Disease Modeling with Kidney Organoids
Abstract
:1. Introduction
2. The Human Kidney vs. Kidney Organoids
2.1. Human Kidney Development
2.2. Protocols to Generate Kidney Organoids
2.3. How They Stack Up: Kidney Organoids vs. Human Kidnies
3. Kidney Organoids as Model Systems
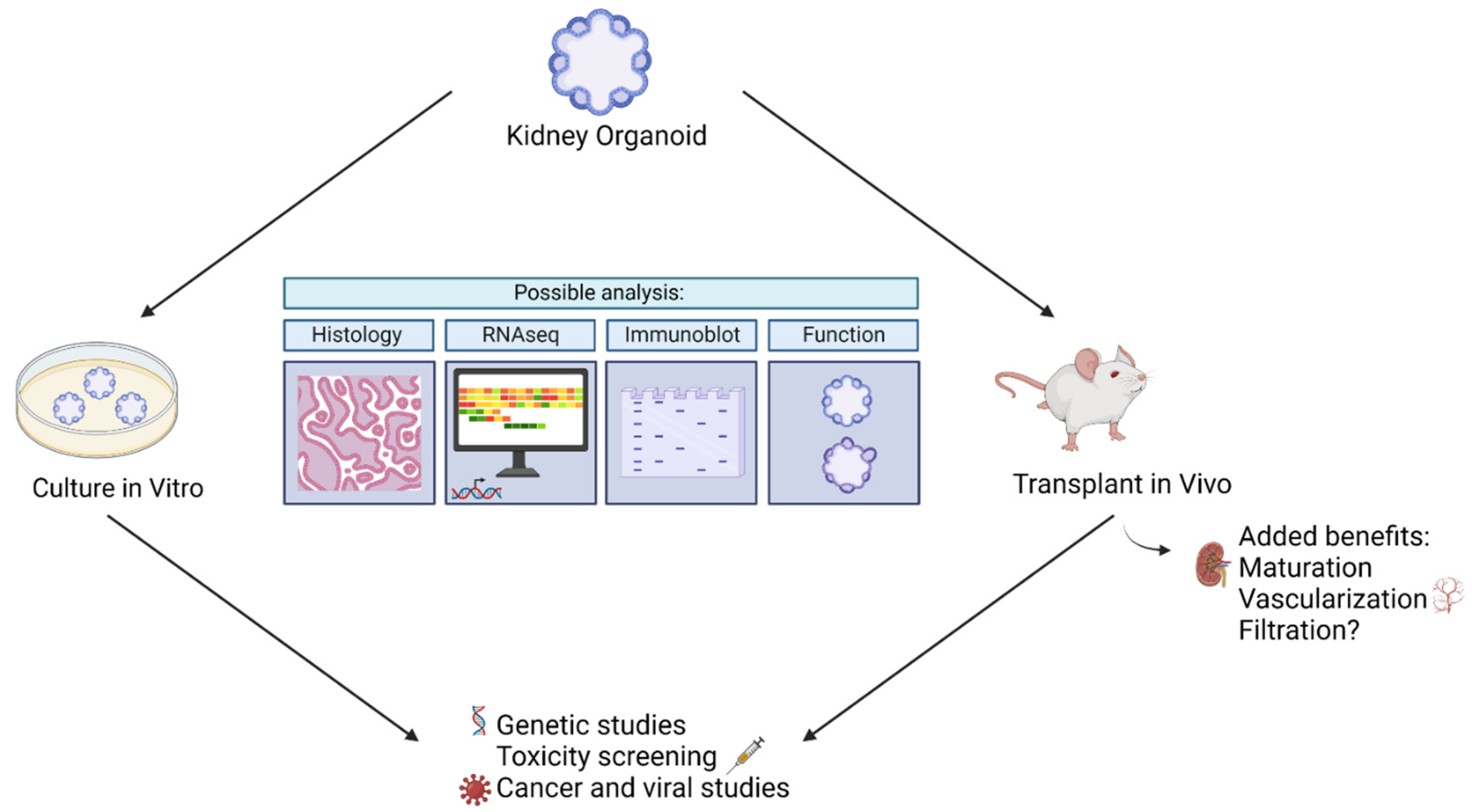
3.1. Kidney Organoid Analysis
3.1.1. In Vitro Assays
3.1.2. In Vivo Analysis
3.2. Disease Modeling Studies Conducted Thus Far
3.2.1. Kidney Organoids as Genetic Disease Models
3.2.2. Kidney Organoids as Models for Other Disease
3.2.3. Kidney Organoids in Drug Evaluation
4. Strategies to Improve Kidney Organoid Platforms
4.1. Introducing Gradients
4.2. Perfusing Organoids with Microfluidics
4.3. Advancing Organoid Maturation via Transplantation
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bannier-Hélaouët, M.; Post, Y.; Korving, J.; Trani Bustos, M.; Gehart, H.; Begthel, H.; Bar-Ephraim, Y.E.; van der Vaart, J.; Kalmann, R.; Imhoff, S.M.; et al. Exploring the Human Lacrimal Gland Using Organoids and Single-Cell Sequencing. Cell Stem Cell 2021, 28, 1221–1232.e7. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Rabbani, C.C.; Gao, H.; Steinhart, M.R.; Woodruff, B.M.; Pflum, Z.E.; Kim, A.; Heller, S.; Liu, Y.; Shipchandler, T.Z.; et al. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature 2020, 582, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Global Facts: About Kidney Disease. Available online: https://www.kidney.org/kidneydisease/global-facts-about-kidney-disease#:~:text=10%25%20of%20the%20population%20worldwide,have%20access%20to%20affordable%20treatment (accessed on 6 July 2022).
- Rehman, S.; Ahmed, D. Embryology, Kidney, Bladder, and Ureter; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Reidy, K.J.; Rosenblum, N.D. Cell and molecular biology of kidney development. Semin. Nephrol. 2009, 29, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.; Sutherland, M.; Flores, T.J.; Kent, A.L.; Dahlstrom, J.; Puelles, V.; Bertram, J.; McMahon, A.P.; Little, M.H.; Moore, L.; et al. Development of the human fetal kidney from mid to late gestation in male and female infants. eBioMedicine 2017, 27, 275–283. [Google Scholar] [CrossRef]
- Al-Awqati, Q.; Oliver, J.A. Stem cells in the kidney. Kidney Int. 2002, 61, 387–395. [Google Scholar] [CrossRef]
- Tran, F.; Klein, C.; Arlt, A.; Imm, S.; Knappe, E.; Simmons, A.; Rosenstiel, P.; Seibler, P. Stem cells and organoid technology in precision medicine in inflammation: Are we there yet? Front. Immunol. 2020, 11, 573562. [Google Scholar] [CrossRef]
- Drost, J.; Clevers, H. Translational applications of adult stem cell-derived organoids. Development 2017, 144, 968–975. [Google Scholar] [CrossRef]
- Hishikawa, K.; Takase, O.; Yoshikawa, M.; Tsujimura, T.; Nangaku, M.; Takato, T. Adult stem-like cells in kidney. World J. Stem Cells 2015, 7, 490–494. [Google Scholar] [CrossRef]
- Takasato, M.; Er, P.X.; Chiu, H.S.; Little, M.H. Generation of kidney organoids from human pluripotent stem cells. Nat. Protoc. 2016, 11, 1681–1692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morizane, R.; Lam, A.Q.; Freedman, B.S.; Kishi, S.; Valerius, M.T.; Bonventre, J.V. nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 2015, 33, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Freedman, B.S.; Brooks, C.R.; Lam, A.Q.; Fu, H.; Morizane, R.; Agrawal, V.; Saad, A.F.; Li, M.K.; Hughes, M.R.; Werff, R.V.; et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 2015, 6, 8715. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Uchimura, K.; Donnelly, E.; Kirita, Y.; Morris, S.A.; Humphreys, B.D. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single cell transcriptomics. Cell Stem Cell 2018, 23, 869–881. [Google Scholar] [CrossRef]
- Garreta, E.; Prado, P.; Tarantino, C.; Oria, R.; Fanlo, L.; Martí, E.; Zalvidea, D.; Trepat, X.; Roca-Cusachs, P.; Gavaldà-Navarro, A.; et al. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat. Mater. 2019, 18, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, A.; Nishinakamura, R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell 2017, 21, 730–746.e6. [Google Scholar] [CrossRef]
- Taguchi, A.; Kaku, Y.; Ohmori, T.; Sharmin, S.; Ogawa, M.; Sasaki, H.; Nishinakamura, R. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 2013, 14, 53–67. [Google Scholar] [CrossRef]
- Przepiorski, A.; Sander, V.; Tran, T.; Hollywood, J.A.; Sorrenson, B.; Shih, J.-H.; Wolvetang, E.J.; McMahon, A.P.; Holm, T.M.; Davidson, A.J. A simple bioreactor-based method to generate kidney organoids from pluripotent stem cells. Stem Cell Rep. 2018, 11, 470–484. [Google Scholar] [CrossRef]
- Takasato, M.; Er, P.X.; Chiu, H.S.; Maier, B.; Baillie, G.J.; Ferguson, C.; Parton, R.G.; Wolvetang, E.J.; Roost, M.S.; Chuva de Sousa Lopes, S.M.; et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 2015, 526, 564–568. [Google Scholar] [CrossRef]
- Subramanian, A.; Sidhom, E.-H.; Emani, M.; Vernon, K.; Sahakian, N.; Zhou, Y.; Kost-Alimova, M.; Slyper, M.; Waldman, J.; Dionne, D.; et al. Single cell census of human kidney organoids shows reproducibility and diminished off-target cells after transplantation. Nat. Commun. 2019, 10, 5462. [Google Scholar] [CrossRef]
- Kit: Home Page. Available online: http://humphreyslab.com/SingleCell/ (accessed on 8 July 2022).
- Nishinakamura, R. Human kidney organoids: Progress and remaining challenges. Nat. Rev. Nephrol. 2019, 15, 613–624. [Google Scholar] [CrossRef]
- Tsujimoto, H.; Kasahara, T.; Sueta, S.-I.; Araoka, T.; Sakamoto, S.; Okada, C.; Mae, S.-I.; Nakajima, T.; Okamoto, N.; Taura, D.; et al. A modular differentiation system maps multiple human kidney lineages from pluripotent stem cells. Cell Rep. 2020, 31, 107476. [Google Scholar] [CrossRef] [PubMed]
- Freedman, B.S. Better being single? Omics improves kidney organoids. Nephron Exp. Nephrol. 2018, 141, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Cruz, N.M.; Song, X.; Czerniecki, S.M.; Gulieva, R.E.; Churchill, A.J.; Kim, Y.K.; Winston, K.; Tran, L.M.; Diaz, M.A.; Fu, H.; et al. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat. Mater. 2017, 16, 1112–1119. [Google Scholar] [CrossRef] [PubMed]
- Morais, M.R.; Tian, P.; Lawless, C.; Murtuza-Baker, S.; Hopkinson, L.; Woods, S.; Mironov, A.; Long, D.A.; Gale, D.P.; Zorn, T.M.; et al. Kidney organoids recapitulate human basement membrane assembly in health and disease. ELife 2022, 11, 73486. [Google Scholar] [CrossRef] [PubMed]
- Ryan, A.R.; England, A.R.; Chaney, C.P.; Cowdin, M.A.; Hiltabidle, M.; Daniel, E.; Gupta, A.K.; Oxburgh, L.; Carroll, T.J.; Cleaver, O. Vascular deficiencies in renal organoids and ex vivo kidney organogenesis. Dev Biol. 2021, 477, 98–116. [Google Scholar] [CrossRef]
- Freedman, B.S. Physiology assays in human kidney organoids. Am. J. Physiol.-Ren. Physiol. 2022, 322, F625–F638. [Google Scholar] [CrossRef]
- Gupta, N.; Matsumoto, T.; Hiratsuka, K.; Saiz, E.G.; Zhang, C.; Galichon, P.; Miyoshi, T.; Susa, K.; Tatsumoto, N.; Yamashita, M.; et al. Modeling injury and repair in kidney organoids reveals that homologous recombination governs tubular intrinsic repair. Sci. Transl. Med. 2022, 14, eabj4772. [Google Scholar] [CrossRef]
- Gupta, A.K.; Coburn, J.M.; Davis-Knowlton, J.; Kimmerling, E.; Kaplan, D.L.; Oxburgh, L. Scaffolding kidney organoids on silk. J. Tissue Eng. Regen. Med. 2019, 13, 812–822. [Google Scholar] [CrossRef]
- Berg, C.W.V.D.; Ritsma, L.; Avramut, M.C.; Wiersma, L.E.; Berg, B.M.V.D.; Leuning, D.G.; Lievers, E.; Koning, M.; Vanslambrouck, J.M.; Koster, A.J.; et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. 2018, 10, 751–765. [Google Scholar] [CrossRef]
- Bantounas, I.; Ranjzad, P.; Tengku, F.; Silajdžić, E.; Forster, D.; Asselin, M.-C.; Lewis, P.; Lennon, R.; Plagge, A.; Wang, Q.; et al. Generation of functioning nephrons by implanting human pluripotent stem cell-derived kidney progenitors. Stem Cell Rep. 2018, 10, 766–779. [Google Scholar] [CrossRef] [Green Version]
- Hale, L.; Howden, S.E.; Phipson, B.; Lonsdale, A.; Er, P.X.; Ghobrial, I.; Hosawi, S.; Wilson, S.; Lawlor, K.; Khan, S.; et al. 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat. Commun. 2018, 9, 5167. [Google Scholar] [CrossRef]
- Kirby, A.; Gnirke, A.; Jaffe, D.B.; Baresova, V.; Pochet, N.; Blumenstiel, B.; Ye, C.; Aird, D.; Stevens, C.; Robinson, J.T.; et al. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat. Genet. 2013, 45, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, T.; Wu, G.; Hayashi, T.; Xenophontos, S.L.; Veldhuisen, B.; Saris, J.J.; Reynolds, D.M.; Cai, Y.; Gabow, P.A.; Pierides, A.; et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 1996, 272, 1339–1342. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.C.; Torres, V.E. Genetic mechanisms and signaling pathways in autosomal dominant polycystic kidney disease. J. Clin. Investig. 2014, 124, 2315–2324. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.; Ward, C.J.; Peral, B.; Aspinwall, R.; Clark, K.; Millán, J.L.S.; Gamble, V.; Harris, P.C. The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat. Genet. 1995, 10, 151–160. [Google Scholar] [CrossRef]
- Low, J.H.; Li, P.; Chew, E.G.Y.; Zhou, B.; Suzuki, K.; Zhang, T.; Lian, M.M.; Liu, M.; Aizawa, E.; Esteban, C.R.; et al. Generation of human PSC-derived kidney organoids with patterned nephron segments and a de novo vascular network. Cell Stem Cell 2019, 25, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.O.R.; Wang, X.; Vazquez-Segoviano, M.; Lopez-Marfil, M.; Sobral-Reyes, M.F.; Moran-Horowich, A.; Sundberg, M.; Lopez-Cantu, D.O.; Probst, C.K.; Ruiz-Esparza, G.U.; et al. A tissue-bioengineering strategy for modeling rare human kidney diseases in vivo. Nat. Commun. 2021, 12, 6496. [Google Scholar] [CrossRef]
- Combes, A.N.; Zappia, L.; Er, P.X.; Oshlack, A.; Little, M.H. Single-cell analysis reveals congruence between kidney organoids and human fetal kidney. Genome Med. 2019, 11, 1–15. [Google Scholar] [CrossRef]
- Dias, T.; Sairam, S.; Kumarasiri, S. Ultrasound diagnosis of fetal renal abnormalities. Best Pract. Res. Clin. Obstet. Gynaecol. 2014, 28, 403–415. [Google Scholar] [CrossRef]
- Jansen, J.; Reimer, K.C.; Nagai, J.S.; Varghese, F.S.; Overheul, G.J.; de Beer, M.; Roverts, R.; Daviran, D.; Fermin, L.A.; Willemsen, B.; et al. SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids. Cell Stem Cell 2021, 29, 217–231.e8. [Google Scholar] [CrossRef]
- Przepiorski, A.; Vanichapol, T.; Espiritu, E.B.; Crunk, A.E.; Parasky, E.; McDaniels, M.D.; Emlet, D.R.; Salisbury, R.; Happ, C.L.; Vernetti, L.A.; et al. Modeling oxidative injury response in human kidney organoids. Stem Cell Res. Ther. 2022, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Perazella, M.A. Drug-induced acute kidney injury. Curr. Opin. Crit. Care 2019, 25, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Czerniecki, S.M.; Cruz, N.M.; Harder, J.L.; Menon, R.; Annis, J.; Otto, E.A.; Gulieva, R.E.; Islas, L.V.; Kim, Y.K.; Tran, L.M.; et al. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 2018, 22, 929–940.e4. [Google Scholar] [CrossRef] [PubMed]
- Costantini, F.; Kopan, R. Patterning a complex organ: Branching morphogenesis and nephron segmentation in kidney development. Dev. Cell 2010, 18, 698–712. [Google Scholar] [CrossRef]
- Ben-Reuven, L.; Reiner, O. Toward spatial identities in human brain organoids-on-chip induced by morphogen-soaked beads. Bioengineering 2020, 7, 164. [Google Scholar] [CrossRef]
- Kamperman, T.; Koerselman, M.; Kelder, C.; Hendriks, J.; Crispim, J.F.; de Peuter, X.; Dijkstra, P.J.; Karperien, M.; Leijten, J. Spatiotemporal material functionalization via competitive supramolecular complexation of avidin and biotin analogs. Nat. Commun. 2019, 10, 4347. [Google Scholar] [CrossRef]
- Cui, K.W.; Engel, L.; Dundes, C.E.; Nguyen, T.C.; Loh, K.M.; Dunn, A.R. Spatially controlled stem cell differentiation via morphogen gradients: A comparison of static and dynamic microfluidic platforms. J. Vac. Sci. Technol. A Vac. Surf. Film. 2020, 38, 033205. [Google Scholar] [CrossRef]
- Cartwright, J.H.E.; Piro, O.; Tuval, I. Chemosensing versus mechanosensing in nodal and Kupffer’s vesicle cilia and in other left–right organizer organs. Philos. Trans. R. Soc. B Biol. Sci. 2019, 375, 20190566. [Google Scholar] [CrossRef]
- Homan, K.A.; Gupta, N.; Kroll, K.T.; Kolesky, D.B.; Skylar-Scott, M.; Miyoshi, T.; Mau, D.; Valerius, M.T.; Ferrante, T.; Bonventre, J.V.; et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 2019, 16, 255–262. [Google Scholar] [CrossRef]
- Nashimoto, Y.; Hayashi, T.; Kunita, I.; Nakamasu, A.; Torisawa, Y.-S.; Nakayama, M.; Takigawa-Imamura, H.; Kotera, H.; Nishiyama, K.; Miura, T.; et al. Integrating perfusable vascular networks with a three-dimensional tissue in a microfluidic device. Integr. Biol. 2017, 9, 506–518. [Google Scholar] [CrossRef]
- Munro, D.A.; Davies, J.A. Vascularizing the kidney in the embryo and organoid: Questioning assumptions about renal vasculogenesis. J. Am. Soc. Nephrol. 2018, 29, 1593–1595. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, A.; Roumans, N.; Rademakers, T.; Joris, V.; Eischen-Loges, M.J.; van Griensven, M.; LaPointe, V.L. Enhanced Microvasculature Formation and Patterning in iPSC–Derived Kidney Organoids Cultured in Physiological Hypoxia. Front. Bioeng. Biotechnol. 2022, 10, 860138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Du, X.; Zhang, X.; Li, K.; Kong, F.; Cheng, G.; Zhao, S. In vitro induction and in vivo engraftment of kidney organoids derived from human pluripotent stem cells. Exp. Ther. Med. 2020, 20, 1307–1314. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karp, S.; Pollak, M.R.; Subramanian, B. Disease Modeling with Kidney Organoids. Micromachines 2022, 13, 1384. https://doi.org/10.3390/mi13091384
Karp S, Pollak MR, Subramanian B. Disease Modeling with Kidney Organoids. Micromachines. 2022; 13(9):1384. https://doi.org/10.3390/mi13091384
Chicago/Turabian StyleKarp, Sophie, Martin R Pollak, and Balajikarthick Subramanian. 2022. "Disease Modeling with Kidney Organoids" Micromachines 13, no. 9: 1384. https://doi.org/10.3390/mi13091384
APA StyleKarp, S., Pollak, M. R., & Subramanian, B. (2022). Disease Modeling with Kidney Organoids. Micromachines, 13(9), 1384. https://doi.org/10.3390/mi13091384





