Identifying Candidate Biomarkers of Ionizing Radiation in Human Pulmonary Microvascular Lumens Using Microfluidics—A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
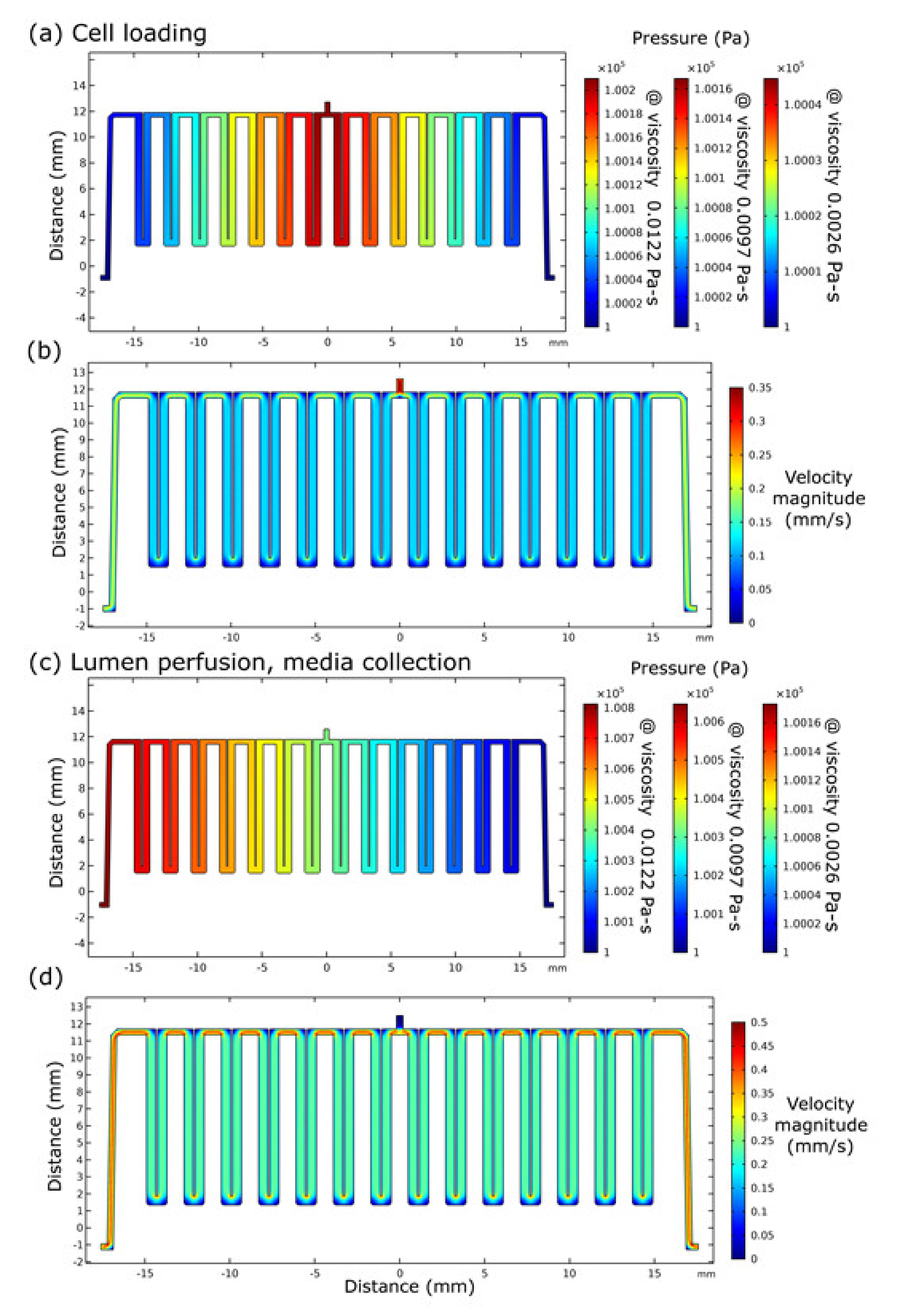
2.1. Microfluidics
2.2. Cell Culture
2.3. Gamma Irradiation Treatment
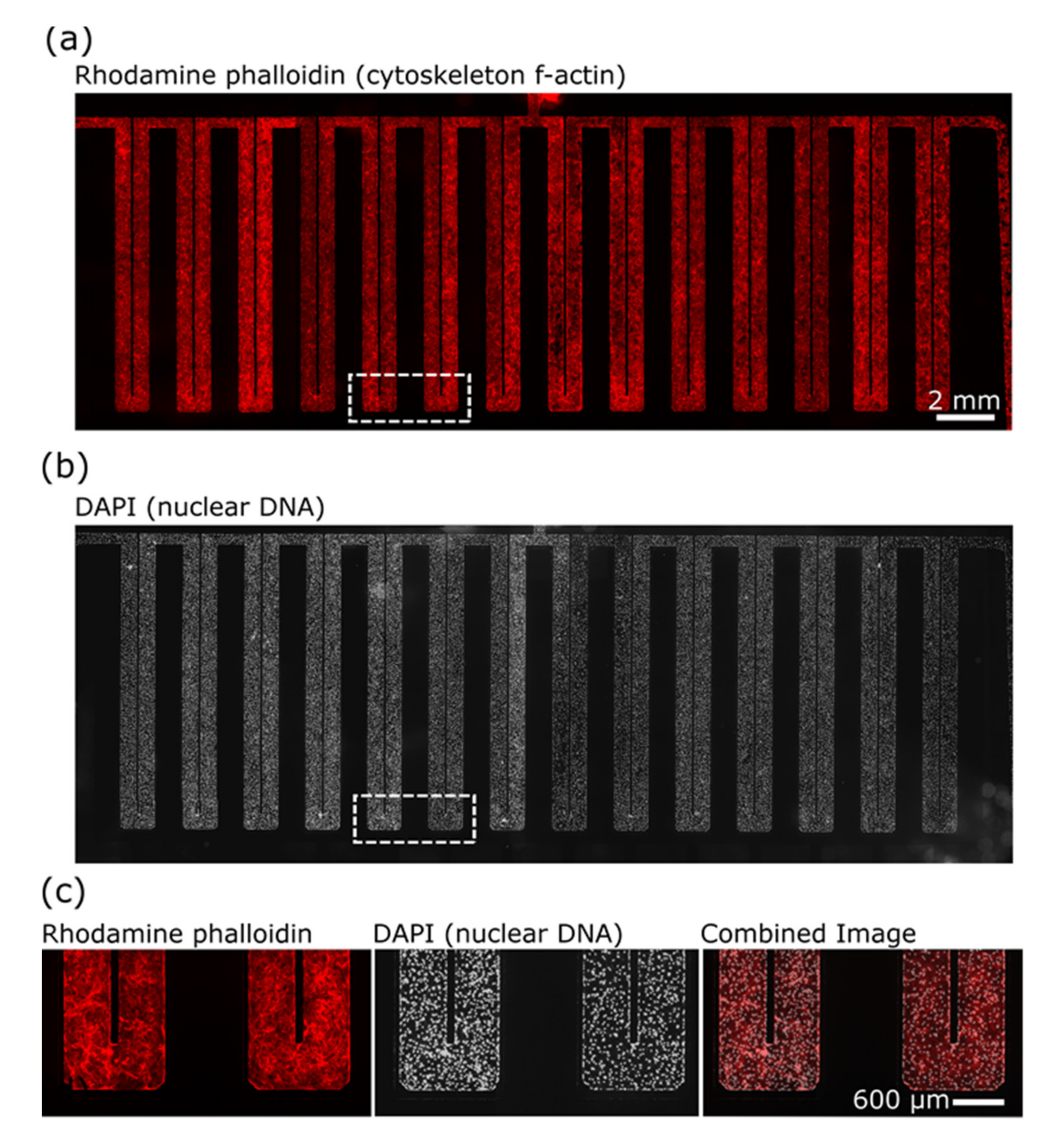
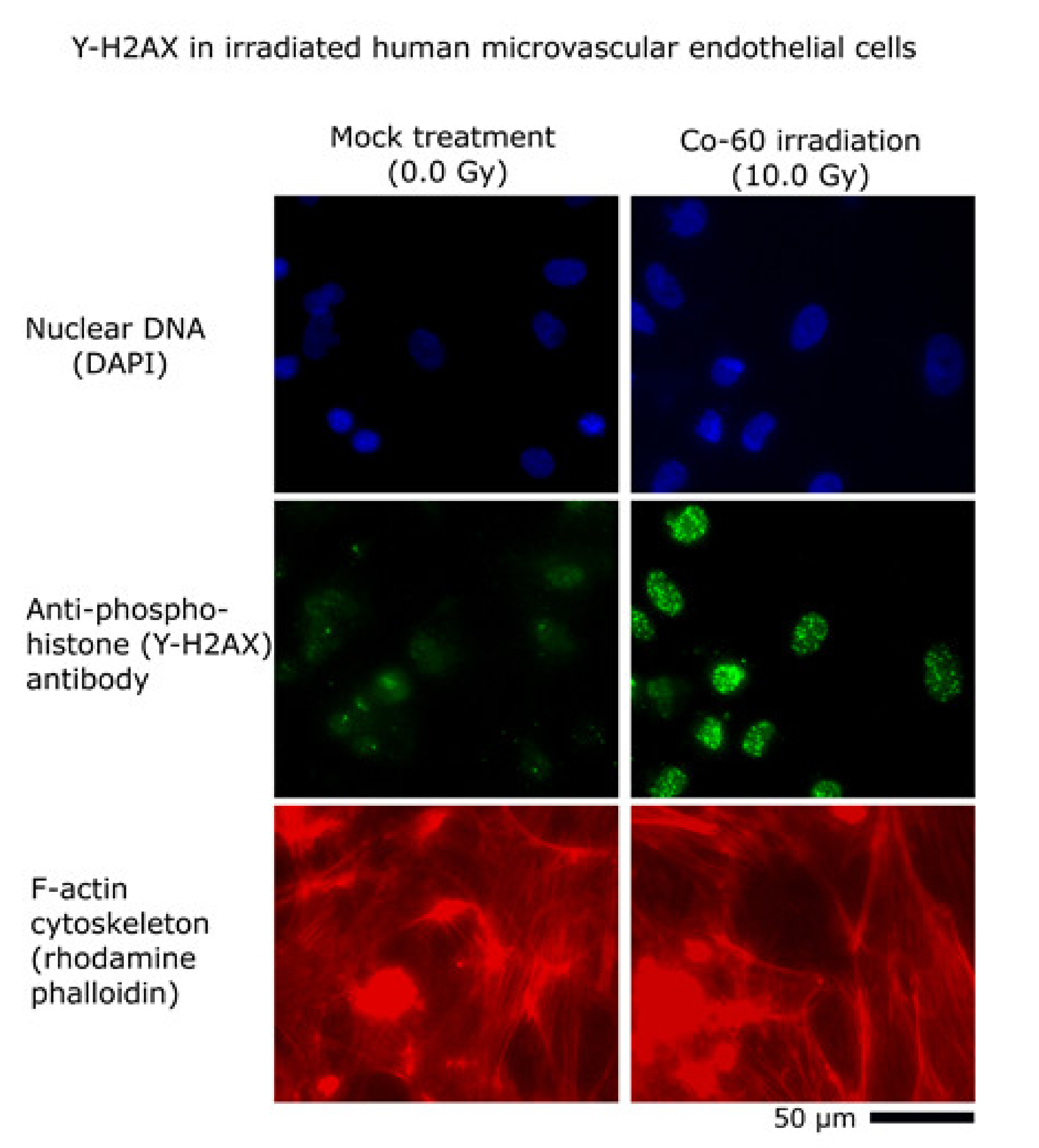
2.4. Immunocytochemistry
2.5. Mass Spectrometry
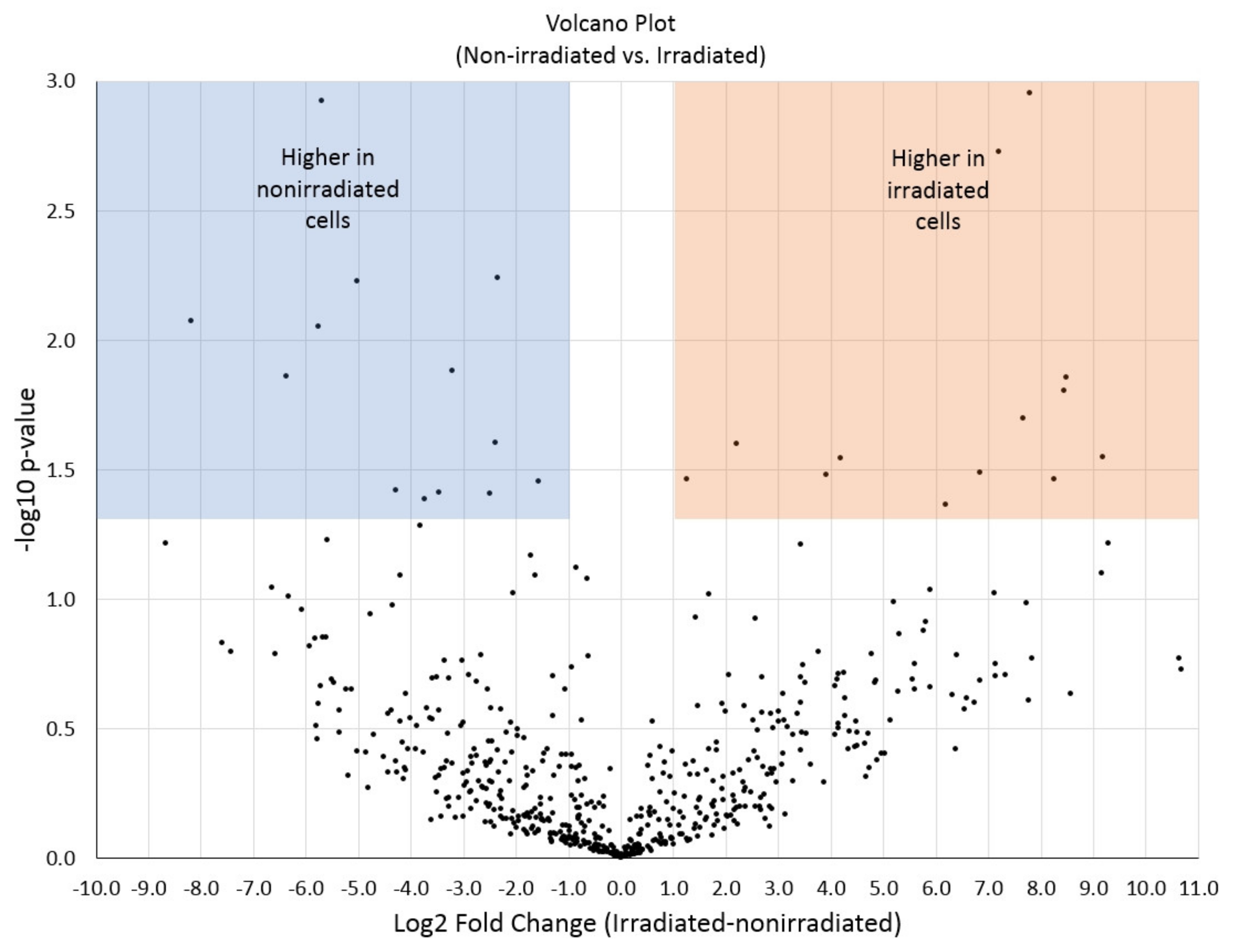
2.6. Data Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fliedner, T.M.; Dörr, H.D.; Meineke, V. Multi-organ involvement as a pathogenetic principle of the radiation syndromes: A study involving 110 case histories documented in SEARCH and classified as the bases of haematopoietic indicators of effect. Br. J. Radiol. 2005, 27, 1–8. [Google Scholar] [CrossRef]
- Akashi, M. Role of infection and bleeding in multiple organ involvement and failure. Br. J. Radiol. 2005, 27, 69–74. [Google Scholar] [CrossRef]
- Kiang, J.G.; Olabisi, A.O. Radiation: A poly-traumatic hit leading to multi-organ injury. Cell Biosci. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- MacVittie, T.J.; Farese, A.M.; Parker, G.A.; Jackson, W. The Time Course of Radiation-induced Lung Injury in a Nonhuman Primate Model of Partial-body Irradiation with Minimal Bone Marrow Sparing: Clinical and Radiographic Evidence and the Effect of Neupogen Administration. Health Phys. 2019, 116, 366–382. [Google Scholar] [CrossRef] [PubMed]
- Yannoutsos, A.; Levy, B.I.; Safar, M.E.; Slama, G.; Blacher, J. Pathophysiology of hypertension. J. Hypertens. 2014, 32, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Milliat, F.; Sabourin, J.-C.; Tarlet, G.; Holler, V.; Deutsch, E.; Buard, V.; Tamarat, R.; Atfi, A.; Benderitter, M.; François, A. Essential Role of Plasminogen Activator Inhibitor Type-1 in Radiation Enteropathy. Am. J. Pathol. 2008, 172, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Citrin, D.E.; Prasanna, P.G.S.; Walker, A.J.; Freeman, M.L.; Eke, I.; Barcellos-Hoff, M.H.; Arankalayil, M.J.; Cohen, E.P.; Wilkins, R.; Ahmed, M.M.; et al. Radiation-Induced Fibrosis: Mechanisms and Opportunities to Mitigate—Report of an NCI Workshop, 19 September 2016. Radiat. Res. 2017, 188, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Satyamitra, M.M.; Dicarlo, A.L.; Taliaferro, L. Understanding the Pathophysiology and Challenges of Development of Medical Countermeasures for Radiation-Induced Vascular/Endothelial Cell Injuries: Report of a NIAID Workshop, 20 August 2015. Radiat. Res. 2016, 186, 99–111. [Google Scholar] [CrossRef]
- Singh, V.K.; Newman, V.L.; Romaine, P.L.; Hauer-Jensen, M.; Pollard, H.B. Use of biomarkers for assessing radiation injury and efficacy of countermeasures. Expert Rev. Mol. Diagn. 2016, 16, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Vicente, E.; Vujaskovic, Z.; Jackson, I.L. A Systematic Review of Metabolomic and Lipidomic Candidates for Biomarkers in Radiation Injury. Metabolites 2020, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Shimura, T.; Yamaguchi, I.; Terada, H.; Okuda, K.; Svendsen, E.; Kunugita, N. Radiation occupational health interventions offered to radiation workers in response to the complex catastrophic disaster at the Fukushima Daiichi Nuclear Power Plant. J. Radiat. Res. 2015, 56, 413–421. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rafii, S.; Ginsberg, M.; Scandura, J.; Butler, J.M.; Ding, B.-S. Transplantation of Endothelial Cells to Mitigate Acute and Chronic Radiation Injury to Vital Organs. Radiat. Res. 2016, 186, 196–202. [Google Scholar] [CrossRef]
- Fish, B.L.; Gao, F.; Narayanan, J.; Bergom, C.; Jacobs, E.R.; Cohen, E.P.; Moulder, J.E.; Orschell, C.M.; Medhora, M. Combined Hydration and Antibiotics with Lisinopril to Mitigate Acute and Delayed High-dose Radiation Injuries to Multiple Organs. Health Phys. 2016, 111, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Nepper-Christensen, S.; Heslet, L.; Bay, C. Acute radiation syndrome (ARS)—Treatment of the reduced host defense. Int. J. Gen. Med. 2012, 5, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.D.; Nowak, M.; Bayles, A.V.; Prabhakarpandian, B.; Karande, P.; Lahann, J.; Helgeson, M.E.; Mitragotri, S. A microfluidic model of human brain (μHuB) for assessment of blood brain barrier. Bioeng. Transl. Med. 2019, 4, e10126. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef] [PubMed]
- McLean, I.C.; Schwerdtfeger, L.A.; Tobet, S.A.; Henry, C.S. Powering ex vivo tissue models in microfluidic systems. Lab Chip 2018, 18, 1399–1410. [Google Scholar] [CrossRef]
- Millet, L.; Gillette, M.U. New perspectives on neuronal development via microfluidic environments. Trends Neurosci. 2012, 35, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Patabadige, D.E.W.; Millet, L.; Aufrecht, J.; Shankles, P.G.; Standaert, R.F.; Retterer, S.T.; Doktycz, M.J. Label-free time- and space-resolved exometabolite sampling of growing plant roots through nanoporous interfaces. Sci. Rep. 2019, 9, 10272. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Luni, C.; Elvassore, N. Microfluidics for secretome analysis under enhanced endogenous signaling. Biochem. Biophys. Res. Commun. 2018, 497, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Pedde, R.D.; Li, H.; Borchers, C.H.; Akbari, M. Microfluidic-Mass Spectrometry Interfaces for Translational Proteomics. Trends Biotechnol. 2017, 35, 954–970. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yi, L.; Mukhitov, N.; Schrell, A.; Dhumpa, R.; Roper, M.G. Microfluidics-to-mass spectrometry: A review of coupling methods and applications. J. Chromatogr. A 2015, 1382, 98–116. [Google Scholar] [CrossRef]
- Tharakan, R.; Tao, D.; Ubaida-Mohien, C.; Dinglasan, R.R.; Graham, D.R. Integrated Microfluidic Chip and Online SCX Separation Allows Untargeted Nanoscale Metabolomic and Peptidomic Profiling. J. Proteome Res. 2015, 14, 1621–1626. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.; Reis, R.L.; Silva-Correia, J.; Oliveira, J.M. Microfluidics for Angiogenesis Research. In Advances in Experimental Medicine and Biology; Springer Science and Business Media LLC: Cham, Switzerland, 2020; Volume 1230, pp. 97–119. [Google Scholar]
- Haase, K.; Kamm, R.D. Advances in on-chip vascularization. Regen. Med. 2017, 12, 285–302. [Google Scholar] [CrossRef]
- Moses, S.R.; Adorno, J.J.; Palmer, A.F.; Song, J.W. Vessel-on-a-chip models for studying microvascular physiology, transport, and function in vitro. Am. J. Physiol. Physiol. 2020, 320, C92–C105. [Google Scholar] [CrossRef]
- Yang, F.; Cohen, R.N.; Brey, E.M. Optimization of Co-Culture Conditions for a Human Vascularized Adipose Tissue Model. Bioengineering 2020, 7, 114. [Google Scholar] [CrossRef]
- Park, T.-E.; Mustafaoglu, N.; Herland, A.; Hasselkus, R.; Mannix, R.; Fitzgerald, E.A.; Prantil-Baun, R.; Watters, A.; Henry, O.; Benz, M.; et al. Hypoxia-enhanced Blood-Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat. Commun. 2019, 10, 2621. [Google Scholar] [CrossRef]
- Whisler, J.A.; Chen, M.B.; Kamm, R.D. Control of Perfusable Microvascular Network Morphology Using a Multiculture Microfluidic System. Tissue Eng. Part C Methods 2014, 20, 543–552. [Google Scholar] [CrossRef]
- Song, H.G.; Lammers, A.; Sundaram, S.; Rubio, L.; Chen, A.X.; Li, L.; Eyckmans, J.; Bhatia, S.N.; Chen, C.S. Transient Support from Fibroblasts is Sufficient to Drive Functional Vascularization in Engineered Tissues. Adv. Funct. Mater. 2020, 30, 202003777. [Google Scholar] [CrossRef]
- Tefft, J.B.; Chen, C.S.; Eyckmans, J. Reconstituting the dynamics of endothelial cells and fibroblasts in wound closure. APL Bioeng. 2021, 5, 016102. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Q.; Pei, J. Microfluidic-Based 3D Engineered Microvascular Networks and Their Applications in Vascularized Microtumor Models. Micromachines 2018, 9, 493. [Google Scholar] [CrossRef]
- Nashimoto, Y.; Hayashi, T.; Kunita, I.; Nakamasu, A.; Torisawa, Y.-S.; Nakayama, M.; Takigawa-Imamura, H.; Kotera, H.; Nishiyama, K.; Miura, T.; et al. Integrating perfusable vascular networks with a three-dimensional tissue in a microfluidic device. Integr. Biol. 2017, 9, 506–518. [Google Scholar] [CrossRef]
- Tsuyama, N.; Mizuno, H.; Masujima, T. Molecular and functional analysis of cellular phenomena using single-cell mass spectrometry. Biol. Pharm. Bull. 2012, 35, 1425–1431. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abouleila, Y.; Onidani, K.; Ali, A.; Shoji, H.; Kawai, T.; Lim, C.T.; Kumar, V.; Okaya, S.; Kato, K.; Hiyama, E.; et al. Live single cell mass spectrometry reveals cancer-specific metabolic profiles of circulating tumor cells. Cancer Sci. 2018, 110, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Vertes, A. Single-Cell Mass Spectrometry Approaches to Explore Cellular Heterogeneity. Angew. Chem. Int. Ed. 2018, 57, 4466–4477. [Google Scholar] [CrossRef]
- Millet, L.J.; Lucheon, J.D.; Standaert, R.; Retterer, S.; Doktycz, M. Modular microfluidics for point-of-care protein purifications. Lab Chip 2015, 15, 1799–1811. [Google Scholar] [CrossRef]
- Standaert, R.F.; Giannone, R.J.; Michener, J.K. Identification of parallel and divergent optimization solutions for homologous metabolic enzymes. Metab. Eng. Commun. 2018, 6, 56–62. [Google Scholar] [CrossRef]
- Taverner, T.; Karpievitch, Y.V.; Polpitiya, A.D.; Brown, J.N.; Dabney, A.R.; Anderson, G.A.; Smith, R. DanteR: An extensible R-based tool for quantitative analysis of -omics data. Bioinformatics 2012, 28, 2404–2406. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Tyanova, S.; Cox, J. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. In Methods in Molecular Biology; Springer Science and Business Media LLC: Berlin, Germany, 2018; Volume 1711, pp. 133–148. [Google Scholar]
- Zhong, J.-J.; Seki, T.; Kinoshita, S.-I.; Yoshida, T. Rheological characteristics of cell suspension and cell culture ofPerilla frutescens. Biotechnol. Bioeng. 1992, 40, 1256–1262. [Google Scholar] [CrossRef]
- Kajiume, T.; Yuge, L.; Kawahara, Y.; Yoshimoto, R.; Sasaki, A.; Ide, T.; Asashima, M.; Kataoka, K.; Kobayashi, M. Floating culture promotes the maintenance of hematopoietic stem cells. FEBS Lett. 2007, 581, 4645–4650. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, D.; Birukov, K. Endothelial Cell Mechano-Metabolomic Coupling to Disease States in the Lung Microvasculature. Front. Bioeng. Biotechnol. 2019, 7, 172. [Google Scholar] [CrossRef] [PubMed]
- Lipowsky, H.H. Shear Stress in the Circulation. In Flow-Dependent Regulation of Vascular Function; Springer Science and Business Media LLC: Berlin, Germany, 1995; pp. 28–45. [Google Scholar]
- Paszkowiak, J.J.; Dardik, A.; Haven, N. Basic Science Review Arterial Wall Shear Stress: Observations from the Bench to the Bedside; Westminster Publications: Glen Head, NY, USA, 2003; Volume 37. [Google Scholar]
- Jarvis, M.R.; Arnold, M.; Ott, J.; Krishnan, V.; Pant, K.; Prabhakarpandian, B.; Mitragotri, S. Detachment of ligands from nanoparticle surface under flow and endothelial cell contact: Assessment using microfluidic devices. Bioeng. Transl. Med. 2018, 3, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mackay, S.; Gordon, D.M.; Anderson, J.; Haithcock, D.W.; Garson, C.J.; Tearney, G.J.; Solomon, G.M.; Pant, K.; Prabhakarpandian, B.; et al. Co-cultured microfluidic model of the airway optimized for microscopy and micro-optical coherence tomography imaging. Biomed. Opt. Express 2019, 10, 5414–5430. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Soroush, F.; Sheffield, J.B.; Wang, B.; Prabhakarpandian, B.; Kiani, M.F. A Biomimetic Microfluidic Tumor Microenvironment Platform Mimicking the EPR Effect for Rapid Screening of Drug Delivery Systems. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Naviaux, R.K.; Naviaux, J.C.; Li, K.; Bright, A.T.; Alaynick, W.A.; Wang, L.; Baxter, A.; Nathan, N.; Anderson, W.; Gordon, E. Metabolic features of chronic fatigue syndrome. Proc. Natl. Acad. Sci. USA 2016, 113, E5472–E5480. [Google Scholar] [CrossRef]
- Noble, W.S.; MacCoss, M.J. Computational and Statistical Analysis of Protein Mass Spectrometry Data. PLoS Comput. Biol. 2012, 8, e1002296. [Google Scholar] [CrossRef] [PubMed]
- Samiei, E.; Tabrizian, M.; Hoorfar, M. A review of digital microfluidics as portable platforms for lab-on a-chip applications. Lab Chip 2016, 16, 2376–2396. [Google Scholar] [CrossRef]
- Lacombe, J.; Phillips, S.L.; Zenhausern, F. Microfluidics as a new tool in radiation biology. Cancer Lett. 2016, 371, 292–300. [Google Scholar] [CrossRef]
- Croushore, C.A.; Supharoek, S.; Lee, C.Y.; Jakmunee, J.; Sweedler, J.V. Microfluidic Device for the Selective Chemical Stimulation of Neurons and Characterization of Peptide Release with Mass Spectrometry. Anal. Chem. 2012, 84, 9446–9452. [Google Scholar] [CrossRef]
- Tillmaand, E.G.; Sweedler, J.V. Integrating mass spectrometry with microphysiological systems for improved neurochemical studies. Microphysiol. Syst. 2018, 2. [Google Scholar] [CrossRef] [PubMed]
- Brengues, M.; Paap, B.; Bittner, M.; Amundson, S.; Seligmann, B.; Korn, R.; Lenigk, R.; Zenhausern, F. Biodosimetry on small blood volume using gene expression assay. Health Phys. 2010, 98, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Pope, I.; Barber, P.; Horn, S.; Ainsbury, E.; Rothkamm, K.; Vojnovic, B. A portable microfluidic fluorescence spectrometer device for γ-H2AX-based biological dosimetry. Radiat. Meas. 2011, 46, 907–911. [Google Scholar] [CrossRef]
- Wang, J.; Song, W.; Song, Y.; Xu, D.; Zhang, M.; Pan, X.; Sun, Y.; Li, D. Quantitative evaluation of radiation dose by γ-H2AX on a microfluidic chip in a miniature fluorescence cytometer. Radiat. Meas. 2014, 62, 71–77. [Google Scholar] [CrossRef]
- Brengues, M.; Gu, J.; Zenhausern, F. Microfluidic module for blood cell separation for gene expression radiobiological assays. Radiat. Prot. Dosim. 2015, 166, 306–310. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Z.; Lan, X. Microfluidic radiobioassays: A radiometric detection tool for understanding cellular physiology and pharmacokinetics. Lab Chip 2019, 19, 2315–2339. [Google Scholar] [CrossRef]
- Jalili-Firoozinezhad, S.; Prantil-Baun, R.; Jiang, A.; Potla, R.; Mammoto, T.; Weaver, J.C.; Ferrante, T.C.; Kim, H.J.; Cabral, J.M.; Levy, O.; et al. Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Torisawa, Y.-S.; Mammoto, T.; Jiang, E.; Jiang, A.; Mammoto, A.; Watters, A.L.; Bahinski, A.; Ingber, D.E. Modeling Hematopoiesis and Responses to Radiation Countermeasures in a Bone Marrow-on-a-Chip. Tissue Eng. Part C Methods 2016, 22, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Maoz, B.M.; Herland, A.; Fitzgerald, E.A.; Grevesse, T.; Vidoudez, C.; Pacheco, A.; Sheehy, S.P.; Park, T.-E.; Dauth, S.; Mannix, R.; et al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat. Biotechnol. 2018, 36, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Lacouture, T.; Grimm, J.; Goldman, H.W.; Ibbott, G.S.; Yorke, E.; Kubicek, G.J. Overview of dosimetric and biological perspectives on radiosurgery of multiple brain metastases in comparison with whole brain radiotherapy. J. Radiosurg. SBRT 2015, 3, 271–279. [Google Scholar]
- Siva, S.; Bressel, M.; Kron, T.; Mai, T.; Le, H.; Montgomery, R.; Hardcastle, N.; Rezo, A.; Gill, S.; Higgs, B.; et al. Stereotactic Ablative Fractionated Radiotherapy versus Radiosurgery for Oligometastatic Neoplasia to the Lung: A Randomized Phase II Trial. Int. J. Radiat. Oncol. 2020, 108, S3–S4. [Google Scholar] [CrossRef]
- Wilson, J.D.; Hammond, E.M.; Higgins, G.S.; Petersson, K. Ultra-High Dose Rate (FLASH) Radiotherapy: Silver Bullet or Fool’s old? Front. Oncol. 2019, 9, 1563. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Gao, F.; Yang, Y.; Wu, D.; Zhang, Y.; Feng, G.; Dai, T.; Du, X. FLASH Radiotherapy: History and Future. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zheng, J.; Yu, Q.; Chen, W.; Xing, J.; Chen, C.; Tian, R. High throughput and accurate serum proteome profiling by integrated sample preparation technology and single-run data independent mass spectrometry analysis. J. Proteom. 2018, 174, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Kim, H.-J.; Liebler, D.C. Efficient Microscale Basic Reverse Phase Peptide Fractionation for Global and Targeted Proteomics. J. Proteome Res. 2016, 15, 2346–2354. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Millet, L.J.; Giannone, R.J.; Greenwood, M.S.; Foster, C.M.; O’Neil, K.M.; Braatz, A.D.; Davern, S.M. Identifying Candidate Biomarkers of Ionizing Radiation in Human Pulmonary Microvascular Lumens Using Microfluidics—A Pilot Study. Micromachines 2021, 12, 904. https://doi.org/10.3390/mi12080904
Millet LJ, Giannone RJ, Greenwood MS, Foster CM, O’Neil KM, Braatz AD, Davern SM. Identifying Candidate Biomarkers of Ionizing Radiation in Human Pulmonary Microvascular Lumens Using Microfluidics—A Pilot Study. Micromachines. 2021; 12(8):904. https://doi.org/10.3390/mi12080904
Chicago/Turabian StyleMillet, Larry J., Richard J. Giannone, Michael S. Greenwood, Carmen M. Foster, Kathleen M. O’Neil, Alexander D. Braatz, and Sandra M. Davern. 2021. "Identifying Candidate Biomarkers of Ionizing Radiation in Human Pulmonary Microvascular Lumens Using Microfluidics—A Pilot Study" Micromachines 12, no. 8: 904. https://doi.org/10.3390/mi12080904
APA StyleMillet, L. J., Giannone, R. J., Greenwood, M. S., Foster, C. M., O’Neil, K. M., Braatz, A. D., & Davern, S. M. (2021). Identifying Candidate Biomarkers of Ionizing Radiation in Human Pulmonary Microvascular Lumens Using Microfluidics—A Pilot Study. Micromachines, 12(8), 904. https://doi.org/10.3390/mi12080904





