A Microfluidic Approach for Probing Heterogeneity in Cytotoxic T-Cells by Cell Pairing in Hydrogel Droplets
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Isolation and Preparation
2.2. Microfluidic Device Fabrication
2.3. Production of Temperature Regulation Device
2.4. Droplet Production, Cell Encapsulation and CTL Stimulation
2.5. Droplet Characterization
2.6. Cell/Microgel Retrieval and Flow Cytometric Measurement
2.7. Statistical Analysis
3. Results
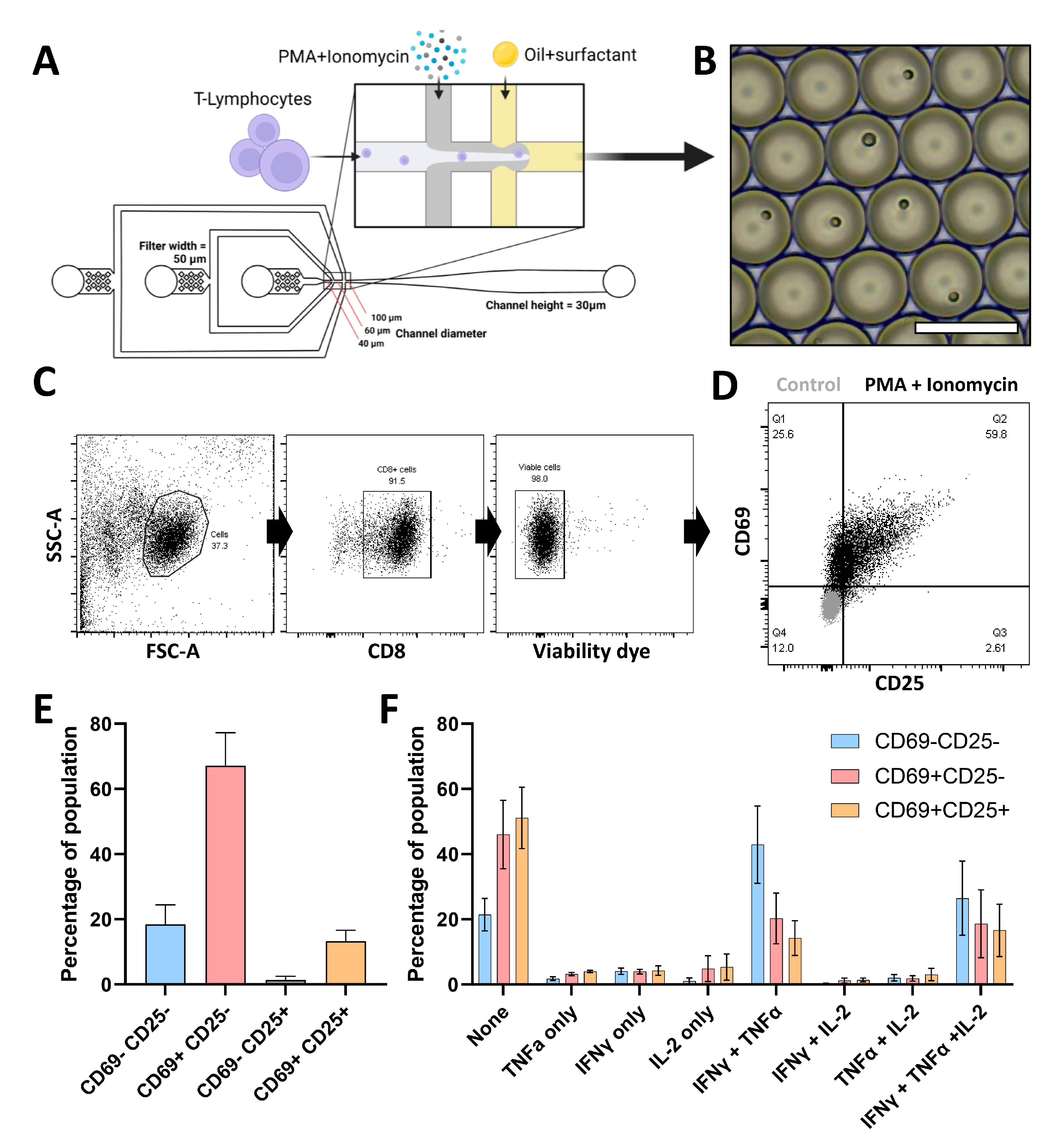
3.1. Single CTL Activation in Droplets Reveals Highly Heterogeneous Responses
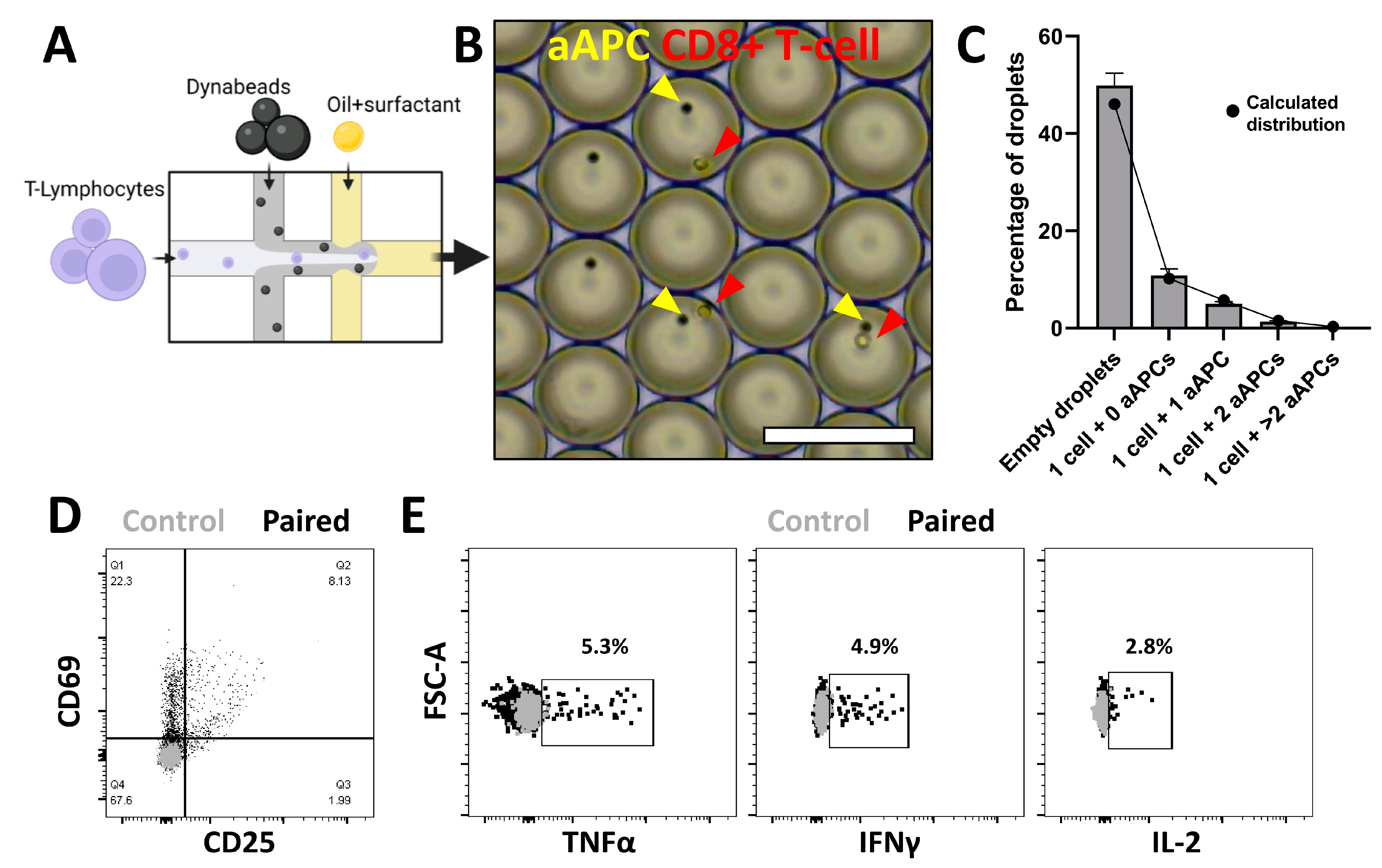
3.2. Spatial Pairing Data Are Lost in Aqueous Droplets upon CTL and aAPC Interactions
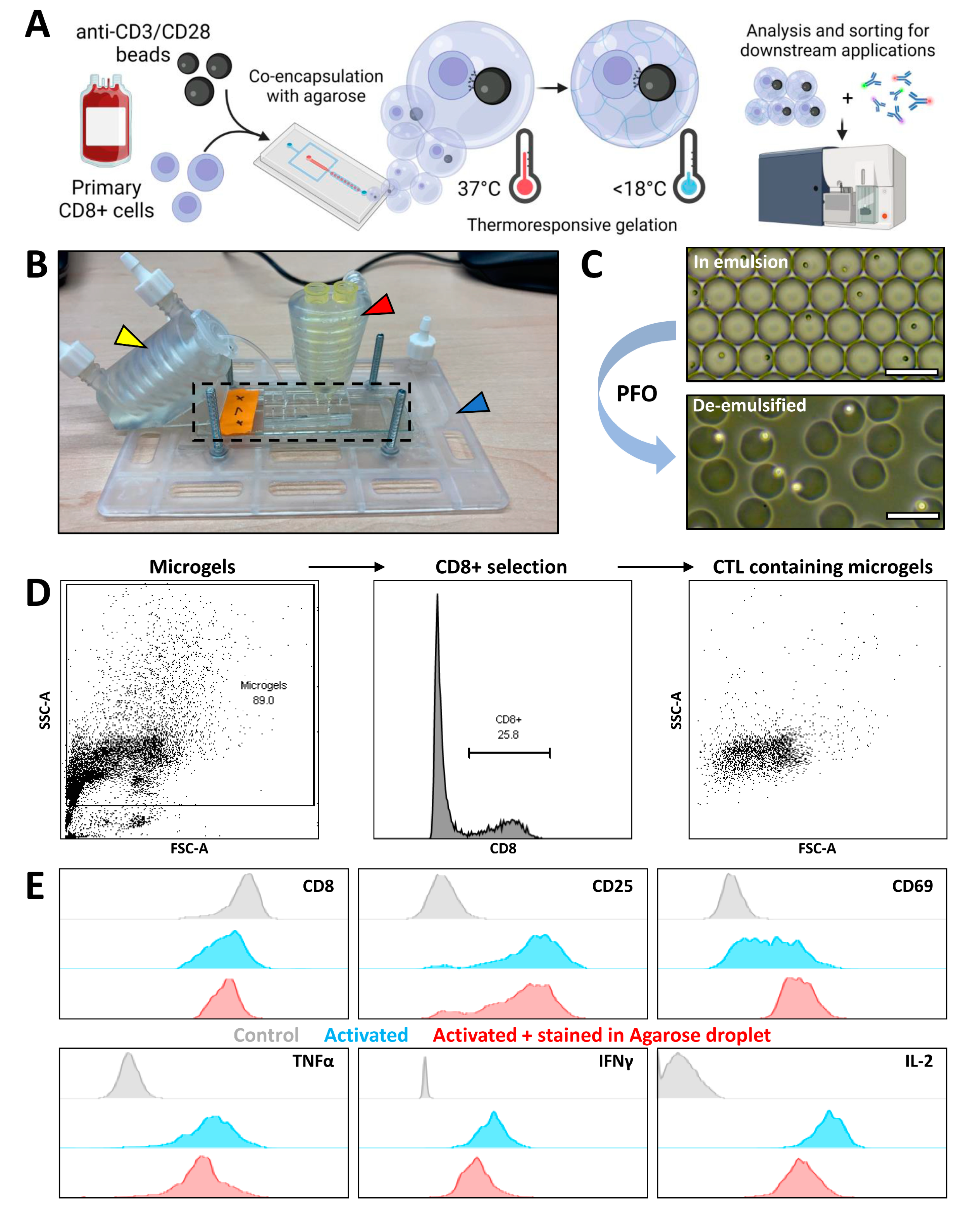
3.3. Agarose Microgels as Bioreactors for Subsequent Flow Cytometry Measurement
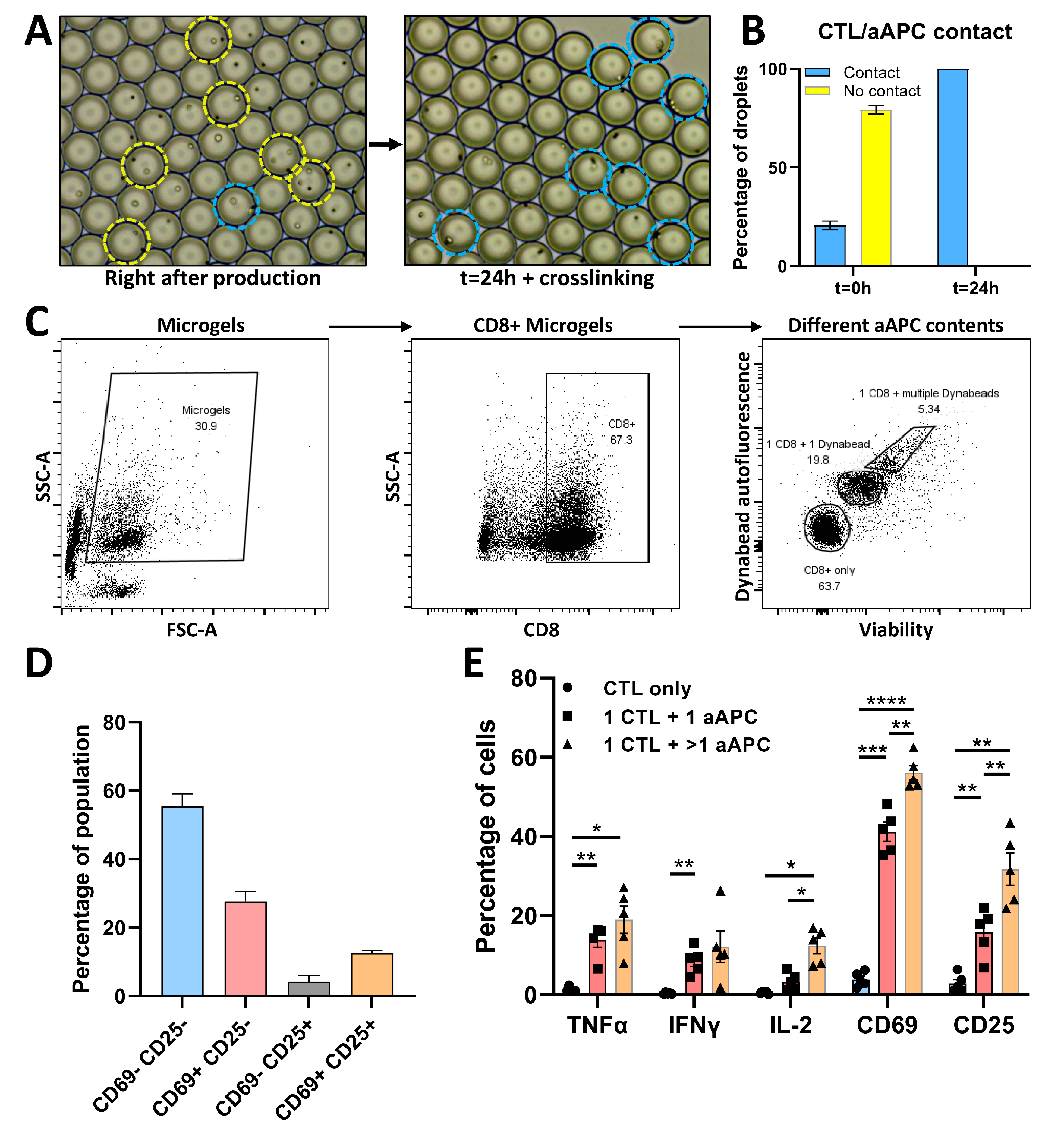
3.4. Phenotypic and Functional Analysis of CTL/aAPC Pairs in Microgels
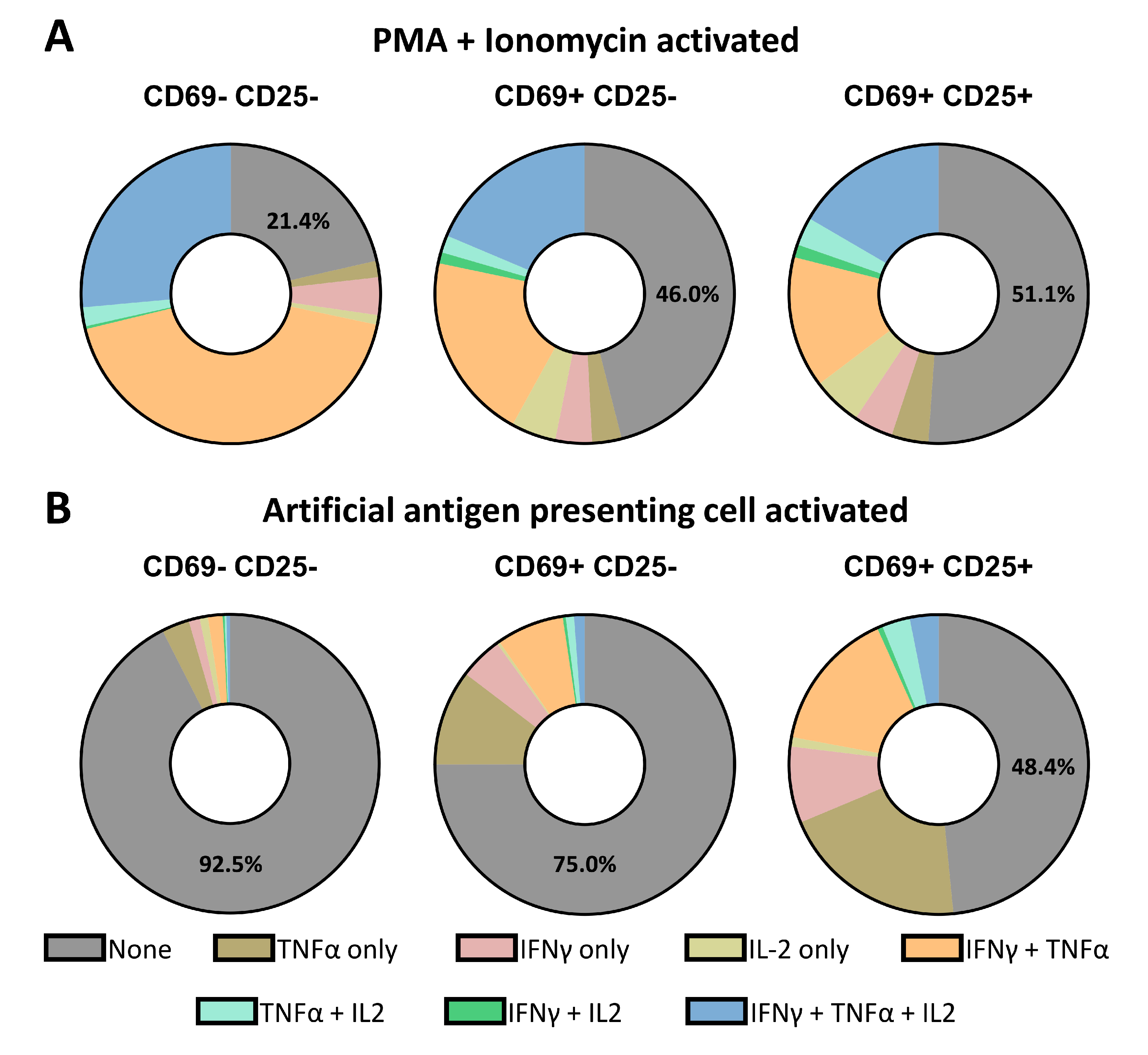
3.5. Single-Cell Decoding of CTL Activation and Secretion Based on Different Stimuli
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Betts, M.R.; Nason, M.C.; West, S.M.; De Rosa, S.C.; Migueles, S.A.; Abraham, J.; Lederman, M.M.; Benito, J.M.; Goepfert, P.A.; Connors, M.; et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 2006, 107, 4781–4789. [Google Scholar] [CrossRef] [PubMed]
- Freel, S.A.; Lamoreaux, L.; Chattopadhyay, P.K.; Saunders, K.; Zarkowsky, D.; Overman, R.G.; Ochsenbauer, C.; Edmonds, T.G.; Kappes, J.C.; Cunningham, C.K.; et al. Phenotypic and Functional Profile of HIV-Inhibitory CD8 T Cells Elicited by Natural Infection and Heterologous Prime/Boost Vaccination. J. Virol. 2010, 84, 4998–5006. [Google Scholar] [CrossRef] [PubMed]
- Zimmerli, S.C.; Harari, A.; Cellerai, C.; Vallelian, F.; Bart, P.-A.; Pantaleo, G. HIV-1-specific IFN-γ/IL-2-secreting CD8 T cells support CD4-independent proliferation of HIV-1-specific CD8 T cells. Proc. Natl. Acad. Sci. USA 2005, 102, 7239–7244. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.S.; Ohashi, P.S. The Roles of CD8+ T Cell Subsets in Antitumor Immunity. Trends Cell Biol. 2020, 30, 695–704. [Google Scholar] [CrossRef]
- Woodland, D.L.; Dutton, R.W. Heterogeneity of CD4+ and CD8+ T cells. Curr. Opin. Immunol. 2003, 15, 336–342. [Google Scholar] [CrossRef]
- Chattopadhyay, P.; Gierahn, T.M.; Roederer, M.; Love, J.C. Single-cell technologies for monitoring immune systems. Nat. Immunol. 2014, 15, 128–135. [Google Scholar] [CrossRef]
- Shalek, A.K.; Satija, R.; Shuga, J.; Trombetta, J.J.; Gennert, D.; Lu, D.; Chen, P.; Gertner, R.S.; Gaublomme, J.T.; Yosef, N.; et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 2014, 510, 363–369. [Google Scholar] [CrossRef]
- Satija, R.; Shalek, A.K. Heterogeneity in immune responses: From populations to single cells. Trends Immunol. 2014, 35, 219–229. [Google Scholar] [CrossRef]
- Altschuler, S.J.; Wu, L.F. Cellular Heterogeneity: Do Differences Make a Difference? Cell 2010, 141, 559–563. [Google Scholar] [CrossRef]
- Villani, A.C.; Satija, R.; Reynolds, G.; Sarkizova, S.; Shekhar, K.; Fletcher, J.; Griesbeck, M.; Butler, A.; Zheng, S.; Lazo, S.; et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017, 356, eaah4573. [Google Scholar] [CrossRef]
- Luo, X.; Chen, J.-Y.; Ataei, M.; Lee, A. Microfluidic Compartmentalization Platforms for Single Cell Analysis. Biosensors 2022, 12, 58. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shao, N.; de Castro, R.B.; Zhang, P.; Ma, Y.; Liu, X.; Huang, F.; Wang, R.-F.; Qin, L. Evaluation of Single-Cell Cytokine Secretion and Cell-Cell Interactions with a Hierarchical Loading Microwell Chip. Cell Rep. 2020, 31, 107574. [Google Scholar] [CrossRef] [PubMed]
- Chokkalingam, V.; Tel, J.; Wimmers, F.; Liu, X.; Semenov, S.; Thiele, J.; Figdor, C.G.; Huck, W.T.S. Probing cellular heterogeneity in cytokine-secreting immune cells using droplet-based microfluidics. Lab Chip 2013, 13, 4740–4744. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Liu, X.; Li, P.; Liu, F.; Kojima, M.; Huang, Q.; Arai, T. On-Chip Cell-Cell Interaction Monitoring at Single-Cell Level by Efficient Immobilization of Multiple Cells in Adjustable Quantities. Anal. Chem. 2020, 92, 11607–11616. [Google Scholar] [CrossRef] [PubMed]
- Kemna, E.W.M.; Schoeman, R.M.; Wolbers, F.; Vermes, I.; Weitz, D.A.; Van Den Berg, A. High-yield cell ordering and deterministic cell-in-droplet encapsulation using Dean flow in a curved microchannel. Lab Chip 2012, 12, 2881–2887. [Google Scholar] [CrossRef]
- Lagus, T.P.; Edd, J.F. High-throughput co-encapsulation of self-ordered cell trains: Cell pair interactions in microdroplets. RSC Adv. 2013, 3, 20512–20522. [Google Scholar] [CrossRef]
- Sinha, N.; Subedi, N.; Tel, J. Integrating Immunology and Microfluidics for Single Immune Cell Analysis. Front. Immunol. 2018, 9, 2373. [Google Scholar] [CrossRef]
- Shembekar, N.; Chaipan, C.; Utharala, R.; Merten, C.A. Droplet-based microfluidics in drug discovery, transcriptomics and high-throughput molecular genetics. Lab Chip 2016, 16, 1314–1331. [Google Scholar] [CrossRef]
- Segaliny, A.I.; Li, G.; Kong, L.; Ren, C.; Chen, X.; Wang, J.K.; Baltimore, D.; Wu, G.; Zhao, W. Functional TCR T cell screening using single-cell droplet microfluidics. Lab Chip 2018, 18, 3733–3749. [Google Scholar] [CrossRef]
- Sarkar, S.; Sabhachandani, P.; Stroopinsky, D.; Palmer, K.; Cohen, N.; Rosenblatt, J.; Avigan, D.; Konry, T. Dynamic analysis of immune and cancer cell interactions at single cell level in microfluidic droplets. Biomicrofluidics 2016, 10, 054115. [Google Scholar] [CrossRef]
- Tiemeijer, B.M.; Tel, J. Hydrogels for Single-Cell Microgel Production: Recent Advances and Applications. Front. Bioeng. Biotechnol. 2022, 10, 891461. [Google Scholar] [CrossRef] [PubMed]
- Rossi, J.; Paczkowski, P.; Shen, Y.-W.; Morse, K.; Flynn, B.; Kaiser, A.; Ng, C.; Gallatin, K.; Cain, T.; Fan, R.; et al. Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells are associated with clinical outcomes in NHL. Blood 2018, 132, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Wimmers, F.; Aarntzen, E.H.J.G.; Duiveman-Deboer, T.; Figdor, C.G.; Jacobs, J.F.M.; Tel, J.; de Vries, I.J.M. Long-lasting multifunctional CD8+T cell responses in end-stage melanoma patients can be induced by dendritic cell vaccination. Oncoimmunology 2015, 5, e1067745. [Google Scholar] [CrossRef] [PubMed]
- Sinha, N.; Subedi, N.; Wimmers, F.; Soennichsen, M.; Tel, J. A Pipette-Tip Based Method for Seeding Cells to Droplet Microfluidic Platforms. J. Vis. Exp. 2019, 144, e57848. [Google Scholar] [CrossRef] [PubMed]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; Dezonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Tiemeijer, B.M.; Sweep, M.W.D.; Sleeboom, J.J.F.; Steps, K.J.; van Sprang, J.F.; De Almeida, P.; Hammink, R.; Kouwer, P.H.J.; Smits, A.I.P.M.; Tel, J. Probing Single-Cell Macrophage Polarization and Heterogeneity Using Thermo-Reversible Hydrogels in Droplet-Based Microfluidics. Front. Bioeng. Biotechnol. 2021, 9, 715408. [Google Scholar] [CrossRef]
- Wimmers, F.; Subedi, N.; Van Buuringen, N.; Heister, D.; Vivié, J.; Beeren-Reinieren, I.; Woestenenk, R.; Dolstra, H.; Piruska, A.; Jacobs, J.F.M.; et al. Single-cell analysis reveals that stochasticity and paracrine signaling control interferon-alpha production by plasmacytoid dendritic cells. Nat. Commun. 2018, 9, 3317. [Google Scholar] [CrossRef]
- van Eyndhoven, L.C.; Chouri, E.; Subedi, N.; Tel, J. Phenotypical Diversification of Early IFNα-Producing Human Plasmacytoid Dendritic Cells Using Droplet-Based Microfluidics. Front. Immunol. 2021, 12, 1592. [Google Scholar] [CrossRef]
- Ai, W.; Li, H.; Song, N.; Li, L.; Chen, H. Optimal Method to Stimulate Cytokine Production and Its Use in Immunotoxicity Assessment. Int. J. Environ. Res. Public Health 2013, 10, 3834–3842. [Google Scholar] [CrossRef]
- Collins, D.J.; Neild, A.; Demello, A.; Liu, A.-Q.; Ai, Y. The Poisson distribution and beyond: Methods for microfluidic droplet production and single cell encapsulation. Lab Chip 2015, 15, 3439–3459. [Google Scholar] [CrossRef]
- Cambiaggi, C.; Scupoli, M.T.; Cestari, T.; Gerosa, F.; Carra, G.; Tridente, G.; Accolla, R.S. Constitutive Expression of CD69 in Interspecies T-Cell Hybrids and Locus Assignment to Human Chromosome 12. Immunogenetics 1992, 36, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Kmieciak, M.; Gowda, M.; Graham, L.; Godder, K.; Bear, H.D.; Marincola, F.M.; Manjili, M.H. Human T cells express CD25 and Foxp3 upon activation and exhibit effector/memory phenotypes without any regulatory/suppressor function. J. Transl. Med. 2009, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Bakdash, G.; Sittig, S.P.; van Dijk, T.; Figdor, C.G.; de Vries, I.J.M. The nature of activatory and tolerogenic dendritic cell-derived signal II. Front. Immunol. 2013, 4, 53. [Google Scholar] [CrossRef]
- Viola, A.; Lanzavecchia, A. T Cell Activation Determined by T Cell Receptor Number and Tunable Thresholds. Science 1996, 273, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Acuto, O.; Michel, F.M. CD28-mediated co-stimulation: A quantitative support for TCR signalling. Nat. Rev. Immunol. 2003, 3, 939–951. [Google Scholar] [CrossRef]
- Kaliński, P.; Hilkens, C.; Wierenga, E.A.; Kapsenberg, M.L. T-cell priming by type-1and type-2 polarized dendritic cells: The concept of a third signal. Immunol. Today 1999, 20, 561–567. [Google Scholar] [CrossRef]
- Dura, B.; Dougan, S.K.; Barisa, M.; Hoehl, M.M.; Lo, C.T.; Ploegh, H.L.; Voldman, J. Profiling lymphocyte interactions at the single-cell level by microfluidic cell pairing. Nat. Commun. 2015, 6, 5940. [Google Scholar] [CrossRef]
- Li, Y.; Kurlander, R.J. Comparison of anti-CD3 and anti-CD28-coated beads with soluble anti-CD3 for expanding human T cells: Differing impact on CD8 T cell phenotype and responsiveness to restimulation. J. Transl. Med. 2010, 8, 104. [Google Scholar] [CrossRef]
- Xiong, J.-Y.; Narayanan, J.; Liu, X.-Y.; Chong, T.K.; Chen, S.B.; Chung, T.-S. Topology Evolution and Gelation Mechanism of Agarose Gel. J. Phys. Chem. B 2005, 109, 5638–5643. [Google Scholar] [CrossRef]
- Narayanan, J.; Xiong, J.-Y.; Liu, X.-Y. Determination of agarose gel pore size: Absorbance measurements vis a vis other techniques. J. Physics: Conf. Ser. 2006, 28, 83–86. [Google Scholar] [CrossRef]
- Ritter, S.; Hiller, R.G.; Wrench, P.M.; Welte, W.; Diederichs, K. Crystal Structure of a Phycourobilin-Containing Phycoerythrin at 1.90-Å Resolution. J. Struct. Biol. 1999, 126, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Antona, S.; Platzman, I.; Spatz, J.P. Droplet-Based Cytotoxicity Assay: Implementation of Time-Efficient Screening of Antitumor Activity of Natural Killer Cells. ACS Omega 2020, 5, 24674–24683. [Google Scholar] [CrossRef] [PubMed]
- Subedi, N.; Van Eyndhoven, L.C.; Hokke, A.M.; Houben, L.; Van Turnhout, M.C.; Bouten, C.V.C.; Eyer, K.; Tel, J. An automated real-time microfluidic platform to probe single NK cell heterogeneity and cytotoxicity on-chip. Sci. Rep. 2021, 11, 17084. [Google Scholar] [CrossRef] [PubMed]
- Antona, S.; Abele, T.; Jahnke, K.; Dreher, Y.; Göpfrich, K.; Platzman, I.; Spatz, J.P. Droplet-Based Combinatorial Assay for Cell Cytotoxicity and Cytokine Release Evaluation. Adv. Funct. Mater. 2020, 30, 2003479. [Google Scholar] [CrossRef]
- Hondroulis, E.; Movila, A.; Sabhachandani, P.; Sarkar, S.; Cohen, N.; Kawai, T.; Konry, T. A Droplet-Merging Platform for Comparative Functional Analysis of M1 and M2 Macrophages in Response to E. coli-Induced Stimuli. Biotechnol. Bioeng. 2017, 114, 705–709. [Google Scholar] [CrossRef]
- Gérard, A.; Woolfe, A.; Mottet, G.; Reichen, M.; Castrillon, C.; Menrath, V.; Ellouze, S.; Poitou, A.; Doineau, R.; Briseno-Roa, L.; et al. High-throughput single-cell activity-based screening and sequencing of antibodies using droplet microfluidics. Nat. Biotechnol. 2020, 38, 715–721. [Google Scholar] [CrossRef]
- Madrigal, J.L.; Schoepp, N.G.; Xu, L.; Powell, C.S.; Delley, C.L.; Siltanen, C.A.; Danao, J.; Srinivasan, M.; Cole, R.H.; Abate, A.R. Characterizing cell interactions at scale with made-to-order droplet ensembles (MODEs). Proc. Natl. Acad. Sci. USA 2022, 119, e2110867119. [Google Scholar] [CrossRef]
- Sarkar, S. T Cell Dynamic Activation and Functional Analysis in Nanoliter Droplet Microarray. J. Clin. Cell Immunol. 2015, 6, 334. [Google Scholar] [CrossRef]
- Bounab, Y.; Eyer, K.; Dixneuf, S.; Rybczynska, M.; Chauvel, C.; Mistretta, M.; Tran, T.; Aymerich, N.; Chenon, G.; Llitjos, J.-F.; et al. Dynamic single-cell phenotyping of immune cells using the microfluidic platform DropMap. Nat. Protoc. 2020, 15, 2920–2955. [Google Scholar] [CrossRef]
- Xue, Q.; Bettini, E.; Paczkowski, P.; Qiong, X.; Kaiser, A.; McConnell, T.; Kodrasi, O.; Quigley, M.F.; Heath, J.; Fan, R.; et al. Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. J. Immunother. Cancer 2017, 5, 85. [Google Scholar] [CrossRef] [PubMed]
- Yanakieva, D.; Elter, A.; Bratsch, J.; Friedrich, K.; Becker, S.; Kolmar, H. FACS-Based Functional Protein Screening via Microfluidic Co-encapsulation of Yeast Secretor and Mammalian Reporter Cells. Sci. Rep. 2020, 10, 10182. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Chu, T.H.; Ackerman, M.E.; Griswold, K.E. Going native: Direct high throughput screening of secreted full-length IgG antibodies against cell membrane proteins. mAbs 2017, 9, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jenkins, G.; Zou, Y.; Zhu, Z.; Yang, C.J. Massively Parallel Single-Molecule and Single-Cell Emulsion Reverse Transcription Polymerase Chain Reaction Using Agarose Droplet Microfluidics. Anal. Chem. 2012, 84, 3599–3606. [Google Scholar] [CrossRef]
- Li, M.; Liu, H.; Zhuang, S.; Goda, K. Droplet flow cytometry for single-cell analysis. RSC Adv. 2021, 11, 20944–20960. [Google Scholar] [CrossRef]
- Bai, Y.; Weibull, E.; Joensson, H.N.; Andersson-Svahn, H. Interfacing picoliter droplet microfluidics with addressable microliter compartments using fluorescence activated cell sorting. Sens. Actuators B Chem. 2014, 194, 249–254. [Google Scholar] [CrossRef]
- White, A.M.; Zhang, Y.; Shamul, J.G.; Xu, J.; Kwizera, E.A.; Jiang, B.; He, X. Deep Learning-Enabled Label-Free On-Chip Detection and Selective Extraction of Cell Aggregate-Laden Hydrogel Microcapsules. Small 2021, 17, 2100491. [Google Scholar] [CrossRef]
- Fu, X.T.; Kim, S.M. Agarase: Review of Major Sources, Categories, Purification Method, Enzyme Characteristics and Applications. Mar. Drugs 2010, 8, 200–218. [Google Scholar] [CrossRef]
- Ahmed, H.; Stokke, B.T. Fabrication of monodisperse alginate microgel beads by microfluidic picoinjection: A chelate free approach. Lab Chip 2021, 21, 2232–2243. [Google Scholar] [CrossRef]
- Mao, A.S.; Shin, J.-W.; Utech, S.; Wang, H.; Uzun, O.; Li, W.; Cooper, M.; Hu, E.; Zhang, L.; Weitz, D.A.; et al. Deterministic encapsulation of single cells in thin tunable microgels for niche modelling and therapeutic delivery. Nat. Mater. 2016, 16, 236–243. [Google Scholar] [CrossRef]
- Håti, A.G.; Bassett, D.C.; Ribe, J.M.; Sikorski, P.; Weitz, D.A.; Stokke, B.T. Versatile, cell and chip friendly method to gel alginate in microfluidic devices. Lab Chip 2016, 16, 3718–3727. [Google Scholar] [CrossRef] [PubMed]
- Dolega, M.E.; Abeille, F.; Picollet-D’Hahan, N.; Gidrol, X. Controlled 3D culture in Matrigel microbeads to analyze clonal acinar development. Biomaterials 2015, 52, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.; Zhang, W.; Wang, C.; Cui, L.; Yang, C.J. Agarose droplet microfluidics for highly parallel and efficient single molecule emulsion PCR. Lab Chip 2010, 10, 2841–2843. [Google Scholar] [CrossRef] [PubMed]
- Novak, R.; Zeng, Y.; Shuga, J.; Venugopalan, G.; Fletcher, D.A.; Smith, M.T.; Mathies, R.A. Single-Cell Multiplex Gene Detection and Sequencing with Microfluidically Generated Agarose Emulsions. Angew. Chem. Int. Ed. 2010, 50, 390–395. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, W.; Leng, X.; Zhang, M.; Guan, Z.; Lu, J.; Yang, C.J. Highly sensitive and quantitative detection of rare pathogens through agarose droplet microfluidic emulsion PCR at the single-cell level. Lab Chip 2012, 12, 3907–3913. [Google Scholar] [CrossRef]
- Huang, H.; Wang, C.; Rubelt, F.; Scriba, T.J.; Davis, M.M. Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat. Biotechnol. 2020, 38, 1194–1202. [Google Scholar] [CrossRef]
- Chiou, S.-H.; Tseng, D.; Reuben, A.; Mallajosyula, V.; Molina, I.S.; Conley, S.; Wilhelmy, J.; McSween, A.M.; Yang, X.; Nishimiya, D.; et al. Global analysis of shared T cell specificities in human non-small cell lung cancer enables HLA inference and antigen discovery. Immunity 2021, 54, 586–602.e8. [Google Scholar] [CrossRef]
- Chatila, T.; Silverman, L.; Miller, R.; Geha, R. Mechanisms of T cell activation by the calcium ionophore ionomycin. J. Immunol. 1989, 143, 1283–1289. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiemeijer, B.M.; Descamps, L.; Hulleman, J.; Sleeboom, J.J.F.; Tel, J. A Microfluidic Approach for Probing Heterogeneity in Cytotoxic T-Cells by Cell Pairing in Hydrogel Droplets. Micromachines 2022, 13, 1910. https://doi.org/10.3390/mi13111910
Tiemeijer BM, Descamps L, Hulleman J, Sleeboom JJF, Tel J. A Microfluidic Approach for Probing Heterogeneity in Cytotoxic T-Cells by Cell Pairing in Hydrogel Droplets. Micromachines. 2022; 13(11):1910. https://doi.org/10.3390/mi13111910
Chicago/Turabian StyleTiemeijer, Bart M., Lucie Descamps, Jesse Hulleman, Jelle J. F. Sleeboom, and Jurjen Tel. 2022. "A Microfluidic Approach for Probing Heterogeneity in Cytotoxic T-Cells by Cell Pairing in Hydrogel Droplets" Micromachines 13, no. 11: 1910. https://doi.org/10.3390/mi13111910
APA StyleTiemeijer, B. M., Descamps, L., Hulleman, J., Sleeboom, J. J. F., & Tel, J. (2022). A Microfluidic Approach for Probing Heterogeneity in Cytotoxic T-Cells by Cell Pairing in Hydrogel Droplets. Micromachines, 13(11), 1910. https://doi.org/10.3390/mi13111910





