1. Introduction
The analysis of cellular behavior by cytometry can arguably provide insight into bacterial behavior more rapidly than most other methods. This can be useful for making quick phenotypic observations, including antibiotic susceptibility. Rapid and portable measurements of antibiotic susceptibility are needed to combat the increasing spread of antibiotic resistance. Bacterial motility can provide some information on bacterial identification, but can also be used to distinguish between live and dead cells and can be used as a measure of antibiotic susceptibility [
1].
Motility has been widely acknowledged as a virulence factor in some pathogenic bacteria and plays a critical role in the formation of biofilm, which increases bacterial tolerance to antibiotics [
2,
3]. For the last several decades, different methods have been developed to differentiate and classify bacteria based on their motility. There are four kinds of different movement of bacteria: darting motility is presented by
Vibrio species; active motility by
Salmonella; sluggish motility by
Bacillus and
Clostridia genus; and thumbing motility by
Listeria. Initial developments of bacterial motility tests included the use of culture medium and naked-eye observation, such as in the Craigie, J. (1993) tube method, which was then advanced by the inclusion of microscopic examination (e.g., the Wet mount method) [
4]. Furthermore, these techniques have been enhanced by the use of dyes, including carbon fuchsin, safranin, and methyl blue or a novel single-tube agar-based technique for motility enhancement of
E. coli O157:H7. Likewise, microscopy-based in situ growth assays have been developed [
5,
6].
Open source hardware has also emerged as a major trend in scientific research. Open source systems make scientific equipment much more accessible not only for research but for a wider range of users [
7]. One powerful digital imaging platform makes use of Raspberry Pi hardware in research, offering low cost and accessibility; indeed, this was originally developed as an educational platform. Raspberry Pi hardware has been incorporated into commercial laboratory instruments (e.g., NuGenius by Syngene) and in many research laboratories for custom laboratory equipment. Many of these are based on imaging systems. One low-cost open source multi-fluorescence imaging system was developed using simply a Raspberry Pi computer and Pi camera [
8]. In addition to being low cost and allowing fully programmable imaging, the flexible Raspberry Pi GPIO output pins simplify addition of robotic sample manipulation or lighting control alongside imaging. Thus, digital imaging capability can be combined with robotics, for example, open source 3D printer hardware motion systems can be used as a platform for Raspberry Pi imaging, such as the Raspberry Pi camera Open source Laboratory Imaging Robot (POLIR) that consists of a 3D-printed 3-axis frame moving a Raspberry Pi camera controlled by an Arduino microcontroller, previously used for time-resolved analytical microbiology, including measuring antimicrobial resistance in bacteria [
9]. Another simple and cost-effective Raspberry Pi system provides controlled temperature and pressure monitoring systems [
10]. Affordable 3D printing has likewise made it easier for the science and engineering community to create rapid prototypes of complex products, paving the way for faster and easier setup of a lab [
11]. Small portable 3D printed structures combined with CMOS digital cameras are ideal for microscopy, illustrated by the OpenFlexure system [
12] and by digital holographic microscope [
13]. Open source hardware allow scientists to share, customise and improve their instrumentation alongside software, ensuring that technological advances in digital imaging can drive rapid scientific progress.
As digital cameras become cheaper, there has been a move from traditional laboratory instruments (e.g., based on film camera, photomultipliers, photodiodes, or CCD cameras) to consumer cameras offering a simplified approach to producing scientific imaging devices. The use of small CMOS cameras for digital microscopy is transforming the cost/performance of microimaging. For example, the Raspberry Pi Camera Module v2 is affordable yet still contains a high-quality CMOS image sensor (3280 × 2464 pixels), with a fixed focus lens that can be reversed and mounted far from the sensor to produce high resolution macro images. This system has been extensively adapted for digital microscopy [
14,
15,
16] by combination with 3D printed x-y-z stages. Matching this trend in microelectronics, recently, microfluidics-based analytical microbiology and diagnostic technology has emerged to offer a promise of improved healthcare systems by allowing the development of low-cost, rapid point-of-care diagnostic devices [
17]. Ideally, microelectronics, such as small digital cameras, can be combined with microfluidics to perform diagnostic assays, such as immunoassays [
18].
Here, we explore for the first time if the OpenFlexure microscope has the resolution to be used for microfluidic microbiology, to directly observe bacterial behaviour and motion. In this study, we combined inexpensive portable components for microbial cytometry, to establish the feasibility of rapidly monitoring bacterial motility in the presence of antibiotics. A novel device is presented that exploits melt-extruded microcapillary film to create panels of microfluidic antibiotic-loaded chambers [
19]. We investigated whether the 3D-printed OpenFlexure microscope using a low-cost Raspberry Pi v2 camera has sufficient magnification and resolution to monitor bacterial motility in microdevices. Adequate magnification and contrast were achieved to view motile bacteria and allowed differences in behavior to be observed in the presence of antibiotics above the organisms’ minimum inhibitory concentration (MIC) for that antibiotic. We suggest that this combination of inexpensive mass-manufactured microfluidic devices with open source 3D-printed digital microscopes permits rapid bacterial cell measurement suitable for portable analytical microbiology tests, such as rapid antibiotic susceptibility testing.
3. Results and Discussion
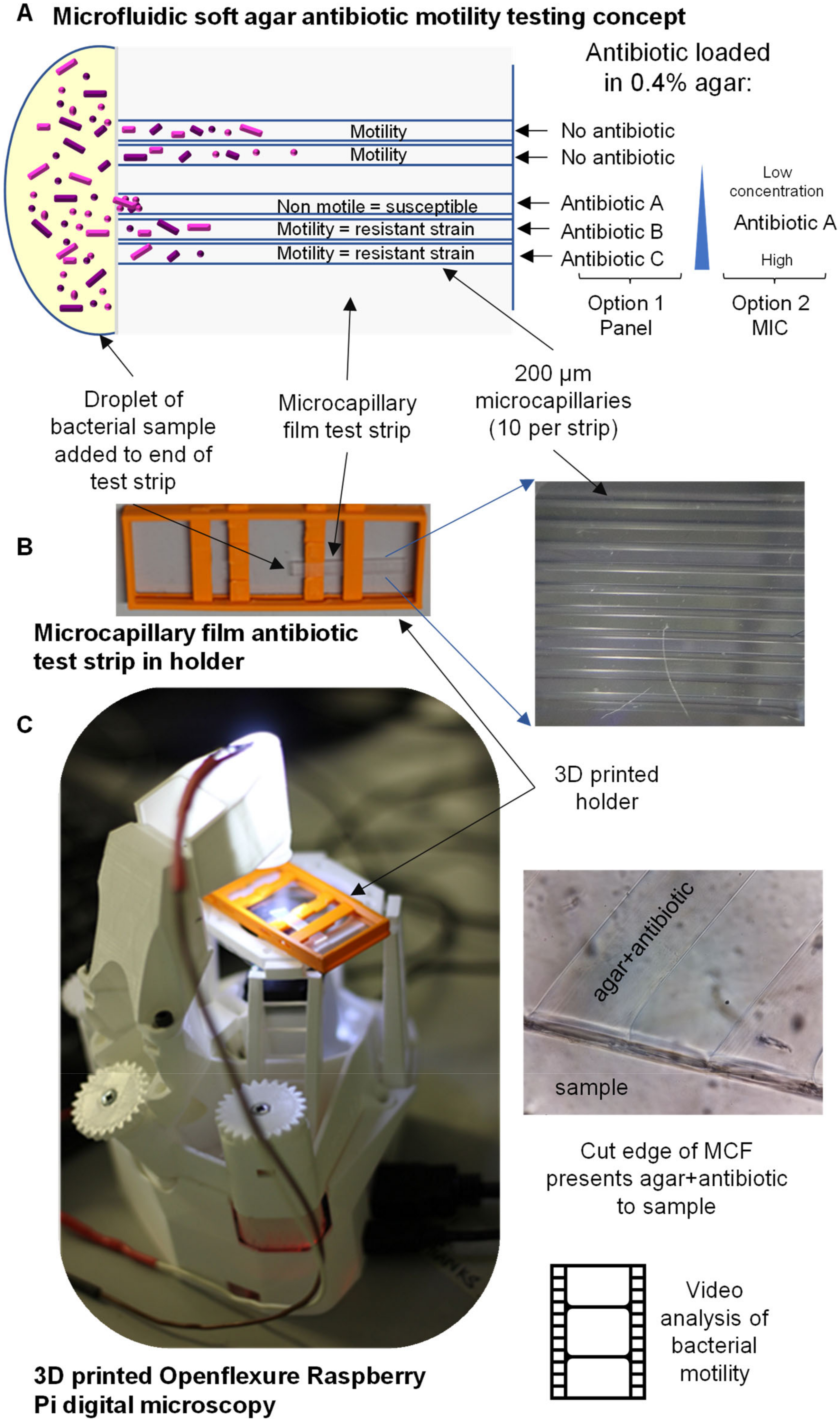
3.1. Concept of Microfluidic Bacterial Cytometry using Microcapillaries Combined with 3D-Printed Microscope
Microfluidic devices allow portable testing, transporting laboratory instruments into near-patient devices, and have been demonstrated for use of antibiotic susceptibility testing (AST). Motility and single-cell imaging provides one of the fastest measurements of phenotypic AST, providing results in as little as 20 min. However, these often require complex and bulky imaging or cytometry systems, such as microscopes or flow cytometers that require a laboratory environment to be operated. Previously, we have demonstrated the use of microcapillary film as a scalable and accurate AST device suited for point-of-care (PoC) applications, using a colour-based indicator to determine growth after several hours [
19]. However, using single-cell imaging, there is a potential to simplify sample preparation and decrease the time to result.
Here, we miniaturised a soft agar motility assay into microcapillaries (
Figure 1). Samples containing bacteria are added to the open end of a microcapillary film test strip, with the capillaries containing soft agar with or without antibiotics. Motile bacteria are able to swim freely into the capillary end, and into the soft agar. However, if susceptible to the antibiotic, a change in motility was observed, including decreased migration into the capillary and after time, bacteria killed from antibiotic exposure stopped moving.
This test device was combined with a 3D-printed holder to fit the stage of the 3D-printed OpenFlexure microscope to record videos and thereby determine motility. In this way, a small and compact bacterial cytometer can be produced for under $200 (based on $100 for the Raspberry Pi computer, $30 for the v2 camera and a small budget for 3D printing and accessory parts). The 3D-printed OpenFlexure microscope is much smaller and lighter than a conventional laboratory microscope. The system is controlled by a Raspberry Pi, and the sample is lit by a single LED, allowing the system to be powered by 5V USB or even by battery rather than requiring mains power, making it ideal for field use or low-resource settings.
3.2. Establishing if System Has Sufficient Resolution to Observe Bacteria for Motility Assays
There is a trade-off between cost of instrument, field of view and resolution. Biological samples often use phase contrast microscopy or differential interference contrast (DIC) microscopy, allowing unstained samples and cellular structures to be more visible. The OpenFlexure lack these modifications, and the sample is lit simply from above using a single white LED. This provides limited contrast for cellular samples, yet the OpenFlexure microscope was originally developed for water quality testing [
12].
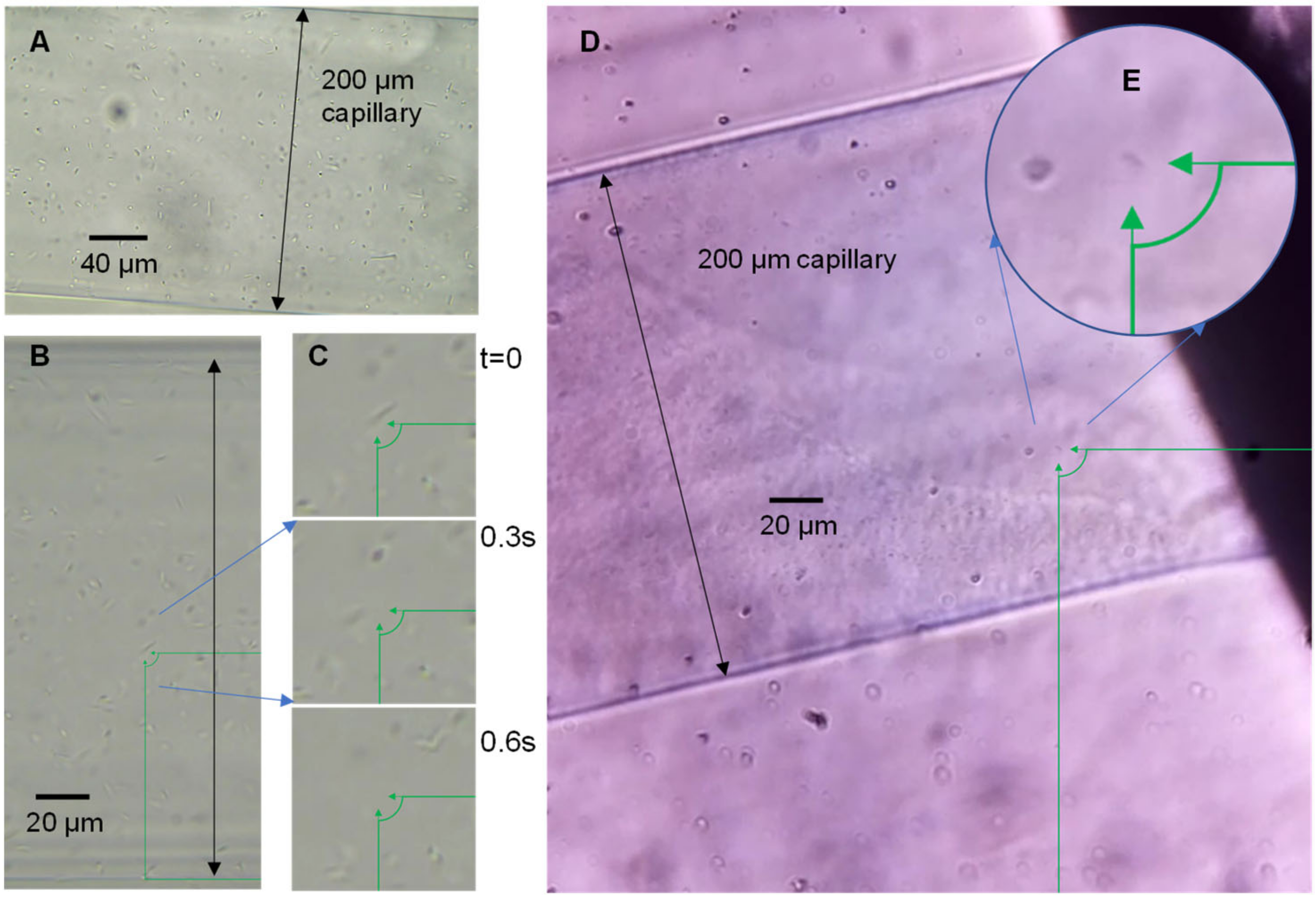
Imaging motility does not necessarily require high resolution or clear structural images; however, to fully observe sample motility and migration into the capillaries, the entire capillary opening needs to be observed. To identify if single bacteria could be imaged using the OpenFlexure microscope in the microcapillaries, a sample of
P. aeruginosa was loaded into the devices and imaged on a conventional laboratory microscope equipped with a digital camera, and these were compared with the OpenFlexure system (
Figure 2). At magnification in which the full diameter of the 200 µm diameter capillary is in view, individual
P. aeruginosa are clearly visible, and the motile rod-shaped bacteria move quickly in the capillary (
Figure 2C). Images and videos collected by the OpenFlexure microscope also show clear individual bacteria (
Figure 2D), and both systems can identify individual bacteria with the full capillary diameter in view. We conclude that the OpenFlexure microscope can be used to image single bacterial cells and screen motility in unstained samples within microfluidic devices.
The focal plane of the microscope is smaller than the diameter of the microcapillaries, and the fixed focal length of the raspberry pi camera means the system has to be adjusted correctly to ensure that motility of bacteria inside the capillaries are observed. The system was focused on the widest part of the capillary, but this can mean that bacteria move in and out of focus as they swim above and below the focal plane. The system may be improved using smaller diameter capillaries or devices with flatter channels, or with robotic z-stage added to the OpenFlexure microscope that can scan and image the full depth of the microdevice.
3.3. Confirming Antibiotic Susceptibility Can Be Tested by Conventional Motility Assays
Motility assays are typically used to measure motility, and in contrast, different standardised antibiotic susceptibility testing (AST) methods, such as disc diffusion, broth microdilution or agar dilution, are used to measure antibiotic susceptibility [
22]. We therefore checked whether motility measurement assays could be adapted to perform AST by combining conventional agar dilution AST with soft agar motility measurement. When checked using reference strains of
E. coli and
P. aeruginosa, this combination of conventional method allowed us to observe motility macroscopically with a range of antibiotic concentrations. Soft agar (0.4%) containing TTC and doubling dilutions of ceftazidime and gentamicin were poured into 96-well plate format microtitre stripwells. Cultures of
E. coli and
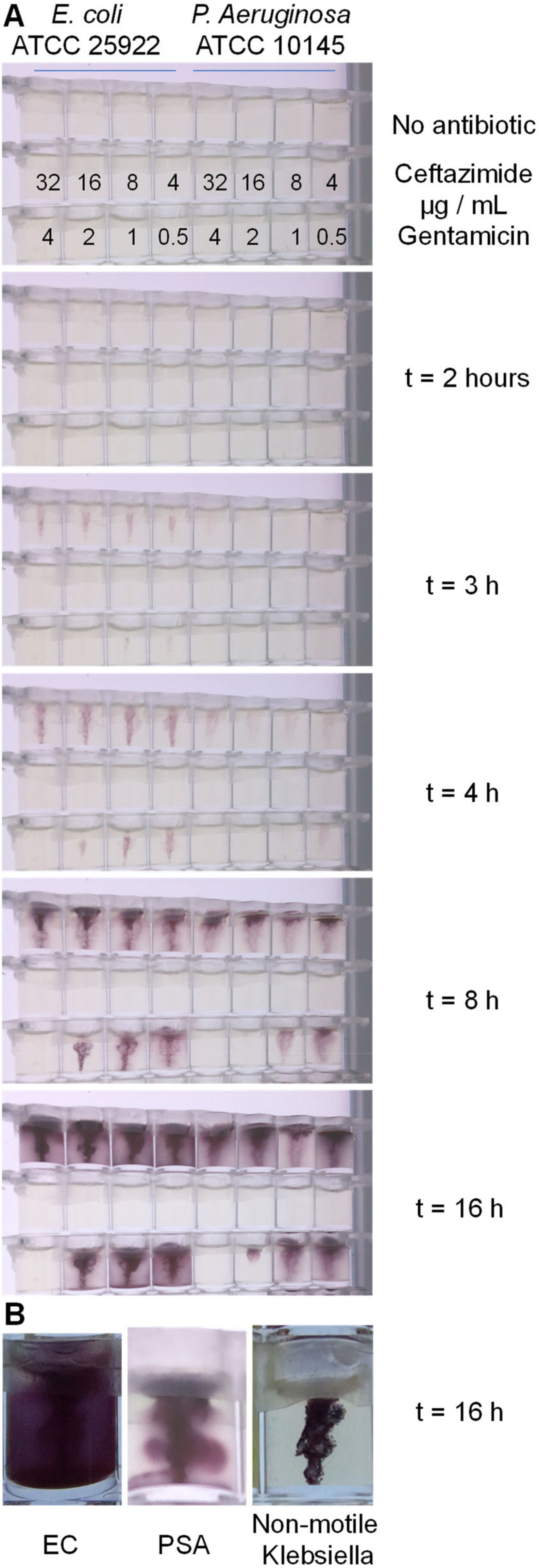
P. aeruginosa were stabbed into each well, and movement of bacteria away from the stab site was monitored during overnight incubation at 37 °C using Piramid, a time-lapse Raspberry Pi imaging box recently developed by our group [Long et al., HardwareX, accepted manuscript]. The MIC can be recorded as the lowest concentration of antibiotic that fully inhibits growth (
Figure 3). The stab culture can clearly identify motile versus non-motile bacteria; the motile bacteria were able to swim into the soft agar, and live bacteria convert TTC, forming a diffuse bloom of purple dye. The reduction of TTC forms an insoluble dye that does not diffuse in the soft agar, making bacterial movement clearly recordable with the digital camera. In contrast, non-motile bacteria, such as
K. pneumoniae, were still able to grow and convert the TTC, but the colour change is strictly limited to the original stab location (
Figure 3B). The motile
E. coli and
P. aeruginosa isolates both showed clear spread without antibiotic, which was inhibited by antibiotics at expected concentrations. We observed an MIC of 4 µg/mL for gentamicin and <4 µg/mL for ceftazidime for both organisms. We conclude that conventional macroscopic methods demonstrate that a bacterial motility readout can be used to quantify antibiotic susceptibility.
3.4. OpenFlexure Microscope Plus Microcapillary Devices Allow Rapid Direct Microfluidic Bacterial Cytometry to Measure Antibiotics Inhibiting Motility
The feasibility of motility-based AST was then assessed in the low-cost, portable combined microfluidic cytometry system, with bacterial movement recorded directly at the inlet of microcapillaries using the OpenFlexure digital microscope. Results were recorded for two bacteria and two antibiotics at concentrations where motility was known to be inhibited in the soft agar-filled capillaries. Videos of 6 min duration were recorded using a frame-rate of 15 frames per second, which gave the best compromise between image resolution and stable video capture by the Raspberry Pi computer, resulting in movies that run at double speed of real-time microbial (
Supplementary Materials listed in
Table 1). The video files clearly show motile bacteria are able to migrate from the end along the capillary length in agar without antibiotics present (PSA no antibiotic) and that the distance travelled is affected by antibiotic.
The results of the microcapillary motility follow the same trends as the stab culture, although as is commonly a challenge for AST, the exact MIC is less clear cut and can be challenging to define. One aspect of this is due to timing. Motility was assessed after just 3 min, compared to 16 h under standard AST conditions. Furthermore, the test involved the bacteria moving into the soft agar capillaries to be exposed to the antibiotics; while motile bacteria can clearly migrate into soft agar, this process is likely to be slower than motility in liquid due to increased viscosity. One adjustment that could be made to this system would be to use liquid media cultures in capillaries containing dried antibiotics; this would expose the bacteria much faster to the antibiotic of interest. The disadvantage of such a liquid system is the risk of rapid mixing or flow of antibiotic solution out of the microcapillary, leading to less confidence in the antibiotic concentration. Another disadvantage of the agar-loaded antibiotic test strips is the limited stability of antibiotic in solution. Our test strips are unlikely to have long shelf-life for many antibiotics. However, the results presented here with this soft agar method clearly demonstrate the concept of low-cost compact 3D-printed microscope combined with melt-extruded microfluidic device
To measure motility in the 200 µm microcapillary film, individual bacteria need to be identifiable, as proven to be possible using the OpenFlexure microscope configured with an approximately 400–500 micron field of view. However, this is not possible with lower magnification that allows monitoring all ten capillaries at once, as the resolution is no longer enough to identify individual bacteria, so capillaries have to be monitored individually, and the stage moved to monitor the next capillary. This makes the system more involved to use, but this compromise between resolution and field of view is also true of conventional microscopes.
4. Conclusions
The OpenFlexure microscope provides a low-cost, portable microscopy that combined with increasingly more affordable portable devices, such as microfluidic systems, can allow field use cytometry, for example as near-patient diagnostics, in this example illustrated by rapid antibiotic resistance detection. The microscope has sufficient resolution to identify individual bacteria under simple single-LED bright field illumination, and video recording can readily capture the motility of unstained bacteria in the microcapillaries used in this study. The increase in availability of microelectronics such as low-cost but high-quality optoelectronics combined with microfluidics, will drive the development of new portable diagnostic devices, improving healthcare needs in especially in rural or low-resource settings.