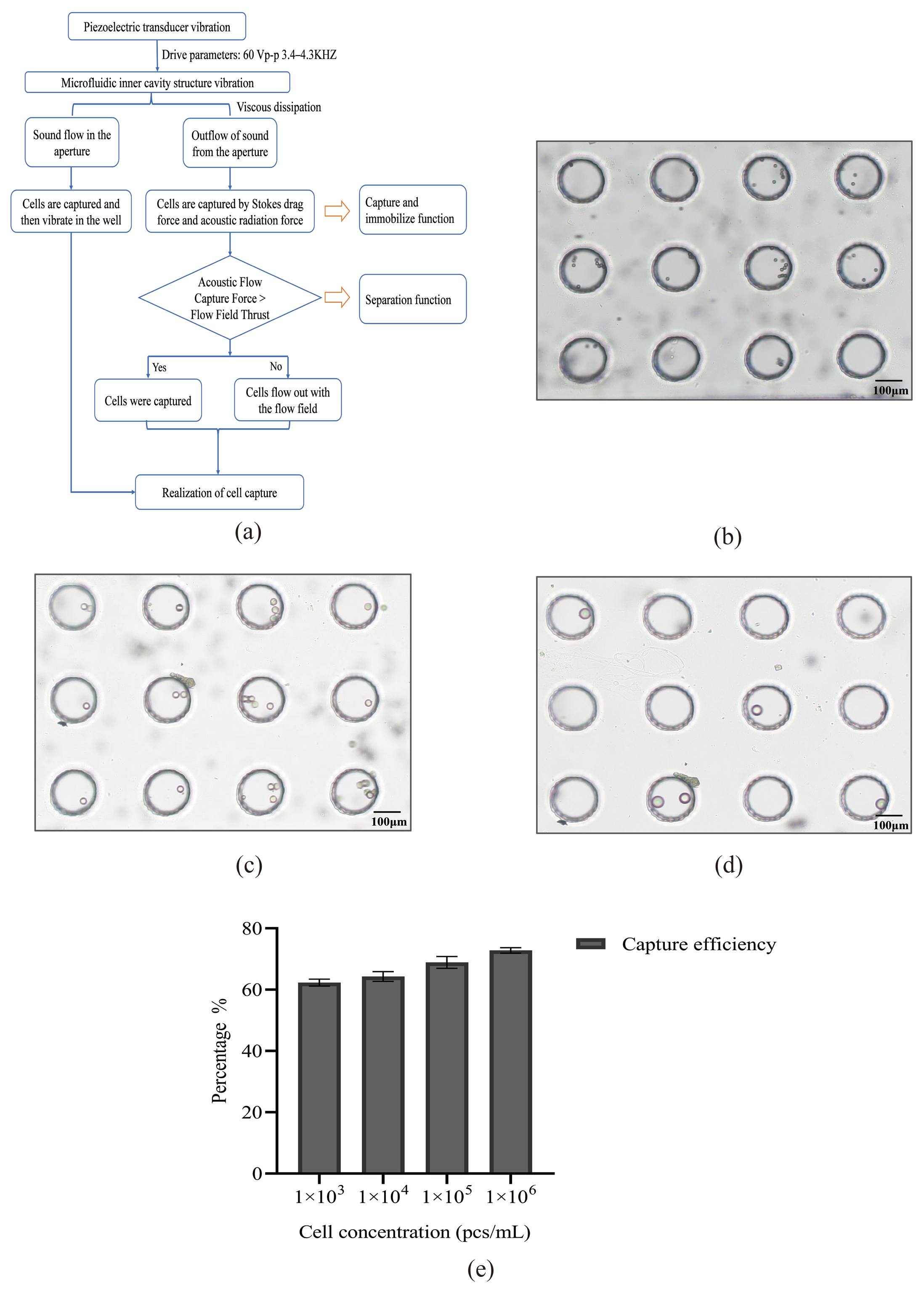
3.1. Model of Acoustic Microfluidic Chip and Mechanism of Capture
3.2. Experiment on Simulating Cells with Polystyrene Microspheres by Using Acoustic Microfluidic Technology
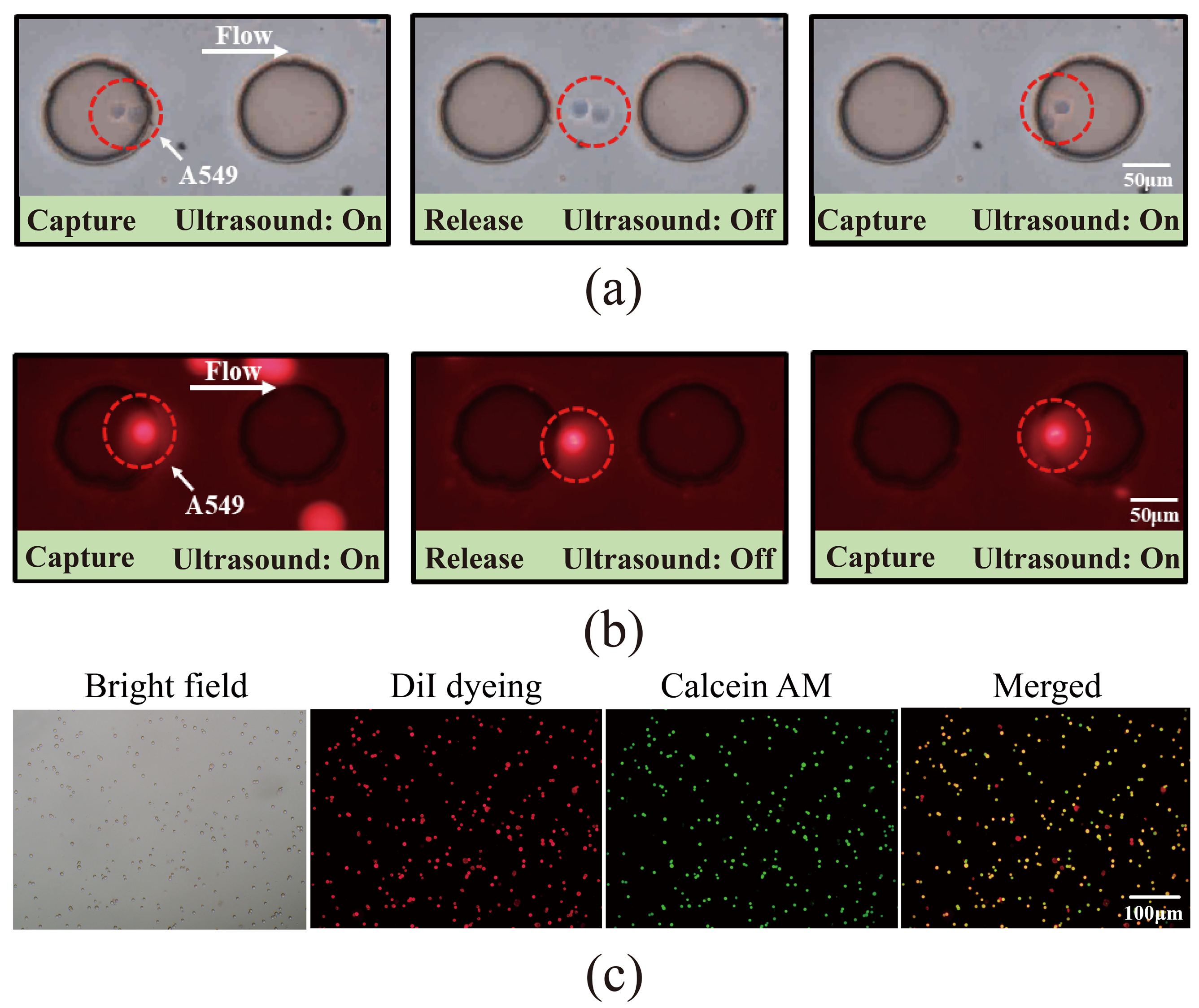
3.3. Study on the Cell Capture Efficiency of Lung Cancer A549 Cells by Acoustic Microfluidic Technology
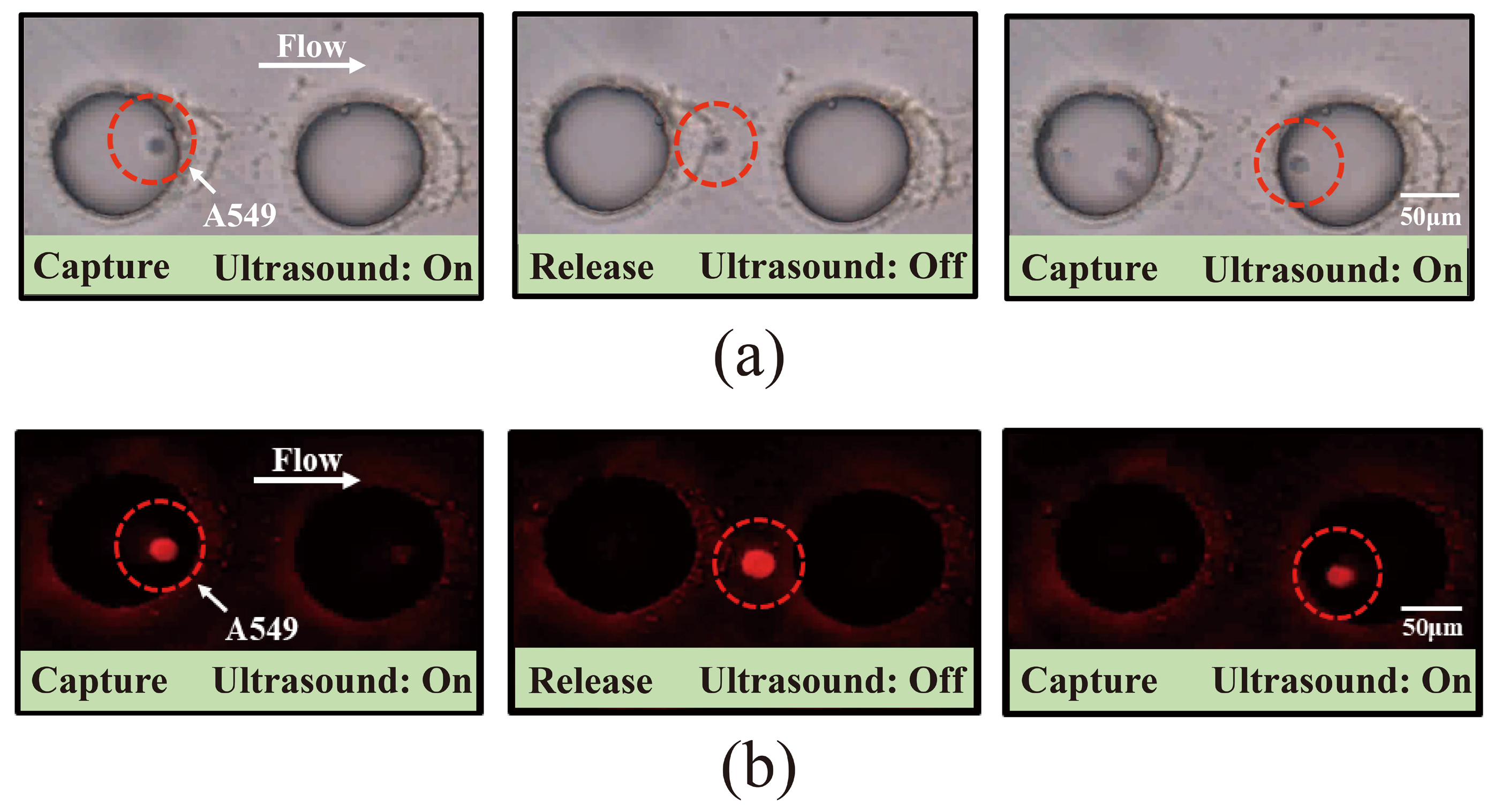
3.4. Investigation of the Capture of Pre-Labeled A549 Cells Using Acoustic Microfluidic Technology
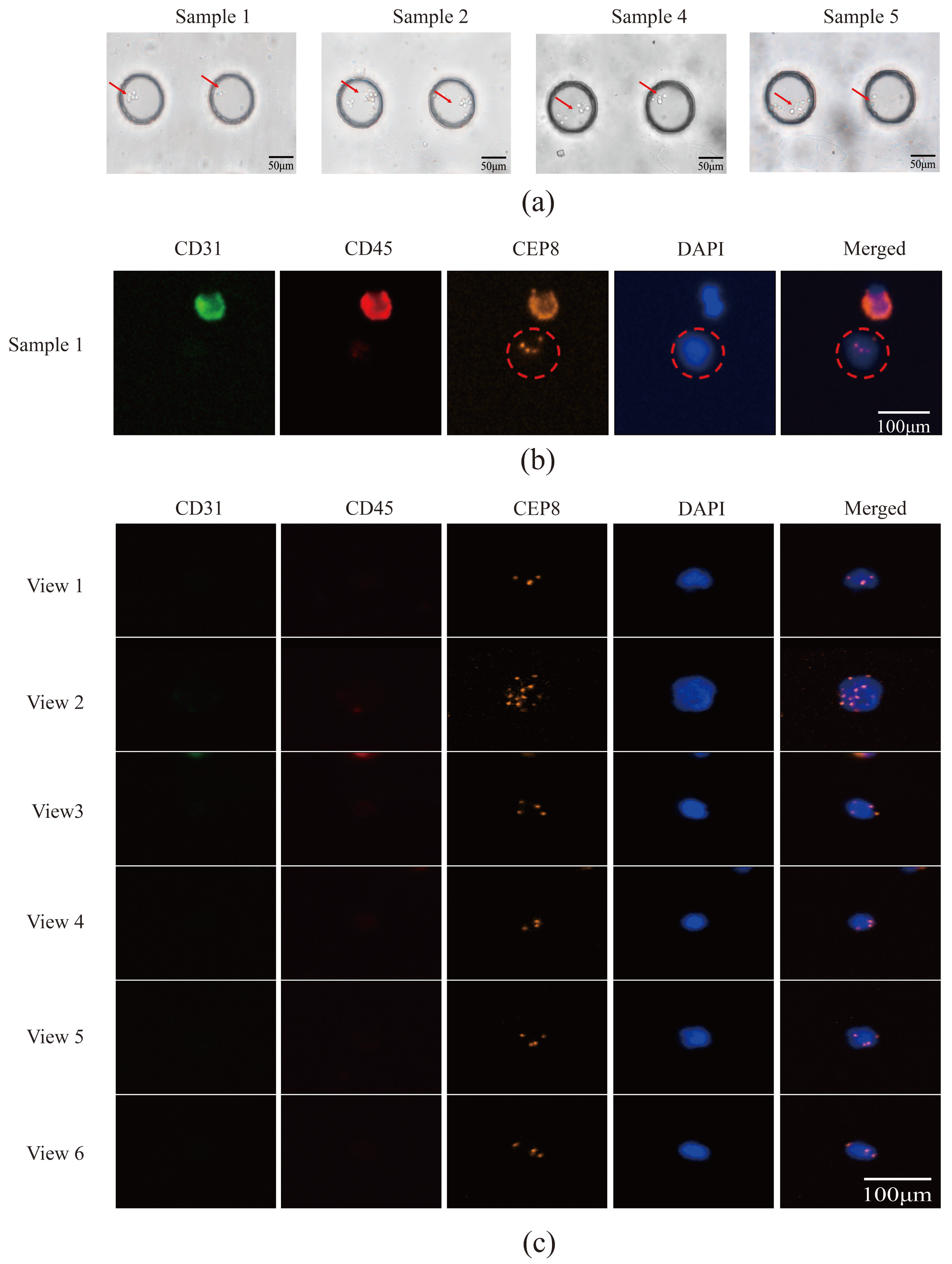
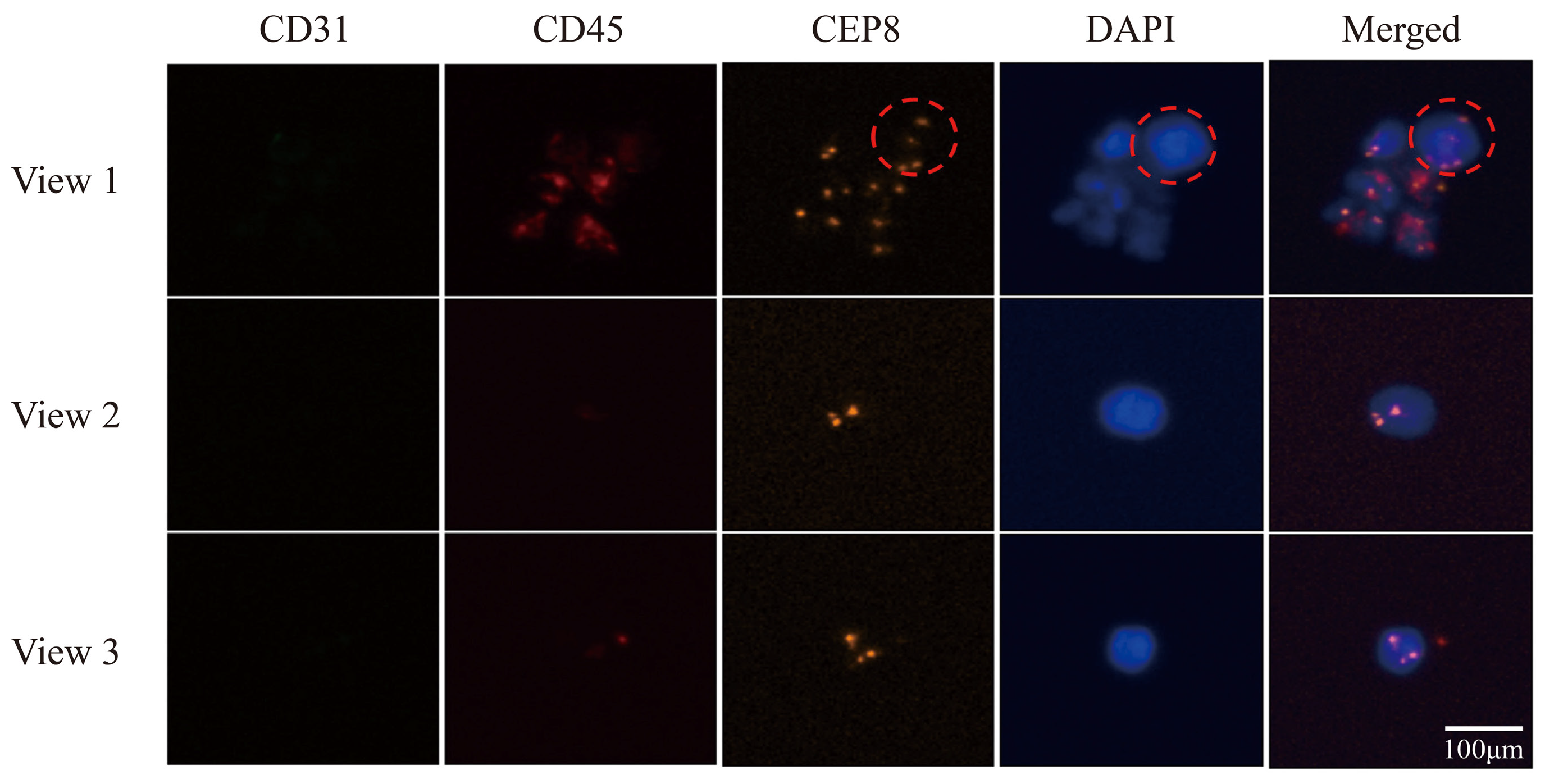
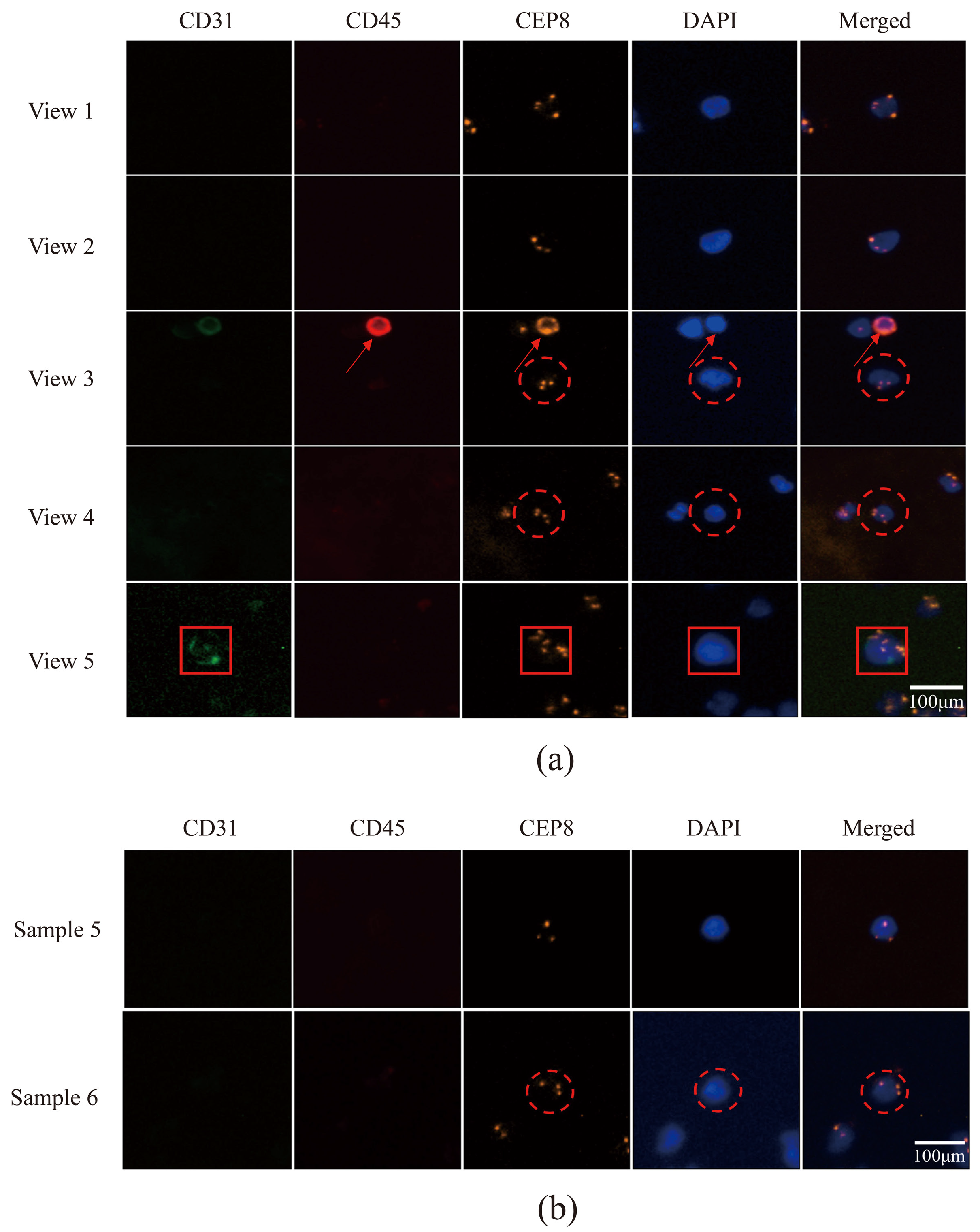
3.5. Utilizing Acoustic Microfluidics Technology to Isolate CTCs from Patients with Lung Cancer
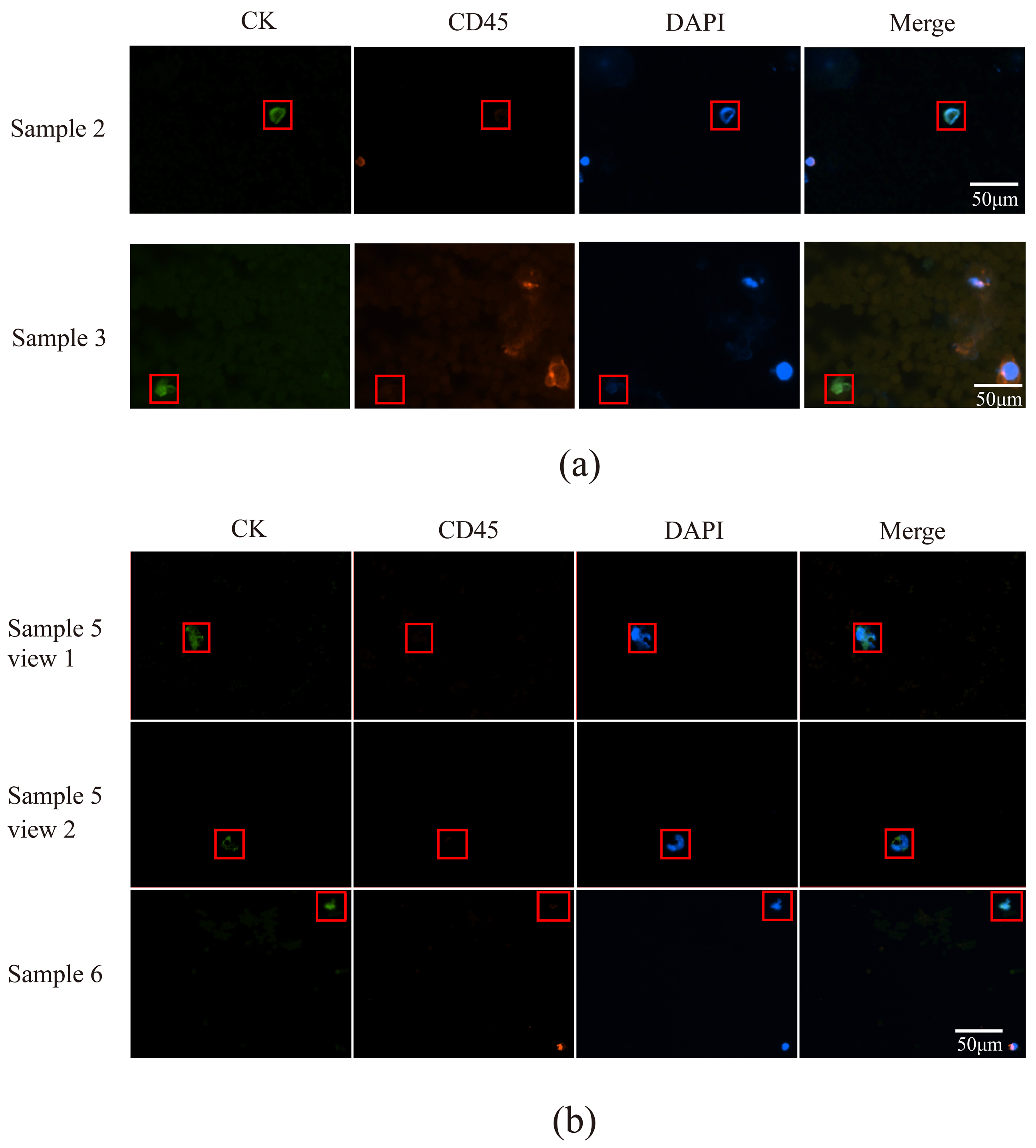
3.6. CTCs in the Peripheral Blood of Patients with Lung Cancer Were Detected Using a Microfluidic Technique Combined with Immunomagnetic Bead Separation and Compared to Acoustic Microfluidic Technology
3.7. Detection of Genetic Mutations in Patients with Lung Cancer Using the ARMS-PCR Method
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CTCs | Circulating Tumor Cells |
| EGFR | Epidermal Growth Factor Receptor |
| NSCLC | Non-Small-Cell Lung Cancer |
| ARMS-PCR | Amplification Refractory Mutation System Polymerase Chain Reaction |
| PDMS | Polydimethylsiloxane |
| PBS | Phosphate-buffered Saline |
| DiI | Cell Plasma Membrane Staining Kit with DiI (Red Fluorescence) |
| Calcein AM | Calcein Acetoxymethyl Ester |
| iFISH | Immunostaining and Fluorescence in Situ Hybridization |
| CD31 | Cluster of Differentiation 31 |
| CD45 | Cluster of Differentiation 45 |
| DAPI | 4′,6-diamidino-2-phenylindole |
| CEP8 | Chromosome 8 Centromere Probe |
| CK | Cytokeratin |
| 19-del | Exon 19 Deletion Mutation |
| L858R | Leucine 858 Arginine Point Mutation |
| S768I | Serine 768 Isoleucine Mutation |
| T790M | Threonine 790 Methionine Mutation |
| cfDNA | Circulating Free DNA |
| EGFR-TKI | Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor |
References
- Mederos, N.; Friedlaender, A.; Peters, S.; Addeo, A. Gender-specific aspects of epidemiology, molecular genetics and outcome: Lung cancer. ESMO Open 2020, 5, e000796. [Google Scholar] [CrossRef]
- Ishiguro, S.; Alhakamy, N.A.; Uppalapati, D.; Delzeit, J.; Berkland, C.J.; Tamura, M. Combined Local Pulmonary and Systemic Delivery of AT2R Gene by Modified TAT Peptide Nanoparticles Attenuates Both Murine and Human Lung Carcinoma Xenografts in Mice. J. Pharm. Sci. 2017, 106, 385–394. [Google Scholar] [CrossRef]
- Cheng, M.; Shao, S.; Xu, W.; Liu, D. Novel causes and assessments of intrapulmonary metastasis. Biochem. Biophys. Rep. 2025, 42, 102004. [Google Scholar] [CrossRef]
- Zhu, M.; Zhai, Z.; Wang, Y.; Chen, F.; Liu, R.; Yang, X.; Zhao, G. Advancements in the application of artificial intelligence in the field of colorectal cancer. Front. Oncol. 2025, 15, 1499223. [Google Scholar] [CrossRef]
- Kresse, S.H.; Brandt-Winge, S.; Pharo, H.; Flatin, B.T.B.; Jeanmougin, M.; Vedeld, H.M.; Lind, G.E. Evaluation of commercial kits for isolation and bisulfite conversion of circulating cell-free tumor DNA from blood. Clin. Epigenetics 2023, 15, 151. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Sun, P.; Xu, H.; Wang, Z. Folic acid-functionalized magnetic nanoprobes via a PAMAM dendrimer/SA-biotin mediated cascade-amplifying system for the efficient enrichment of circulating tumor cells. Biomater. Sci. 2020, 8, 6395–6403. [Google Scholar] [CrossRef]
- Abouleila, Y.; Onidani, K.; Ali, A.; Shoji, H.; Kawai, T.; Lim, C.T.; Kumar, V.; Okaya, S.; Kato, K.; Hiyama, E.; et al. Live single cell mass spectrometry reveals cancer-specific metabolic profiles of circulating tumor cells. Cancer Sci. 2019, 110, 697–706. [Google Scholar] [CrossRef]
- Li, M.; Wu, S.; Zhuang, C.; Shi, C.; Gu, L.; Wang, P.; Guo, F.; Wang, Y.; Liu, Z. Metabolomic analysis of circulating tumor cells derived liver metastasis of colorectal cancer. Heliyon 2023, 9, e12515. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Ewald, A.J. A collective route to metastasis: Seeding by tumor cell clusters. Science 2016, 352, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lu, C.; Shen, M.; Sun, X.; Mo, X.; Yang, G. Fast, Reusable, Cell Uniformly Distributed Membrane Filtration Device for Separation of Circulating Tumor Cells. ACS Omega 2022, 7, 20761–20767. [Google Scholar] [CrossRef]
- Deng, Z.; Wu, S.; Wang, Y.; Shi, D. Circulating tumor cell isolation for cancer diagnosis and prognosis. EBioMedicine 2022, 83, 104237. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.B.; Hou, L.K.; Yu, F.; Zhang, J.; Wu, W.; Tang, X.M.; Sun, F.; Lu, H.M.; Deng, J.; et al. Liquid biopsy in lung cancer: Significance in diagnostics, prediction, and treatment monitoring. Mol. Cancer 2022, 21, 25. [Google Scholar] [CrossRef]
- Chen, H.; Li, Q.; Hu, Q.; Jiao, X.; Ren, W.; Wang, S.; Peng, G. Double spiral chip-embedded micro-trapezoid filters (SMT filters) for the sensitive isolation of CTCs of prostate cancer by spectral detection. Nanoscale Adv. 2022, 4, 5392–5403. [Google Scholar] [CrossRef]
- Xue, M.; Xiang, A.; Guo, Y.; Wang, L.; Wang, R.; Wang, W.; Ji, G.; Lu, Z. Dynamic Halbach array magnet integrated microfluidic system for the continuous-flow separation of rare tumor cells. RSC Adv. 2019, 9, 38496–38504. [Google Scholar] [CrossRef]
- Peng, T.; Qiang, J.; Yuan, S. Sheathless inertial particle focusing methods within microfluidic devices: A review. Front. Bioeng. Biotechnol. 2023, 11, 1331968. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Huang, P.H.; Zhang, R.; Mao, Z.; Chen, C.; Kemeny, G.; Li, P.; Lee, A.V.; Gyanchandani, R.; Armstrong, A.J.; et al. Circulating Tumor Cell Phenotyping via High-Throughput Acoustic Separation. Small 2020, 16, e2004438. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.J.; Kozminsky, M.; Nagrath, S. Emerging role of nanomaterials in circulating tumor cell isolation and analysis. ACS Nano 2014, 8, 1995–2017. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Becker, T.M.; Po, J.W.; Chua, W.; Ma, Y. Single-Circulating Tumor Cell Whole Genome Amplification to Unravel Cancer Heterogeneity and Actionable Biomarkers. Int. J. Mol. Sci. 2022, 23, 8386. [Google Scholar] [CrossRef]
- Moon, G.Y.; Dalkiran, B.; Park, H.S.; Shin, D.; Son, C.; Choi, J.H.; Bang, S.; Lee, H.; Doh, I.; Kim, D.H.; et al. Dual Biomarker Strategies for Liquid Biopsy: Integrating Circulating Tumor Cells and Circulating Tumor DNA for Enhanced Tumor Monitoring. Biosensors 2025, 15, 74. [Google Scholar] [CrossRef]
- Eliahoo, P.; Setayesh, H.; Hoffman, T.; Wu, Y.; Li, S.; Treweek, J.B. Viscoelasticity in 3D Cell Culture and Regenerative Medicine: Measurement Techniques and Biological Relevance. ACS Mater. Au 2024, 4, 354–384. [Google Scholar] [CrossRef]
- Zhao, Y.; Shi, W.; Tang, Q. An eleven-gene risk model associated with lymph node metastasis predicts overall survival in lung adenocarcinoma. Sci. Rep. 2023, 13, 6852. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Wang, C.; Wang, C.; Zhao, K.; Ma, Z.; Hu, S. Inconsistent clinical outcomes following afatinib treatment in NSCLC patients harboring uncommon epidermal growth factor receptor mutation. Front. Oncol. 2022, 12, 999606. [Google Scholar] [CrossRef]
- He, C.; Wei, C.; Wen, J.; Chen, S.; Chen, L.; Wu, Y.; Shen, Y.; Bai, H.; Zhang, Y.; Chen, X.; et al. Comprehensive analysis of NGS and ARMS-PCR for detecting EGFR mutations based on 4467 cases of NSCLC patients. J. Cancer Res. Clin. Oncol. 2022, 148, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; He, Q.; Liang, H.; Cheng, B.; Li, J.; Xiong, S.; Zhao, Y.; Guo, M.; Liu, Z.; He, J.; et al. Diagnostic Accuracy of Droplet Digital PCR and Amplification Refractory Mutation System PCR for Detecting EGFR Mutation in Cell-Free DNA of Lung Cancer: A Meta-Analysis. Front. Oncol. 2020, 10, 290. [Google Scholar] [CrossRef]
- Liang, C.; Wu, Z.; Gan, X.; Liu, Y.; You, Y.; Liu, C.; Zhou, C.; Liang, Y.; Mo, H.; Chen, A.M.; et al. Detection of Rare Mutations in EGFR-ARMS-PCR-Negative Lung Adenocarcinoma by Sanger Sequencing. Yonsei Med. J. 2018, 59, 13–19. [Google Scholar] [CrossRef]
- Bai, X.; Song, B.; Chen, Z.; Zhang, W.; Chen, D.; Dai, Y.; Liang, S.; Zhang, D.; Zhao, Z.; Feng, L. Postoperative evaluation of tumours based on label-free acoustic separation of circulating tumour cells by microstreaming. Lab Chip 2021, 21, 2721–2729. [Google Scholar] [CrossRef]
- Hashemiesfahan, M.; Christiaens, J.W.; Maisto, A.; Gelin, P.; Gardeniers, H.; De Malsche, W. Characterizing Acoustic Behavior of Silicon Microchannels Separated by a Porous Wall. Micromachines 2024, 15, 868. [Google Scholar] [CrossRef]
- Gautam, G.P.; Gurung, R.; Fencl, F.A.; Piyasena, M.E. Separation of sub-micron particles from micron particles using acoustic fluid relocation combined with acoustophoresis. Anal. Bioanal. Chem. 2018, 410, 6561–6571. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.R.; Sarowar, M.T.; Chen, X. Separation of CTCs from WBCs using DEP-assisted inertial manipulation: A numerical study. Electrophoresis 2023, 44, 1781–1794. [Google Scholar] [CrossRef]
- Sonn, C.H.; Cho, J.H.; Kim, J.W.; Kang, M.S.; Lee, J.; Kim, J. Detection of circulating tumor cells in patients with non-small cell lung cancer using a size-based platform. Oncol. Lett. 2017, 13, 2717–2722. [Google Scholar] [CrossRef]
- Cai, S.; Deng, Y.; Wang, Z.; Zhu, J.; Huang, C.; Du, L.; Wang, C.; Yu, X.; Liu, W.; Yang, C.; et al. Development and clinical validation of a microfluidic-based platform for CTC enrichment and downstream molecular analysis. Front. Oncol. 2023, 13, 1238332. [Google Scholar] [CrossRef]
- Lin, A.Y.; Wang, D.D.; Li, L.; Lin, P.P. Identification and Comprehensive Co-Detection of Necrotic and Viable Aneuploid Cancer Cells in Peripheral Blood. Cancers 2021, 13, 5108. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Yi, M.; Li, N.; Luo, S.; Wu, K. Predictive biomarkers of anti-PD-1/PD-L1 therapy in NSCLC. Exp. Hematol. Oncol. 2021, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Liu, L.; Li, P.; Xu, Y.; Hou, Y.; Peng, J.; Song, Y.; Bing, Z.; Wang, Y.; Wang, Y.; et al. Efficient isolation and quantification of circulating tumor cells in non-small cell lung cancer patients using peptide-functionalized magnetic nanoparticles. J. Thorac. Dis. 2020, 12, 4262–4273. [Google Scholar] [CrossRef]
- You, H.S.; Gao, C.X.; Wang, H.B.; Luo, S.S.; Chen, S.Y.; Dong, Y.L.; Lyu, J.; Tian, T. Concordance of Treatment Recommendations for Metastatic Non-Small-Cell Lung Cancer Between Watson for Oncology System and Medical Team. Cancer Manag. Res. 2020, 12, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Pan, S.; Liu, P.; Feng, N.; Lu, P.; Wang, Z.; Huang, C.; Wu, L.; Chen, Y. Polystyrene microsphere-mediated optical sensing strategy for ultrasensitive determination of aflatoxin M(1) in milk. Talanta 2023, 258, 124357. [Google Scholar] [CrossRef]
- Sun, W.; Jia, C.; Huang, T.; Sheng, W.; Li, G.; Zhang, H.; Jing, F.; Jin, Q.; Zhao, J.; Li, G.; et al. High-performance size-based microdevice for the detection of circulating tumor cells from peripheral blood in rectal cancer patients. PLoS ONE 2013, 8, e75865. [Google Scholar] [CrossRef]
- Fu, Q.; Zhang, Y.; Huang, T.; Liu, Y. Measurement of the Compressibility of Cell and Nucleus Based on Acoustofluidic Microdevice. J. Vis. Exp. JoVE 2022, 14, 185. [Google Scholar] [CrossRef]
- Ding, X.; Peng, Z.; Lin, S.C.; Geri, M.; Li, S.; Li, P.; Chen, Y.; Dao, M.; Suresh, S.; Huang, T.J. Cell separation using tilted-angle standing surface acoustic waves. Proc. Natl. Acad. Sci. USA 2014, 111, 12992–12997. [Google Scholar] [CrossRef]
- Ding, X.; Li, P.; Lin, S.C.; Stratton, Z.S.; Nama, N.; Guo, F.; Slotcavage, D.; Mao, X.; Shi, J.; Costanzo, F.; et al. Surface acoustic wave microfluidics. Lab Chip 2013, 13, 3626–3649. [Google Scholar] [CrossRef]
- Lu, C.; Han, J.; Sun, X.; Yang, G. Electrochemical Detection and Point-of-Care Testing for Circulating Tumor Cells: Current Techniques and Future Potentials. Sensors 2020, 20, 6073. [Google Scholar] [CrossRef]
- Topa, J.; Grešner, P.; Żaczek, A.J.; Markiewicz, A. Breast cancer circulating tumor cells with mesenchymal features-an unreachable target? Cell. Mol. Life Sci. CMLS 2022, 79, 81. [Google Scholar] [CrossRef]
- Li, P.; Mao, Z.; Peng, Z.; Zhou, L.; Chen, Y.; Huang, P.H.; Truica, C.I.; Drabick, J.J.; El-Deiry, W.S.; Dao, M.; et al. Acoustic separation of circulating tumor cells. Proc. Natl. Acad. Sci. USA 2015, 112, 4970–4975. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, R.; Zhang, J.; Zhang, J. ARMS for EGFR mutation analysis of cytologic and corresponding lung adenocarcinoma histologic specimens. J. Cancer Res. Clin. Oncol. 2015, 141, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, M.; Peng, J.; Wang, J.; Zhao, Y.; Wu, W.; Lan, X. Novel technologies in cfDNA analysis and potential utility in clinic. Chin. J. Cancer Res. Chung-Kuo Yen Cheng Yen Chiu 2021, 33, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Hong, B.; Wang, S.; Wang, J.; Fang, J.; Sun, R.; Nie, J.; Wang, H. Plasma cell-free DNA methylome-based liquid biopsy for accurate gastric cancer detection. Cancer Sci. 2024, 115, 3426–3438. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, M.; Chen, S.; Weng, L.; He, Z. Efficacy and Safety of First-Generation EGFR-TKIs Combined with Chemotherapy for Treatment-Naïve Advanced Non-Small-Cell Lung Cancer Patients Harboring Sensitive EGFR Mutations: A Single-Center, Open-Label, Single-Arm, Phase II Clinical Trial. J. Inflamm. Res. 2021, 14, 2557–2567. [Google Scholar] [CrossRef]
- Fu, K.; Xie, F.; Wang, F.; Fu, L. Therapeutic strategies for EGFR-mutated non-small cell lung cancer patients with osimertinib resistance. J. Hematol. Oncol. 2022, 15, 173. [Google Scholar] [CrossRef]
- You, Q.; Wang, J.; Yu, Y.; Li, F.; Meng, L.; Chen, M.; Yang, Q.; Xu, Z.; Sun, J.; Zhuo, W.; et al. The histone deacetylase SIRT6 promotes glycolysis through the HIF-1α/HK2 signaling axis and induces erlotinib resistance in non-small cell lung cancer. Apoptosis Int. J. Program. Cell Death 2022, 27, 883–898. [Google Scholar] [CrossRef]
- He, J.; Huang, Z.; Han, L.; Gong, Y.; Xie, C. Mechanisms and management of 3rd-generation EGFR-TKI resistance in advanced non-small cell lung cancer (Review). Int. J. Oncol. 2021, 59, 90. [Google Scholar] [CrossRef]
- Oya, Y.; Yoshida, T.; Kuroda, H.; Shimizu, J.; Horio, Y.; Sakao, Y.; Inaba, Y.; Hida, T.; Yatabe, Y. Association Between EGFR T790M Status and Progression Patterns During Initial EGFR-TKI Treatment in Patients Harboring EGFR Mutation. Clin. Lung Cancer 2017, 18, 698–705.e2. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, J.; Zhang, C.; Han, Z.; Zheng, X.; Cao, D. Application value of CT radiomic nomogram in predicting T790M mutation of lung adenocarcinoma. BMC Pulm. Med. 2023, 23, 339. [Google Scholar] [CrossRef] [PubMed]
- Paliogiannis, P.; Colombino, M.; Sini, M.C.; Manca, A.; Casula, M.; Palomba, G.; Pisano, M.; Doneddu, V.; Zinellu, A.; Santeufemia, D.; et al. Global prognostic impact of driver genetic alterations in patients with lung adenocarcinoma: A real-life study. BMC Pulm. Med. 2022, 22, 32. [Google Scholar] [CrossRef]
- Cui, Q.; Hu, Y.; Cui, Q.; Wu, D.; Mao, Y.; Ma, D.; Liu, H. Osimertinib Rechallenge with Bevacizumab vs. Chemotherapy Plus Bevacizumab in EGFR-Mutant NSCLC Patients with Osimertinib Resistance. Front. Pharmacol. 2021, 12, 746707. [Google Scholar] [CrossRef] [PubMed]
- Trachu, N.; Reungwetwattana, T.; Meanwatthana, J.; Sukasem, C.; Majam, T.; Saengsiwaritt, W.; Jittikoon, J.; Udomsinprasert, W. Leukocytes telomere length as a biomarker of adverse drug reactions induced by Osimertinib in advanced non-small cell lung cancer. Sci. Rep. 2024, 14, 26543. [Google Scholar] [CrossRef]
- Li, Y.; Mao, T.; Wang, J.; Zheng, H.; Hu, Z.; Cao, P.; Yang, S.; Zhu, L.; Guo, S.; Zhao, X.; et al. Toward the next generation EGFR inhibitors: An overview of osimertinib resistance mediated by EGFR mutations in non-small cell lung cancer. Cell Commun. Signal. CCS 2023, 21, 71. [Google Scholar] [CrossRef]
- Wu, S.G.; Yu, C.J.; Yang, J.C.; Shih, J.Y. The effectiveness of afatinib in patients with lung adenocarcinoma harboring complex epidermal growth factor receptor mutation. Ther. Adv. Med. Oncol. 2020, 12, 1758835920946156. [Google Scholar] [CrossRef]


| Living A549 Cells | Dead A549 Cells | Living White Blood Cells | Dead White Blood Cells | |
|---|---|---|---|---|
| Bright-field field of view | yes | yes | yes | yes |
| Red fluorescence field of view | yes | yes | no | no |
| Green fluorescence field of view | yes | no | yes | no |
| Sample Number | Years | Sex | Histological Type | Metastasis | Tumor Staging | Personal History |
|---|---|---|---|---|---|---|
| Number 1 | 72 | Female | Squamous Carcinoma | Bone | cT4N2M1 IV | No history of smoking |
| Number 2 | 74 | Male | Adenocarcinoma | Head | cT1cN2M1b IVA | Previous history of smoking |
| Number 3 | 62 | Male | Adenocarcinoma | None | cT1cN0M0 IA3 | Previous history of smoking |
| Number 4 | 59 | Male | Squamous Carcinoma | None | cT3N3M0 IIIc | No history of smoking |
| Number 5 | 74 | Male | Squamous Carcinoma | Lymphatic Node | cT4N3M1 IV | No history of smoking |
| Number 6 | 64 | Male | Squamous Carcinoma | None | cT4N3M1 IV | Previous history of smoking |
| Number of CTCs Captured by Acoustic Microfluidics Combined with iFISH | Number of CTCs Captured by Microfluidics Combined with the Immunomagnetic Bead Separation Technique | |
|---|---|---|
| Number 1 | 1 | 52 |
| Number 2 | 6 | 17 |
| Number 3 | 3 | 13 |
| Number 4 | 4 | 8 |
| Number 5 | 1 | 9 |
| Number 6 | 1 | 1 |
| Sample Number | Type of EGFR Gene Mutation |
|---|---|
| Number 1 | 19-del, L858R mutation |
| Number 2 | S768I mutation |
| Number 3 | None |
| Number 4 | T790M mutation |
| Number 5 | None |
| Number 6 | L858R mutation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, T.; Ma, S.; Wang, Y.; Yin, H.; Dang, T.; Li, G.; Li, J.; Feng, W.; Tian, M.; Ma, J.; et al. A Novel Microfluidic Platform for Circulating Tumor Cell Identification in Non-Small-Cell Lung Cancer. Micromachines 2025, 16, 1136. https://doi.org/10.3390/mi16101136
Tian T, Ma S, Wang Y, Yin H, Dang T, Li G, Li J, Feng W, Tian M, Ma J, et al. A Novel Microfluidic Platform for Circulating Tumor Cell Identification in Non-Small-Cell Lung Cancer. Micromachines. 2025; 16(10):1136. https://doi.org/10.3390/mi16101136
Chicago/Turabian StyleTian, Tingting, Shanni Ma, Yan Wang, He Yin, Tiantian Dang, Guangqi Li, Jiaming Li, Weijie Feng, Mei Tian, Jinbo Ma, and et al. 2025. "A Novel Microfluidic Platform for Circulating Tumor Cell Identification in Non-Small-Cell Lung Cancer" Micromachines 16, no. 10: 1136. https://doi.org/10.3390/mi16101136
APA StyleTian, T., Ma, S., Wang, Y., Yin, H., Dang, T., Li, G., Li, J., Feng, W., Tian, M., Ma, J., & Zhao, Z. (2025). A Novel Microfluidic Platform for Circulating Tumor Cell Identification in Non-Small-Cell Lung Cancer. Micromachines, 16(10), 1136. https://doi.org/10.3390/mi16101136







