ANN-Based Instantaneous Simulation of Particle Trajectories in Microfluidics
Abstract
1. Introduction
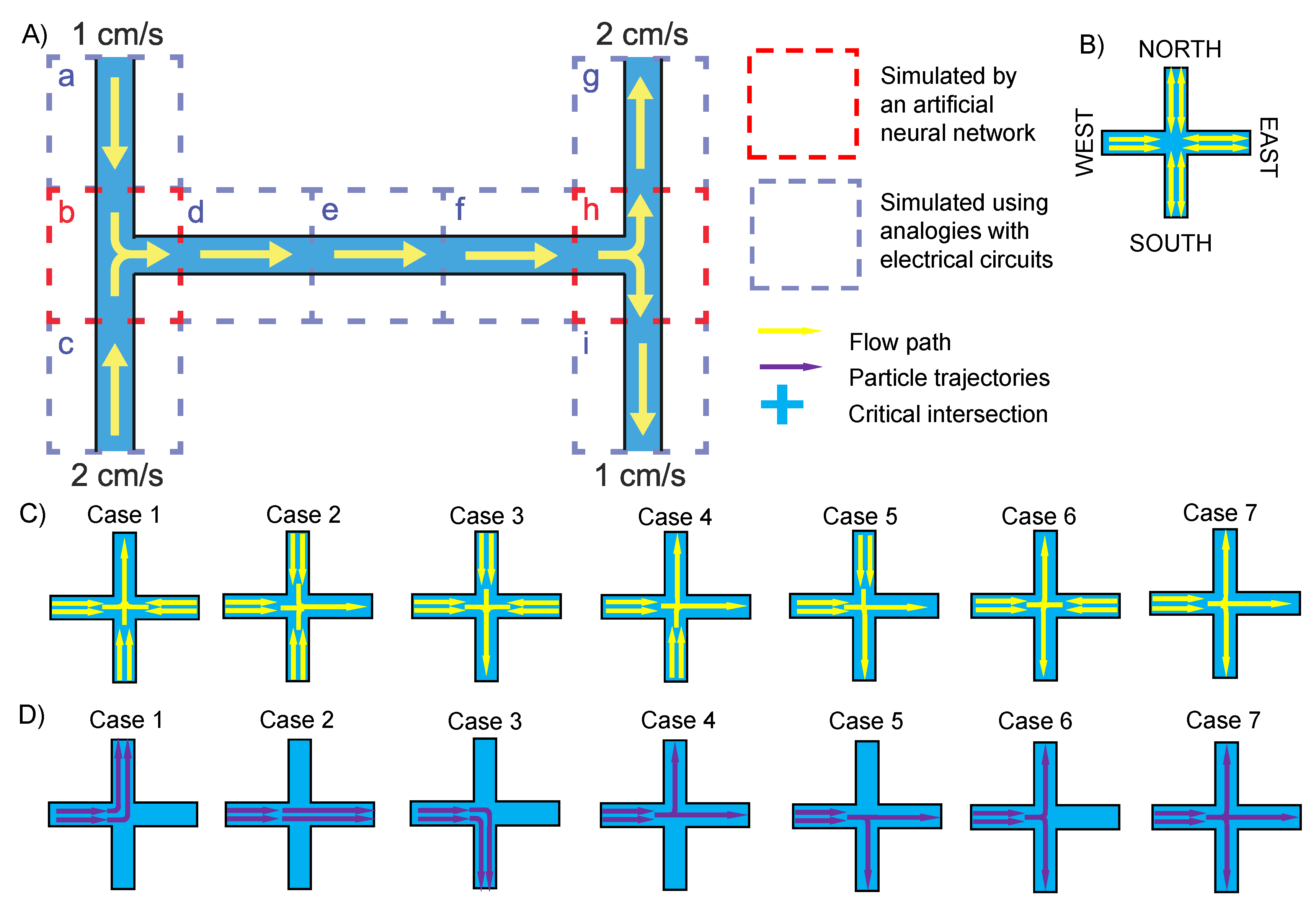
2. Theory of the Design
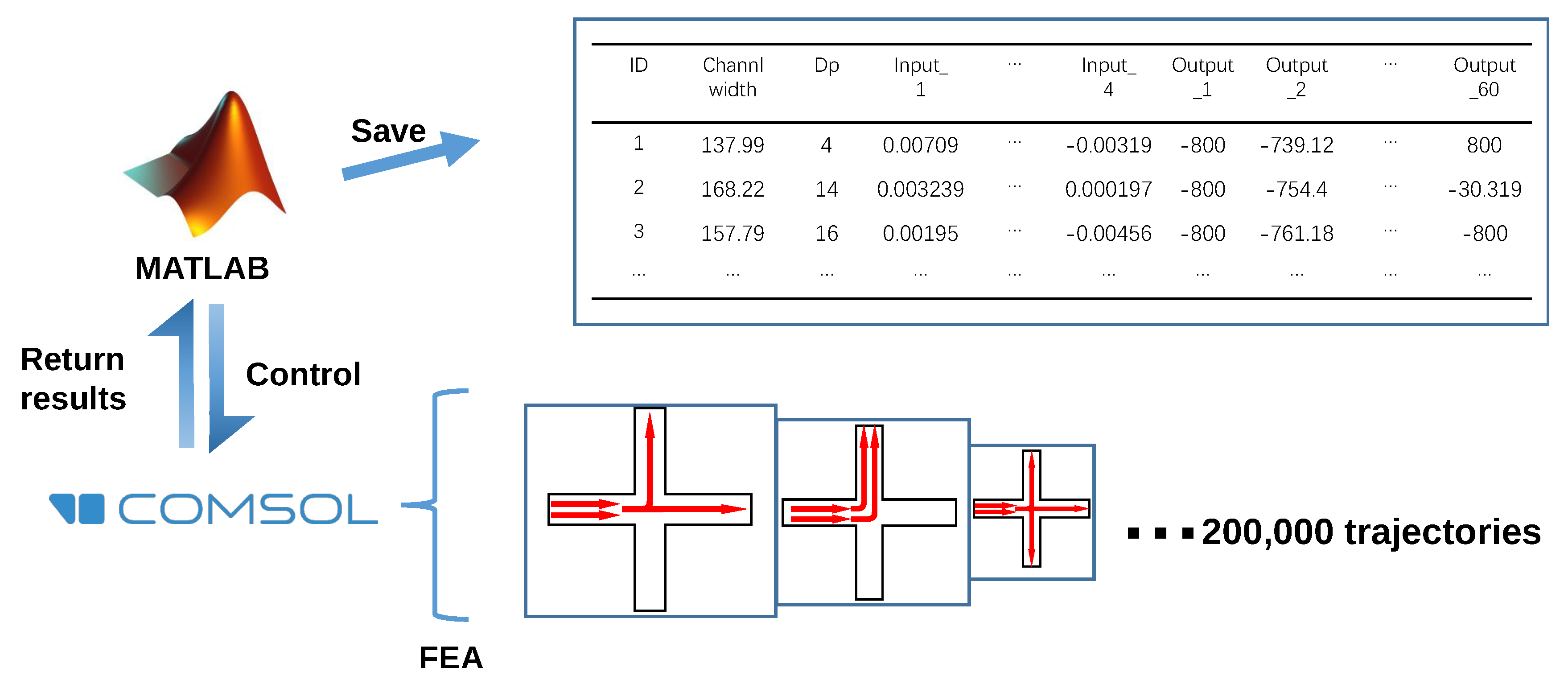
3. Materials and Methods
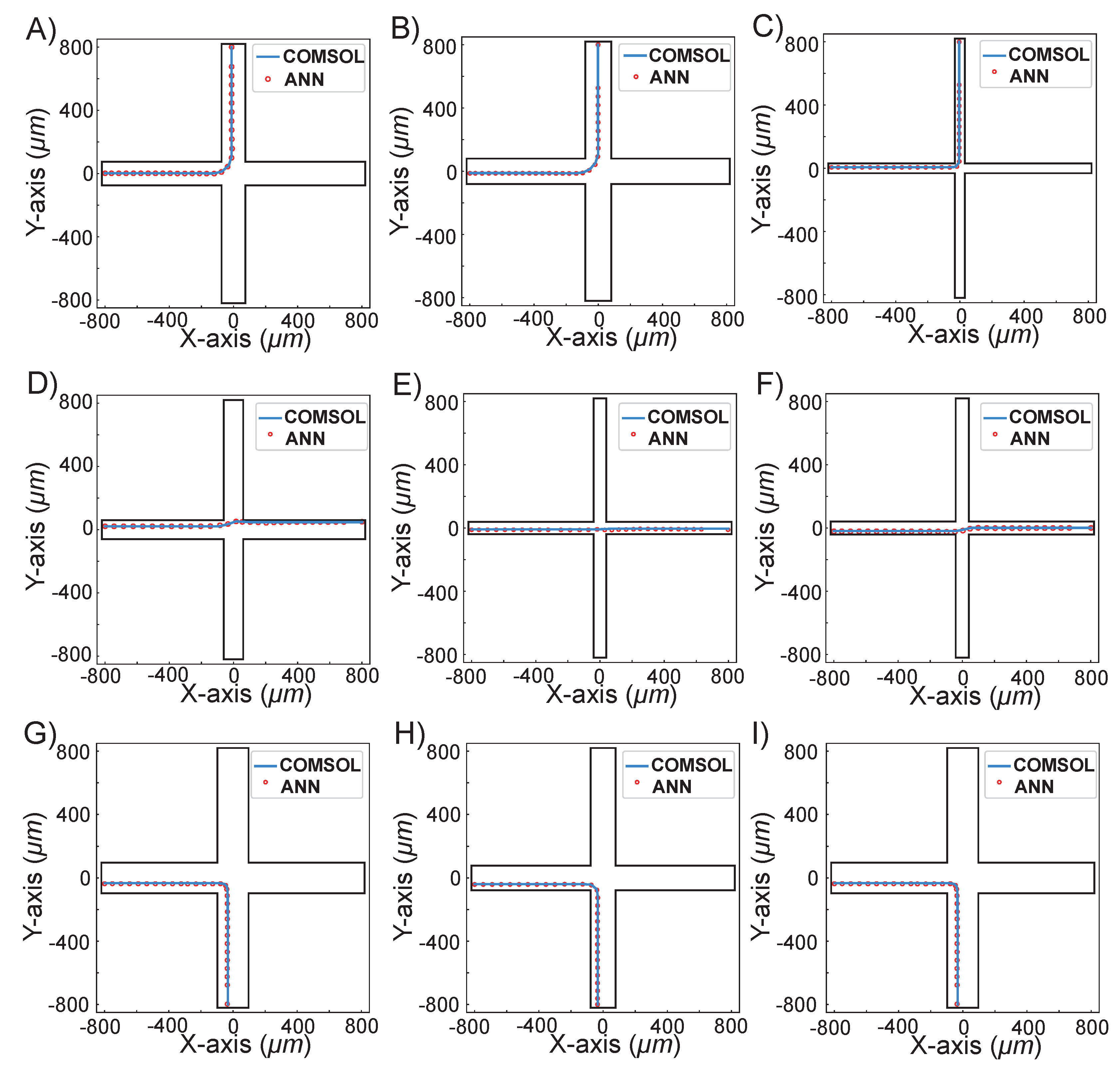
4. Results and Discussion
Potential Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, W.; Jiang, D.; Li, Z.; Zhu, L.; Shi, J.; Yang, J.; Xiang, N. Recent advances in microfluidic cell sorting techniques based on both physical and biochemical principles. Electrophoresis 2019, 40, 930–954. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Jiang, F.; Han, Y.; Xiang, N.; Ni, Z. Microfluidics for label-free sorting of rare circulating tumor cells. Analyst 2020, 145, 7103–7124. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Liu, H.; Tian, Y.; Li, Z.; Cui, X. Sorting technology for circulating tumor cells based on microfluidics. ACS Comb. Sci. 2020, 22, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Laxmi, V.; Joshi, S.S.; Agrawal, A. Effect of various parameters on the distribution and extraction of platelets in a microfluidic system. Microfluid. Nanofluidics 2021, 25, 1–12. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X. Dielectrophoretic microfluidic device for separation of red blood cells and platelets: A model-based study. J. Braz. Soc. Mech. Sci. Eng. 2020, 42, 1–11. [Google Scholar] [CrossRef]
- Richard, C.; Fakhfouri, A.; Colditz, M.; Striggow, F.; Kronstein-Wiedemann, R.; Tonn, T.; Medina-Sánchez, M.; Schmidt, O.G.; Gemming, T.; Winkler, A. Blood platelet enrichment in mass-producible surface acoustic wave (SAW) driven microfluidic chips. Lab Chip 2019, 19, 4043–4051. [Google Scholar] [CrossRef]
- Segaliny, A.I.; Li, G.; Kong, L.; Ren, C.; Chen, X.; Wang, J.K.; Baltimore, D.; Wu, G.; Zhao, W. Functional TCR T cell screening using single-cell droplet microfluidics. Lab Chip 2018, 18, 3733–3749. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, R.; Shen, B.; Li, N.; Zhou, H.; Wang, W.; Zhao, Y.; Huang, M.; Fang, P.; Wang, S.; et al. High-throughput functional screening for next-generation cancer immunotherapy using droplet-based microfluidics. Sci. Adv. 2021, 7, eabe3839. [Google Scholar] [CrossRef]
- Buryk-Iggers, S.; Kieda, J.; Tsai, S.S. Diamagnetic droplet microfluidics applied to single-cell sorting. AIP Adv. 2019, 9, 075106. [Google Scholar] [CrossRef]
- Sesen, M.; Whyte, G. Image-based single cell sorting automation in droplet microfluidics. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Kim, G.Y.; Han, J.I.; Park, J.K. Inertial microfluidics-based cell sorting. BioChip J. 2018, 12, 257–267. [Google Scholar] [CrossRef]
- Nakao, S.; Takeo, T.; Watanabe, H.; Kondoh, G.; Nakagata, N. Successful selection of mouse sperm with high viability and fertility using microfluidics chip cell sorter. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Yuan, X.; Glidle, A.; Fu, Y.; Furusho, H.; Yang, T.; Yin, H. Automated Raman based cell sorting with 3D microfluidics. Lab Chip 2020, 20, 4235–4245. [Google Scholar] [CrossRef] [PubMed]
- Xiang, N.; Ni, Z. Electricity-free hand-held inertial microfluidic sorter for size-based cell sorting. Talanta 2021, 235, 122807. [Google Scholar] [CrossRef] [PubMed]
- Descamps, L.; Audry, M.C.; Howard, J.; Mekkaoui, S.; Albin, C.; Barthelemy, D.; Payen, L.; Garcia, J.; Laurenceau, E.; Le Roy, D.; et al. Self-assembled permanent micro-magnets in a polymer-based microfluidic device for magnetic cell sorting. Cells 2021, 10, 1734. [Google Scholar] [CrossRef] [PubMed]
- Mutafopulos, K.; Spink, P.; Lofstrom, C.; Lu, P.; Lu, H.; Sharpe, J.; Franke, T.; Weitz, D. Traveling surface acoustic wave (TSAW) microfluidic fluorescence activated cell sorter (μFACS). Lab Chip 2019, 19, 2435–2443. [Google Scholar] [CrossRef]
- Cai, K.; Mankar, S.; Ajiri, T.; Shirai, K.; Yotoriyama, T. An integrated high-throughput microfluidic circulatory fluorescence-activated cell sorting system (μ-CFACS) for the enrichment of rare cells. Lab Chip 2021, 21, 3112–3127. [Google Scholar] [CrossRef]
- LaBelle, C.A.; Massaro, A.; Cortés-Llanos, B.; Sims, C.E.; Allbritton, N.L. Image-based live cell sorting. Trends Biotechnol. 2021, 39, 613–623. [Google Scholar] [CrossRef]
- Lee, K.; Kim, S.E.; Doh, J.; Kim, K.; Chung, W.K. User-friendly image-activated microfluidic cell sorting technique using an optimized, fast deep learning algorithm. Lab Chip 2021, 21, 1798–1810. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, R.H.; Zhukov, A.A.; Fullerton, J.N.; Want, A.J.; Hussain, F.; la Cour, M.F.; Bashtanov, M.E.; Gold, R.D.; Hailes, A.; Banham-Hall, E.; et al. Cell sorting actuated by a microfluidic inertial vortex. Lab Chip 2019, 19, 2456–2465. [Google Scholar] [CrossRef]
- Turan, B.; Masuda, T.; Lei, W.; Noor, A.M.; Horio, K.; Saito, T.I.; Miyata, Y.; Arai, F. A pillar-based microfluidic chip for T-cells and B-cells isolation and detection with machine learning algorithm. Robomech J. 2018, 5, 1–9. [Google Scholar] [CrossRef]
- Hung, S.; Hsu, C.H.; Chen, C. Cell sorting in microfluidic systems using dielectrophoresis. In Proceedings of the 2015 IEEE 15th International Conference on Nanotechnology (IEEE-NANO), Rome, Italy, 27–30 July 2015; IEEE: New York, NY, USA, 2015; pp. 872–875. [Google Scholar]
- Lin, Y.T.; Huang, C.S.; Tseng, S.C. How to Control the Microfluidic Flow and Separate the Magnetic and Non-Magnetic Particles in the Runner of a Disc. Micromachines 2021, 12, 1335. [Google Scholar] [CrossRef]
- Myklatun, A.; Cappetta, M.; Winklhofer, M.; Ntziachristos, V.; Westmeyer, G.G. Microfluidic sorting of intrinsically magnetic cells under visual control. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, D.; Lashkaripour, A.; Fordyce, P.; Densmore, D. Machine learning for microfluidic design and control. Lab Chip 2022, 22, 2925–2937. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, N.; Chen, J.; Su, G.; Yao, H.; Ho, T.Y.; Sun, L. Predicting the fluid behavior of random microfluidic mixers using convolutional neural networks. Lab Chip 2021, 21, 296–309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Liu, Z.; Wang, J. Machine-Learning-Enabled Design and Manipulation of a Microfluidic Concentration Gradient Generator. Micromachines 2022, 13, 1810. [Google Scholar] [CrossRef]
- Ji, J.; Zhang, J.; Wang, J.; Huang, Q.; Jiang, X.; Zhang, W.; Sang, S.; Guo, X.; Li, S. Three-dimensional analyses of cells’ positioning on the quadrupole-electrode microfluid chip considering the coupling effect of nDEP, ACEO, and ETF. Biosens. Bioelectron. 2020, 165, 112398. [Google Scholar] [CrossRef]
- Ji, J.; Wang, J.; Wang, L.; Zhang, Q.; Duan, Q.; Sang, S.; Huang, Q.; Li, S.; Zhang, W.; Jiang, X. Dynamic-coupling analyses of cells localization by the negative dielectrophoresis. Proc. Inst. Mech. Eng. Part C J. Mech. Eng. Sci. 2021, 235, 402–411. [Google Scholar] [CrossRef]
- Du, C.J.; Sun, D.W. Learning techniques used in computer vision for food quality evaluation: A review. J. Food Eng. 2006, 72, 39–55. [Google Scholar] [CrossRef]
- Nadkarni, P.M.; Ohno-Machado, L.; Chapman, W.W. Natural language processing: An introduction. J. Am. Med. Inform. Assoc. 2011, 18, 544–551. [Google Scholar] [CrossRef]
- Almeida, L.G.; Backović, M.; Cliche, M.; Lee, S.J.; Perelstein, M. Playing tag with ANN: Boosted top identification with pattern recognition. J. High Energy Phys. 2015, 2015, 1–21. [Google Scholar] [CrossRef]
- Wang, J.; Rodgers, V.G.; Brisk, P.; Grover, W.H. Instantaneous simulation of fluids and particles in complex microfluidic devices. PLoS ONE 2017, 12, e0189429. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fu, L.; Yu, L.; Huang, X.; Brisk, P.; Grover, W.H. Accelerating Simulation of Particle Trajectories in Microfluidic Devices by Constructing a Cloud Database. In Proceedings of the 2018 IEEE Computer Society Annual Symposium on VLSI (ISVLSI), Hong Kong, China, 8–11 July 2018; IEEE: New York, NY, USA, 2018; pp. 666–671. [Google Scholar]


| ID | Layer | Dense 1 (Neurons) | Dense 2 (Neurons) | Dense 3 (Neurons) | Activation |
|---|---|---|---|---|---|
| 1 | Input | 7 | 27 | 27 | Linear |
| 2 | Fully connected | 400 | 400 | 400 | Leaky ReLU |
| 3 | Fully connected | 400 | 400 | 400 | Leaky ReLU |
| 4 | Fully connected | 400 | 400 | 400 | Leaky ReLU |
| 5 | Fully connected | 400 | 400 | 400 | Leaky ReLU |
| 6 | Fully connected | 400 | 400 | 400 | Leaky ReLU |
| 7 | Output | 20 | 20 | 20 | Linear |
| ID | Channel Width (μm) | Particle Diameter (μm) |
|---|---|---|
| A | 190.34 | 13 |
| B | 161.23 | 11 |
| C | 62.36 | 10 |
| D | 120.63 | 17 |
| E | 78.65 | 10 |
| F | 82.36 | 18 |
| G | 194.02 | 13 |
| H | 156.21 | 11 |
| I | 72.94 | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Liang, K.; Liu, Z.; Sun, T.; Wang, J. ANN-Based Instantaneous Simulation of Particle Trajectories in Microfluidics. Micromachines 2022, 13, 2100. https://doi.org/10.3390/mi13122100
Zhang N, Liang K, Liu Z, Sun T, Wang J. ANN-Based Instantaneous Simulation of Particle Trajectories in Microfluidics. Micromachines. 2022; 13(12):2100. https://doi.org/10.3390/mi13122100
Chicago/Turabian StyleZhang, Naiyin, Kaicong Liang, Zhenya Liu, Taotao Sun, and Junchao Wang. 2022. "ANN-Based Instantaneous Simulation of Particle Trajectories in Microfluidics" Micromachines 13, no. 12: 2100. https://doi.org/10.3390/mi13122100
APA StyleZhang, N., Liang, K., Liu, Z., Sun, T., & Wang, J. (2022). ANN-Based Instantaneous Simulation of Particle Trajectories in Microfluidics. Micromachines, 13(12), 2100. https://doi.org/10.3390/mi13122100







