Analysis of the Binding of Analyte-Receptor in a Micro-Fluidic Channel for a Biosensor Based on Brownian Motion
Abstract
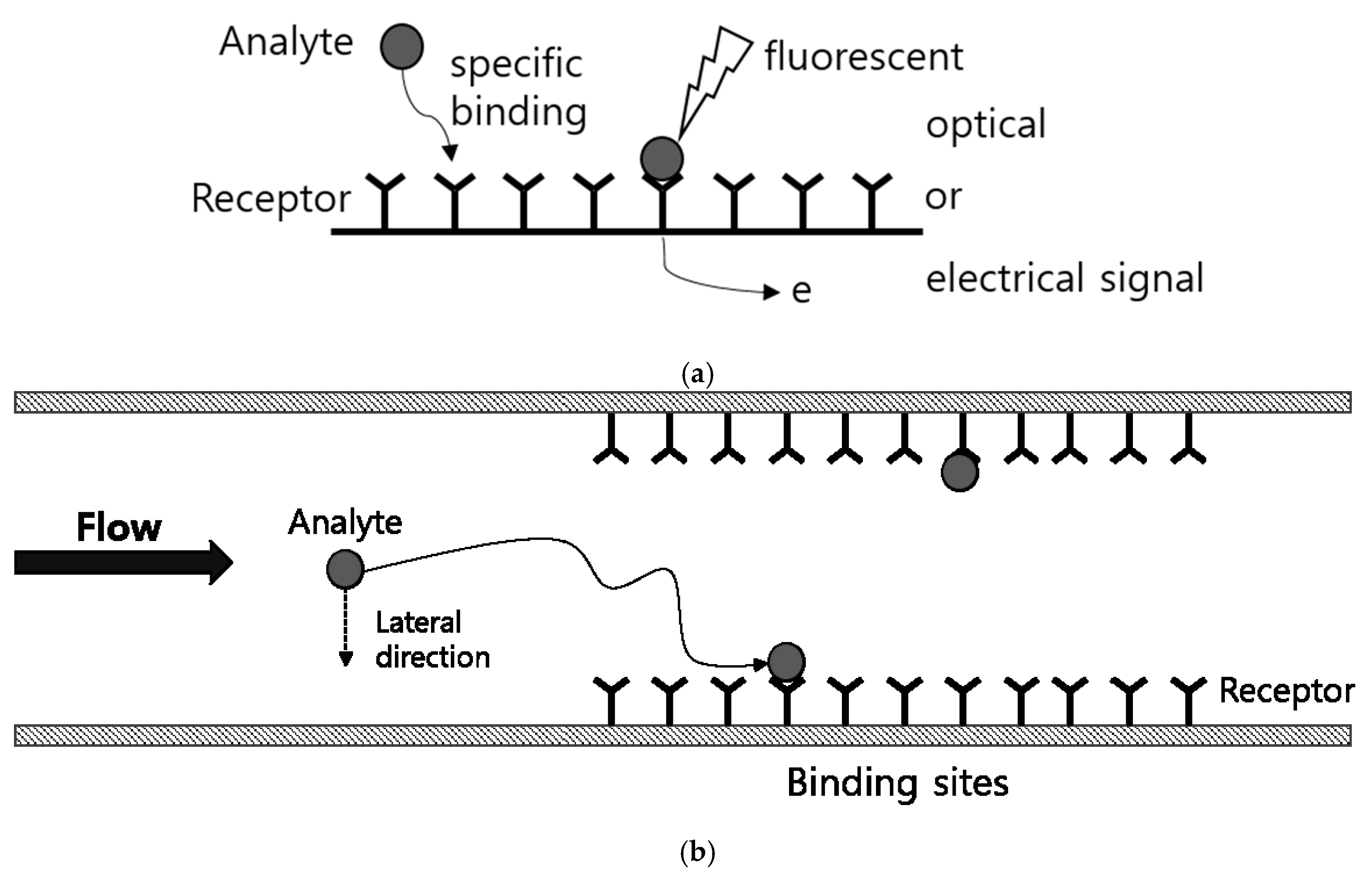
1. Introduction
2. Experiments
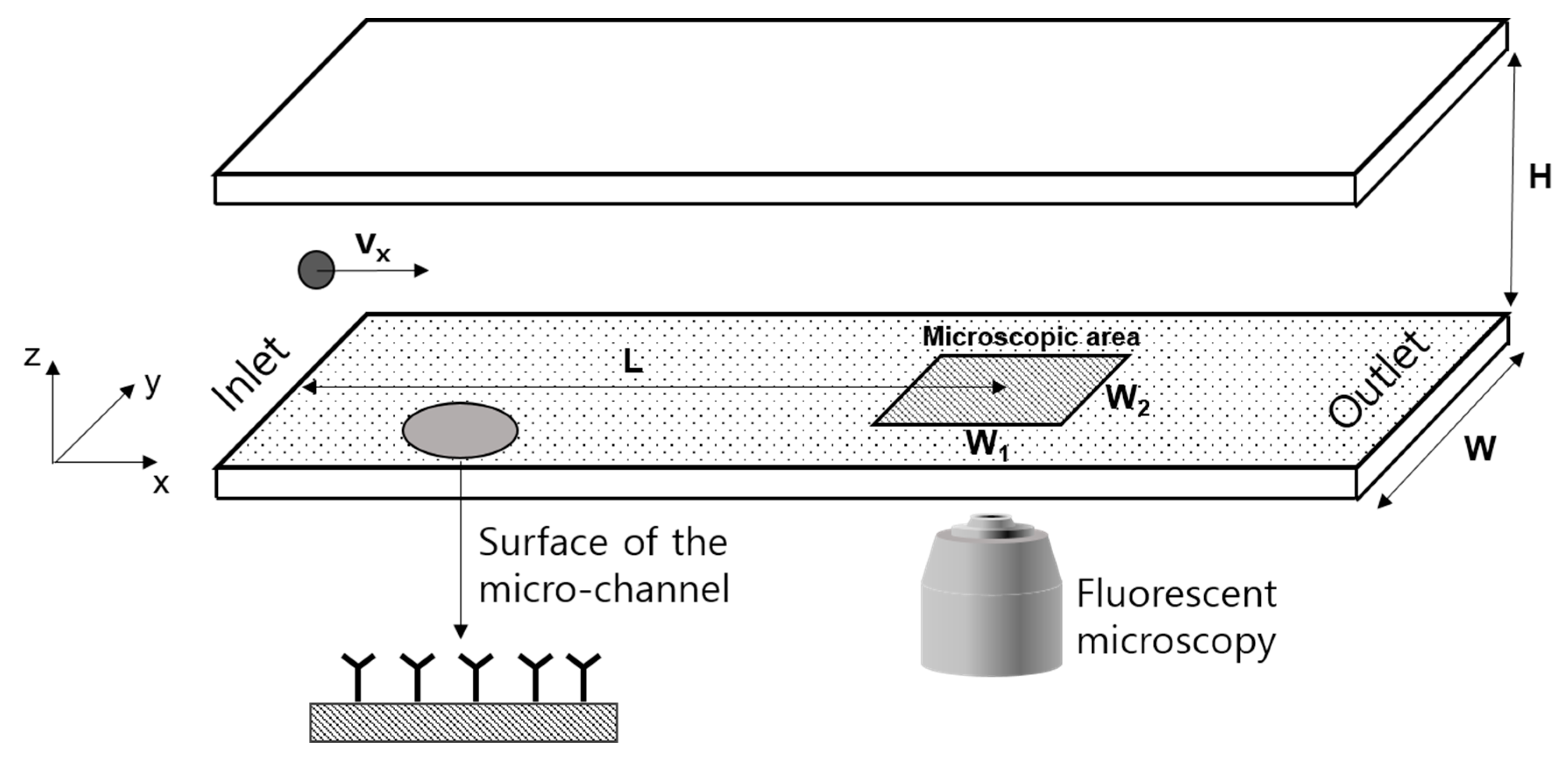
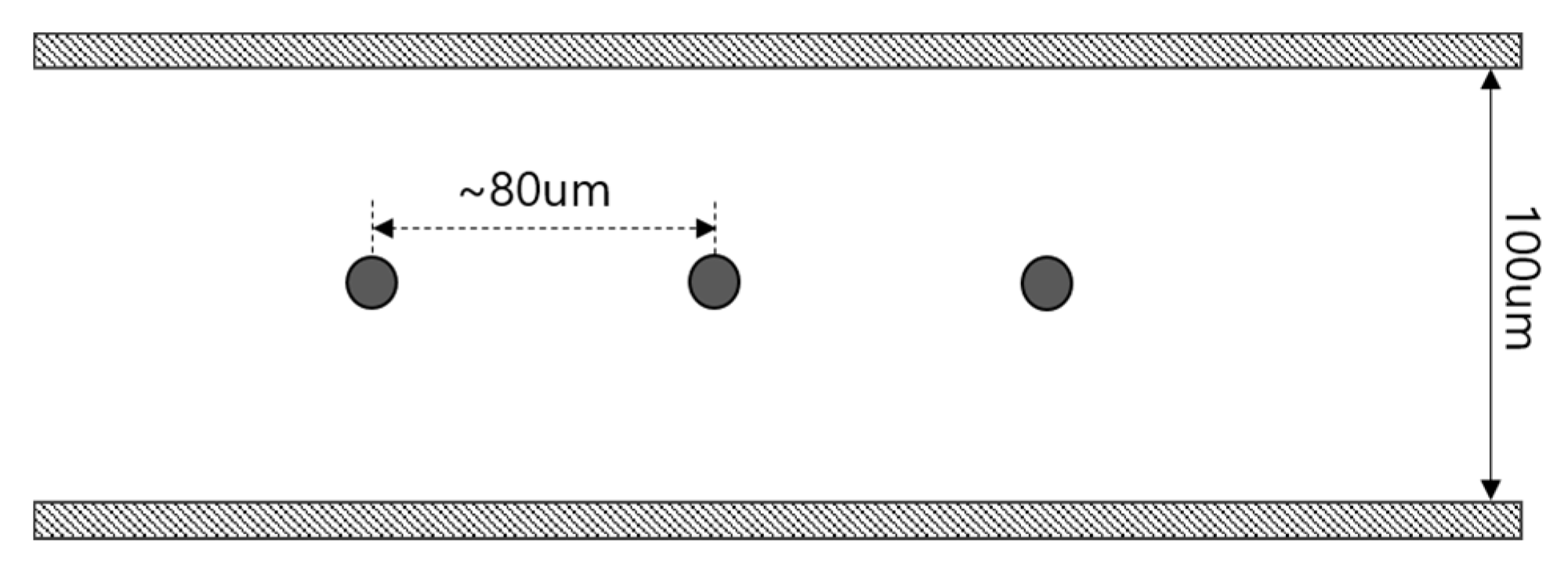
2.1. Microfluidic Channel and Surface Modification
2.2. Investigation for Bound Particles
2.3. Experimental Conditions
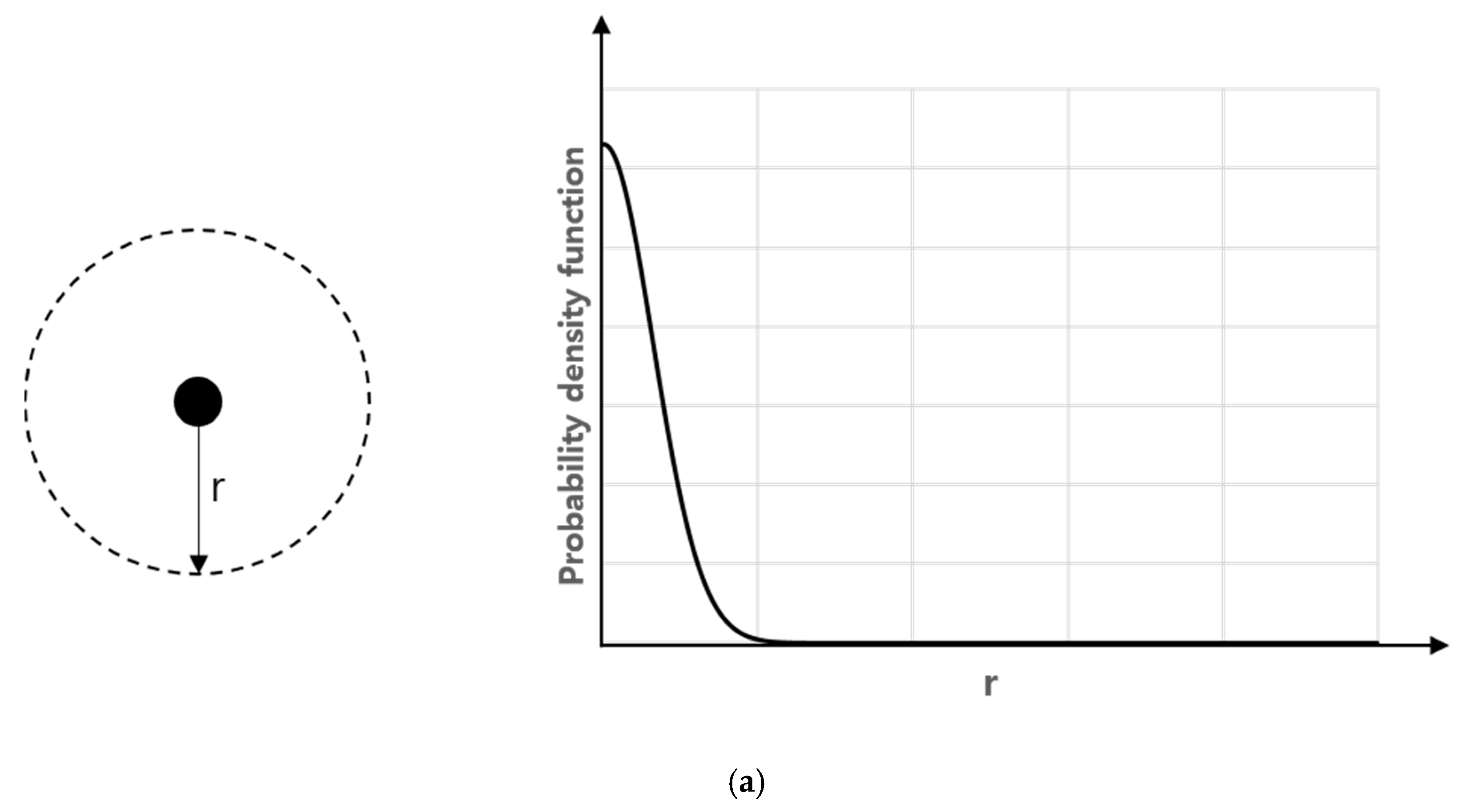
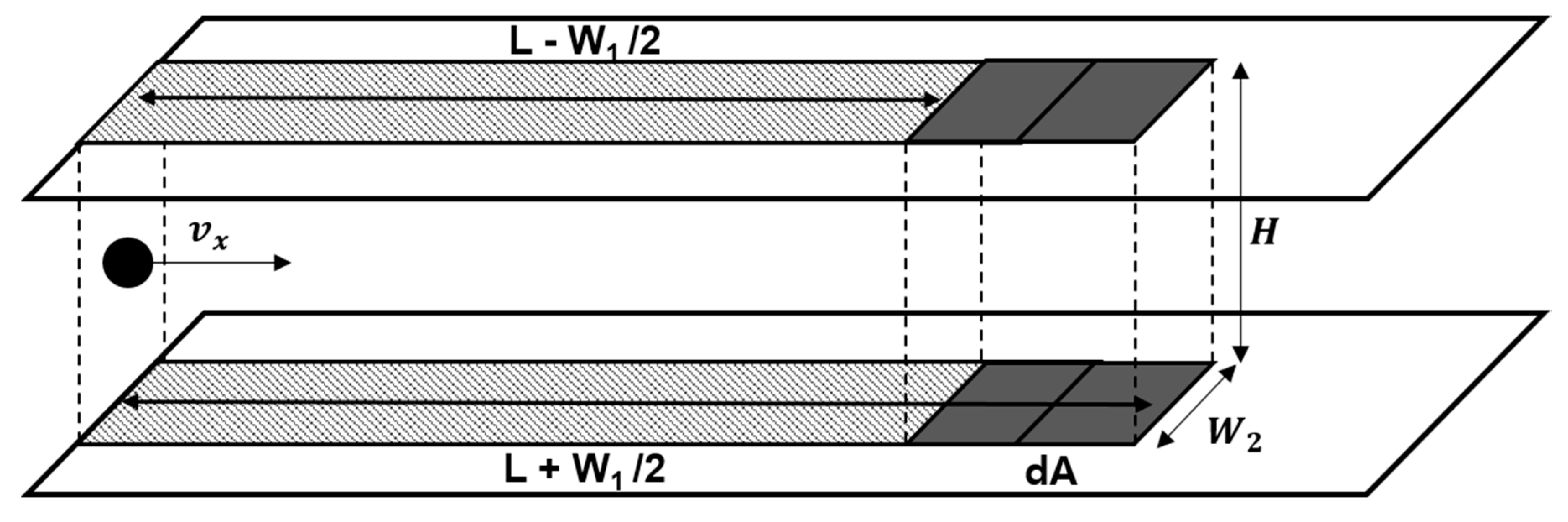
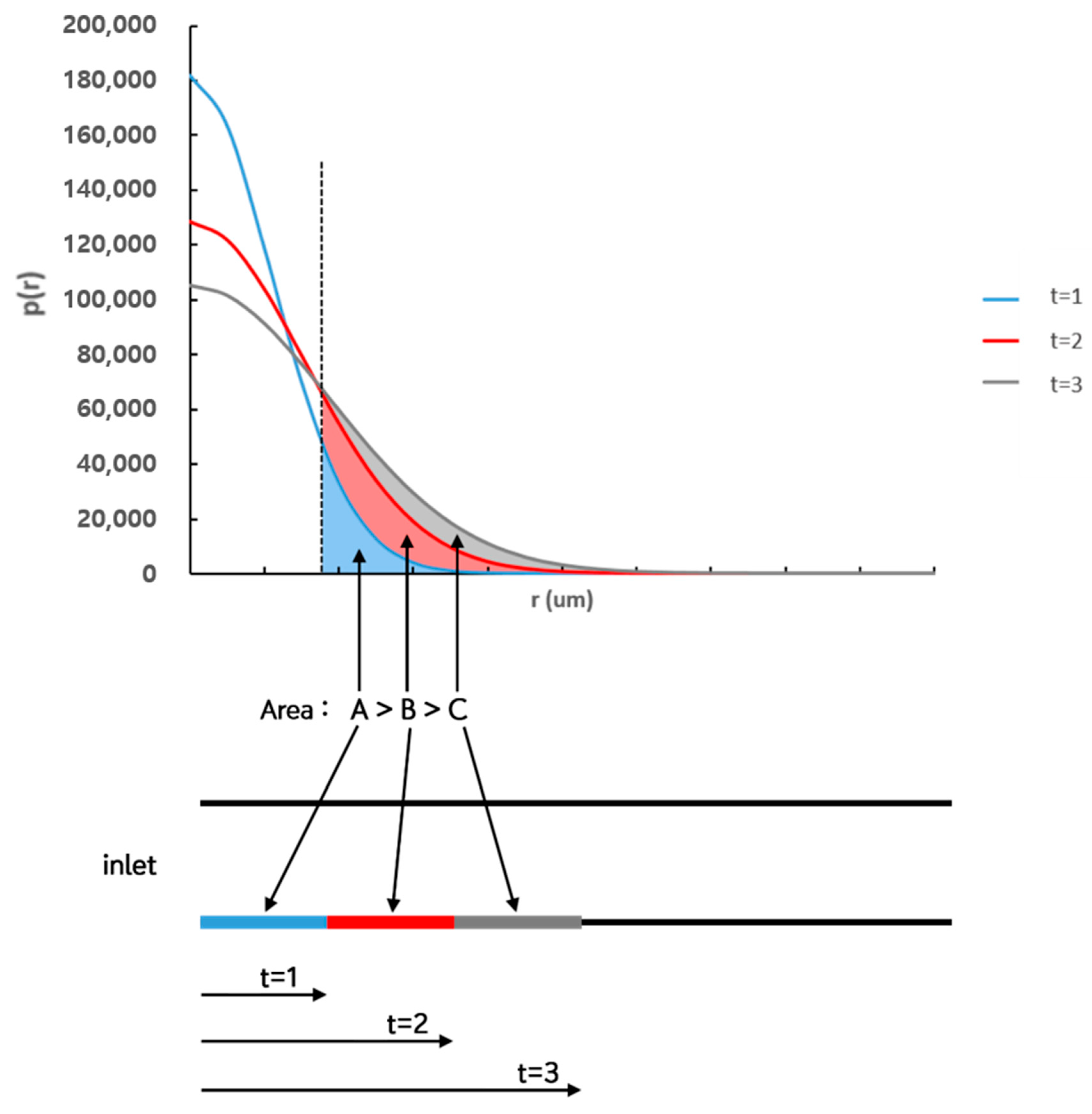
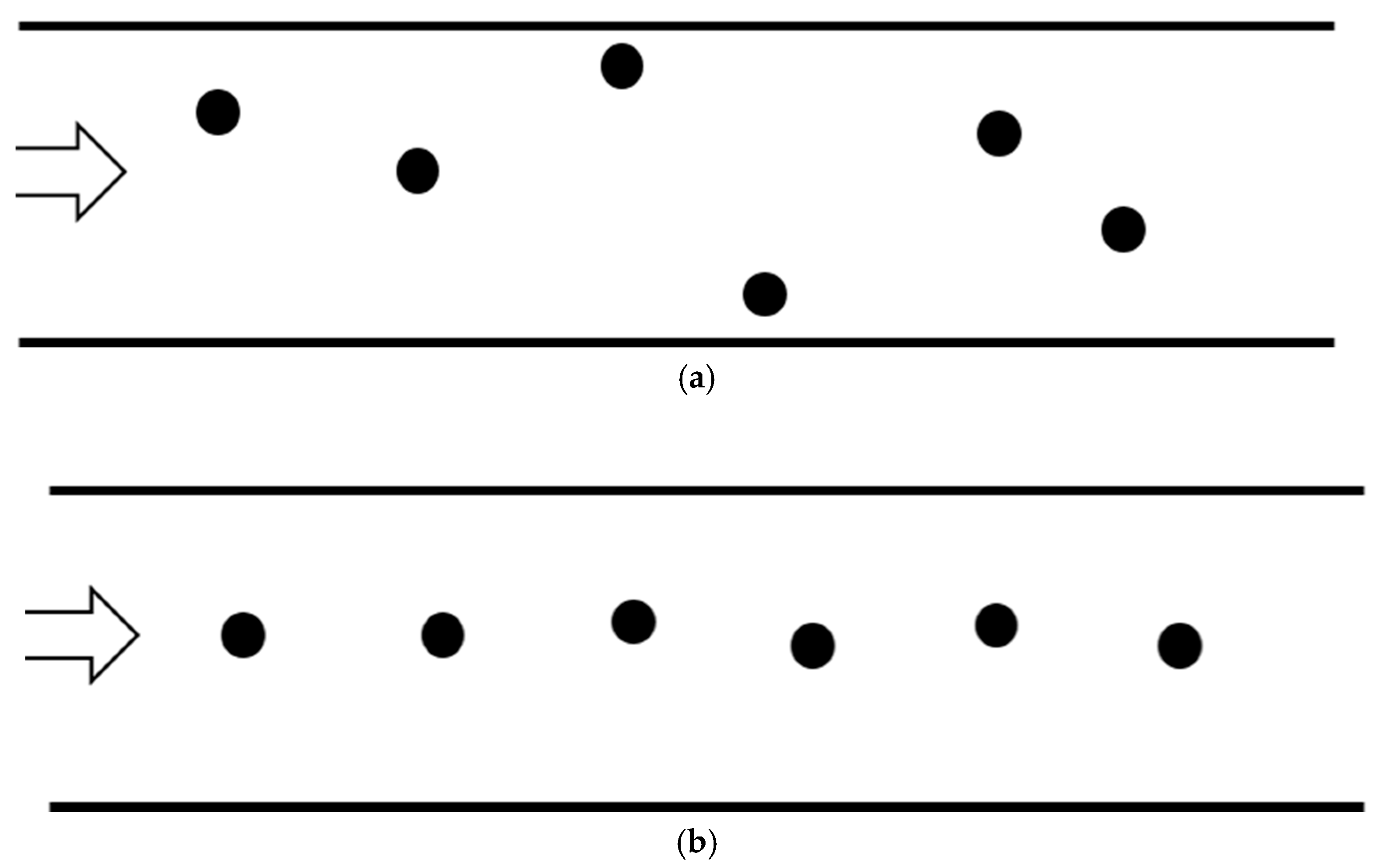
3. Mathematical Model and Analysis
4. Results and Discussion
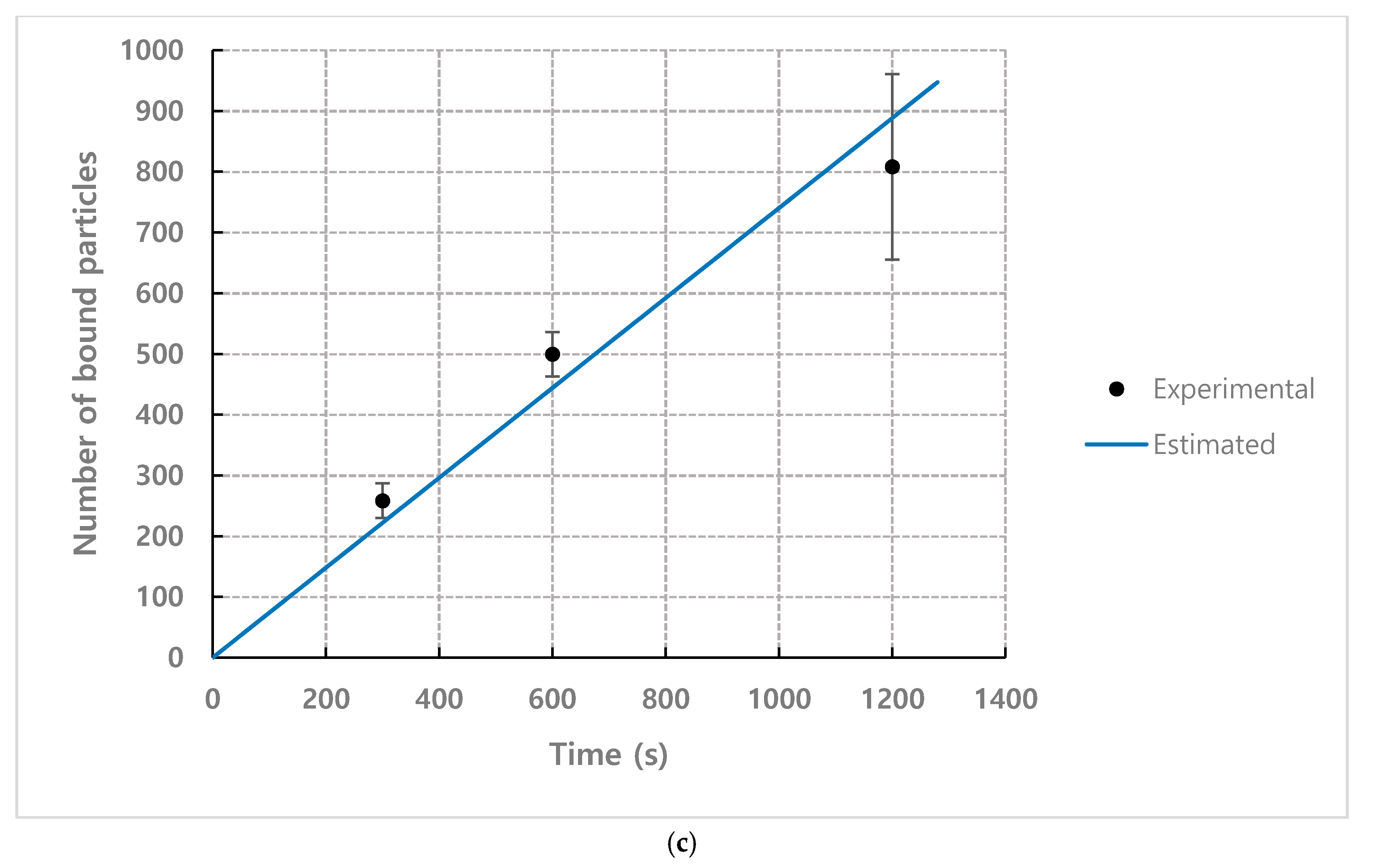
4.1. The Number of Bound Particles
4.2. Numerical Estimation for the Number of Bound Particles based on Mathematical Model
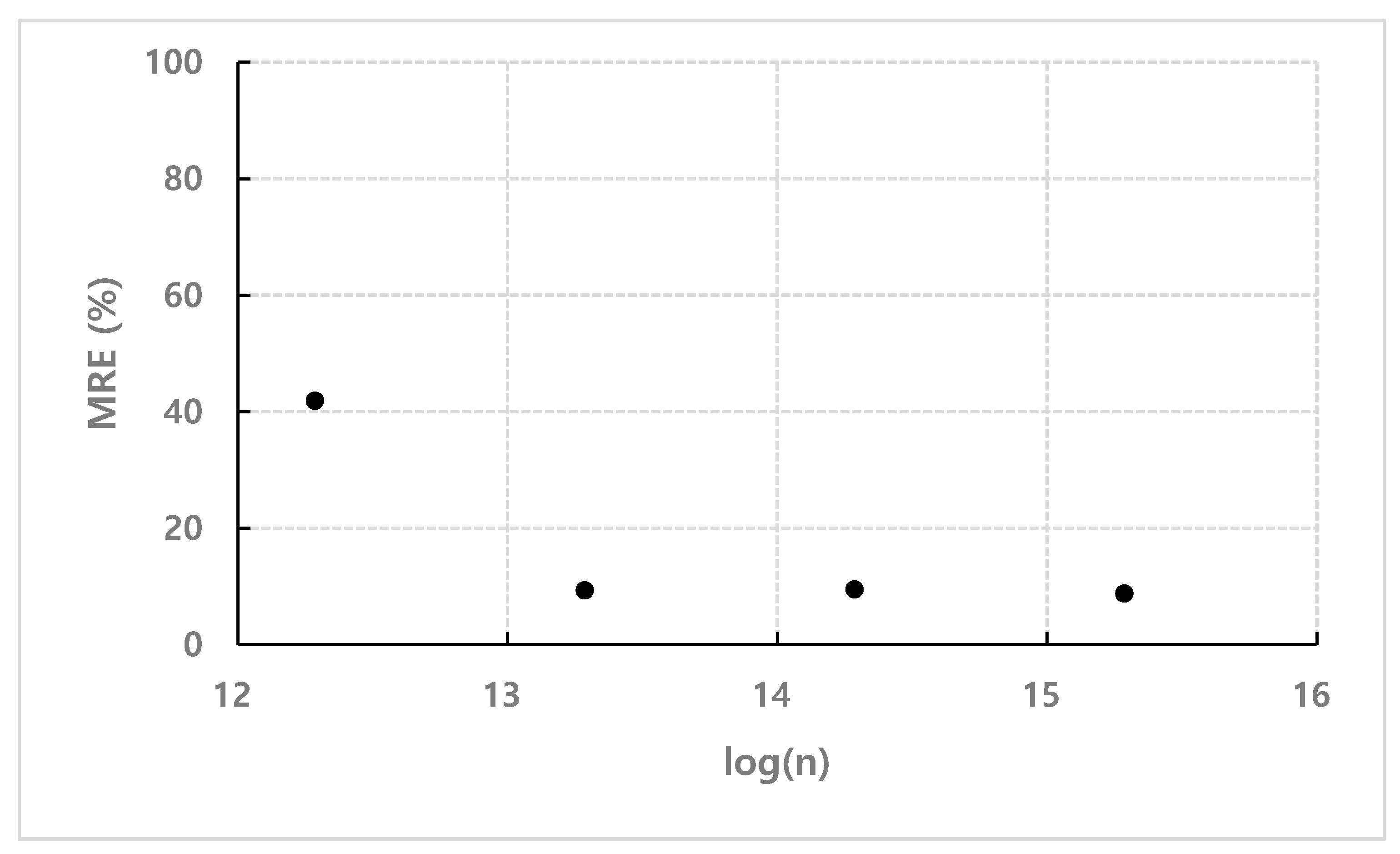
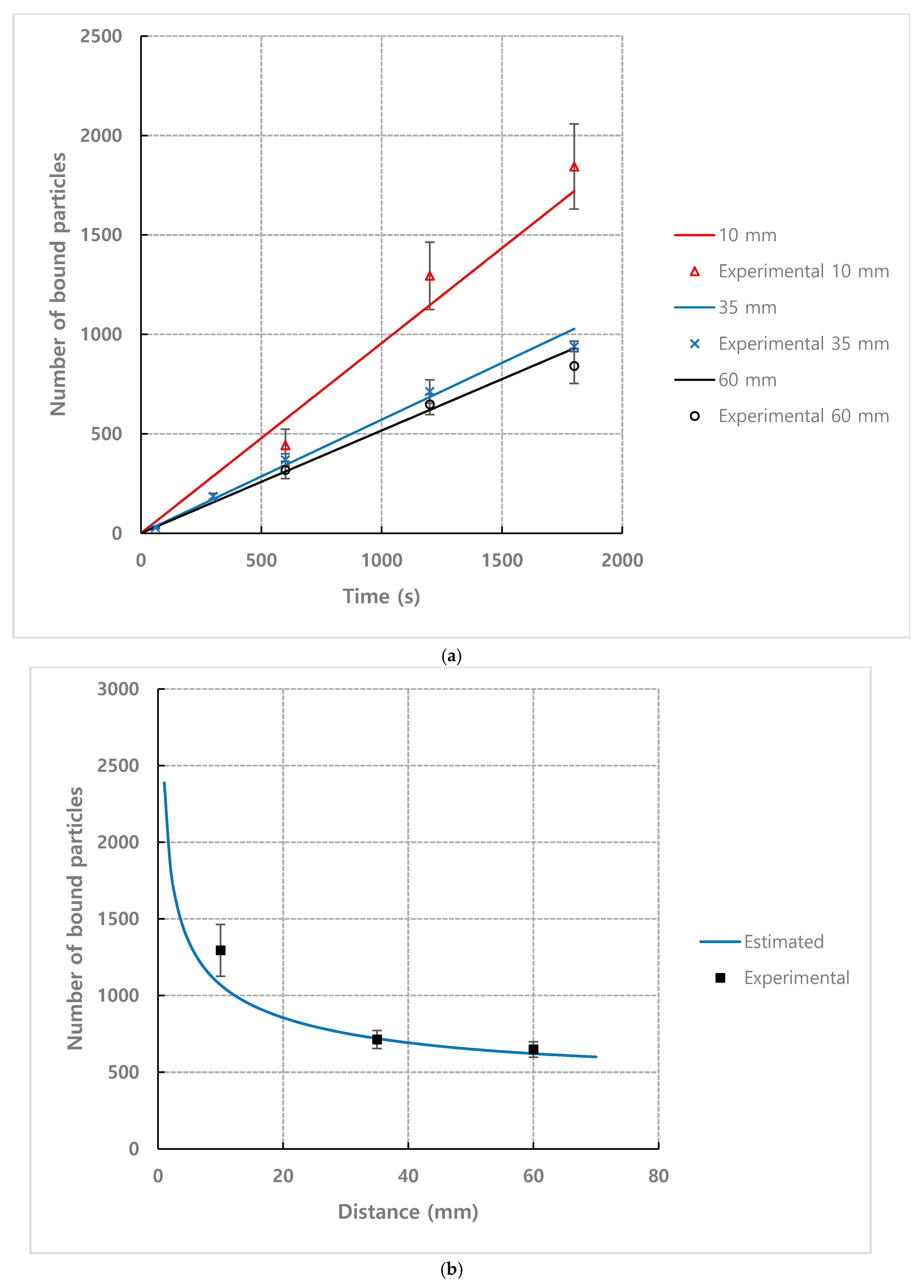
4.3. Empirical Coefficient of Mathematical Model for Bound Particles
4.4. Effect of Observation Position from Inlet and Channel Height
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [PubMed]
- Guo, X. Surface plasmon resonance based biosensor technique: A review. J. Biophotonics 2012, 5, 483–501. [Google Scholar] [CrossRef] [PubMed]
- Vigneshvar, S.; Sudhakumari, C.; Senthilkumaran, B.; Prakash, H. Recent advances in biosensor technology for potential applications—An overview. Front Bioeng. Biotechnol. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Fracchiolla, N.S.; Artuso, S.; Cortelezzi, A. Biosensors in clinical practice: Focus on oncohematology. Sensors 2013, 13, 6423–6447. [Google Scholar] [CrossRef]
- Turner, A.P. Biosensors: Sense and sensibility. Chem. Soc. Rev. 2013, 42, 3184–3196. [Google Scholar] [CrossRef]
- Mascini, M. Biosensors for medical applications. Sens. Actuators B Chem. 1992, 6, 79–82. [Google Scholar] [CrossRef]
- Kerman, K.; Saito, M.; Tamiya, E.; Yamamura, S.; Takamura, Y. Nanomaterial-based electrochemical biosensors for medical applications. TrAC Trends Anal. Chem. 2008, 27, 585–592. [Google Scholar] [CrossRef]
- Alhadrami, H.A. Biosensors: Classifications, medical applications, and future prospective. Biotechnol. Appl. Biochem. 2018, 65, 497–508. [Google Scholar] [CrossRef]
- Anderson, G.P.; Golden, J.P.; Ligler, F.S. A fiber optic biosensor: Combination tapered fibers designed for improved signal acquisition. Biosens. Bioelectron. 1993, 8, 249–256. [Google Scholar] [CrossRef]
- Luo, C.; Tang, H.; Cheng, W.; Yan, L.; Zhang, D.; Ju, H.; Ding, S. A sensitive electrochemical DNA biosensor for specific detection of Enterobacteriaceae bacteria by Exonuclease III-assisted signal amplification. Biosens. Bioelectron. 2013, 48, 132–137. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Zhang, Y.; Chen, Z.; Liu, Y.; Li, Z.; Li, J. Sensitive electrochemical aptamer biosensor for dynamic cell surface N-glycan evaluation featuring multivalent recognition and signal amplification on a dendrimer–graphene electrode interface. Anal. Chem. 2014, 86, 4278–4286. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, Q.; Li, Q.; Yang, X.; Wang, K.; Nie, W. Surface plasmon resonance biosensor for sensitive detection of microRNA and cancer cell using multiple signal amplification strategy. Biosens. Bioelectron. 2017, 87, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.-H.; Ma, Q.-J.; Wu, X.-X.; Zhu, X. Graphene oxide-based biosensor for sensitive fluorescence detection of DNA based on exonuclease III-aided signal amplification. Anal. Chim. Acta 2012, 727, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.M.; Wang, L.; Reipa, V.; Murphy, T.E. Porous silicon biosensor for detection of viruses. Biosens. Bioelectron. 2007, 23, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.K.; Saha, S.; Ramirez-Vick, J.E.; Gupta, V.; Bhansali, S.; Singh, S.P. Recent advances in ZnO nanostructures and thin films for biosensor applications. Anal. Chim. Acta 2012, 737, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Saei, A.A.; Dolatabadi, J.E.N.; Najafi-Marandi, P.; Abhari, A.; de la Guardia, M. Electrochemical biosensors for glucose based on metal nanoparticles. Trac Trends Anal. Chem. 2013, 42, 216–227. [Google Scholar] [CrossRef]
- Luo, X.; Morrin, A.; Killard, A.J.; Smyth, M.R. Application of nanoparticles in electrochemical sensors and biosensors. Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal. 2006, 18, 319–326. [Google Scholar] [CrossRef]
- Lee, G.U.; Kidwell, D.A.; Colton, R.J. Sensing discrete streptavidin-biotin interactions with atomic force microscopy. Langmuir 1994, 10, 354–357. [Google Scholar] [CrossRef]
- Rife, J.; Miller, M.; Sheehan, P.; Tamanaha, C.; Tondra, M.; Whitman, L. Design and performance of GMR sensors for the detection of magnetic microbeads in biosensors. Sens. Actuators A Phys. 2003, 107, 209–218. [Google Scholar] [CrossRef]
- Baselt, D.R.; Lee, G.U.; Natesan, M.; Metzger, S.W.; Sheehan, P.E.; Colton, R.J. A biosensor based on magnetoresistance technology. Biosens. Bioelectron. 1998, 13, 731–739. [Google Scholar] [CrossRef]
- Graham, D.; Ferreira, H.; Bernardo, J.; Freitas, P.; Cabral, J. Single magnetic microsphere placement and detection on-chip using current line designs with integrated spin valve sensors: Biotechnological applications. J. Appl. Phys. 2002, 91, 7786–7788. [Google Scholar] [CrossRef]
- Golden, J.P.; Floyd-Smith, T.M.; Mott, D.R.; Ligler, F.S. Target delivery in a microfluidic immunosensor. Biosens. Bioelectron. 2007, 22, 2763–2767. [Google Scholar] [CrossRef] [PubMed]
- Fürjes, P.; Holczer, E.; Tóth, E.; Iván, K.; Fekete, Z.; Bernier, D.; Dortu, F.; Giannone, D. PDMS microfluidics developed for polymer based photonic biosensors. Microsyst. Technol. 2015, 21, 581–590. [Google Scholar] [CrossRef]
- Lynn Jr, N.S.; Martínez-López, J.-I.; Bocková, M.; Adam, P.; Coello, V.; Siller, H.R.; Homola, J. Biosensing enhancement using passive mixing structures for microarray-based sensors. Biosens. Bioelectron. 2014, 54, 506–514. [Google Scholar] [CrossRef]
- Albrecht, T.; Gibb, T.; Nuttall, P. Ion transport in nanopores. In Engineered Nanopores for Bioanalytical Applications; Elsevier: Amsterdam, The Netherlands, 2013; pp. 1–30. [Google Scholar]
- Kätelhön, E.; Sokolov, S.V.; Compton, R.G. Near-wall hindered diffusion: Implications for surface-based sensors. Sens. Actuators B Chem. 2016, 234, 420–425. [Google Scholar] [CrossRef]
- Uma, B.; Swaminathan, T.; Radhakrishnan, R.; Eckmann, D.; Ayyaswamy, P. Nanoparticle Brownian motion and hydrodynamic interactions in the presence of flow fields. Phys. Fluids 2011, 23, 073602. [Google Scholar] [CrossRef]
- Jomeh, S.; Hoorfar, M. Numerical modeling of mass transport in microfluidic biomolecule-capturing devices equipped with reactive surfaces. Chem. Eng. J. 2010, 165, 668–677. [Google Scholar] [CrossRef]
- Gervais, T.; Jensen, K.F. Mass transport and surface reactions in microfluidic systems. Chem. Eng. Sci. 2006, 61, 1102–1121. [Google Scholar] [CrossRef]
- Sadeghi, A.; Amini, Y.; Saidi, M.H.; Chakraborty, S. Numerical modeling of surface reaction kinetics in electrokinetically actuated microfluidic devices. Anal. Chim. Acta 2014, 838, 64–75. [Google Scholar] [CrossRef]
- Allen, S.; Davies, J.; Dawkes, A.; Davies, M.; Edwards, J.; Parker, M.; Roberts, C.; Sefton, J.; Tendler, S.; Williams, P. In situ observation of streptavidin-biotin binding on an immunoassay well surface using an atomic force microscope. FEBS Lett. 1996, 390, 161–164. [Google Scholar] [CrossRef]
- Yuan, C.; Chen, A.; Kolb, P.; Moy, V.T. Energy landscape of streptavidin− biotin complexes measured by atomic force microscopy. Biochemistry 2000, 39, 10219–10223. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Vaidya, B.; Patel, A.B.; Soper, S.A.; McCarley, R.L. Photochemically patterned poly (methyl methacrylate) surfaces used in the fabrication of microanalytical devices. J. Phys. Chem. B 2005, 109, 16988–16996. [Google Scholar] [CrossRef] [PubMed]
- Vesel, A.; Elersic, K.; Mozetic, M. Immobilization of protein streptavidin to the surface of PMMA polymer. Vacuum 2012, 86, 773–775. [Google Scholar] [CrossRef]
- Zheng, X.; Silber-Li, Z. The influence of Saffman lift force on nanoparticle concentration distribution near a wall. Appl. Phys. Lett. 2009, 95, 124105. [Google Scholar] [CrossRef]
- Goldman, A.; Cox, R.; Brenner, H. Slow viscous motion of a sphere parallel to a plane wall—II Couette flow. Chem. Eng. Sci. 1967, 22, 653–660. [Google Scholar] [CrossRef]




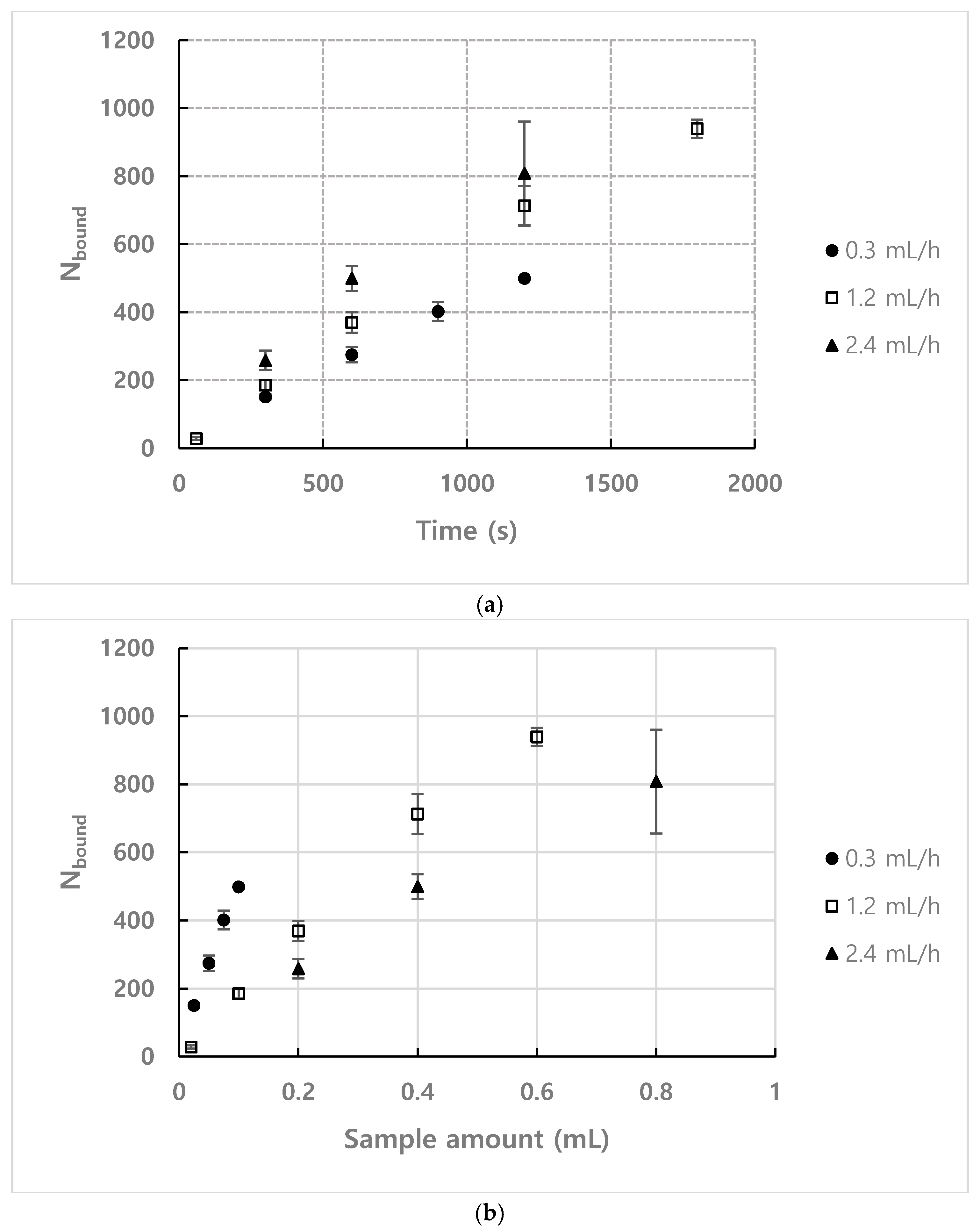
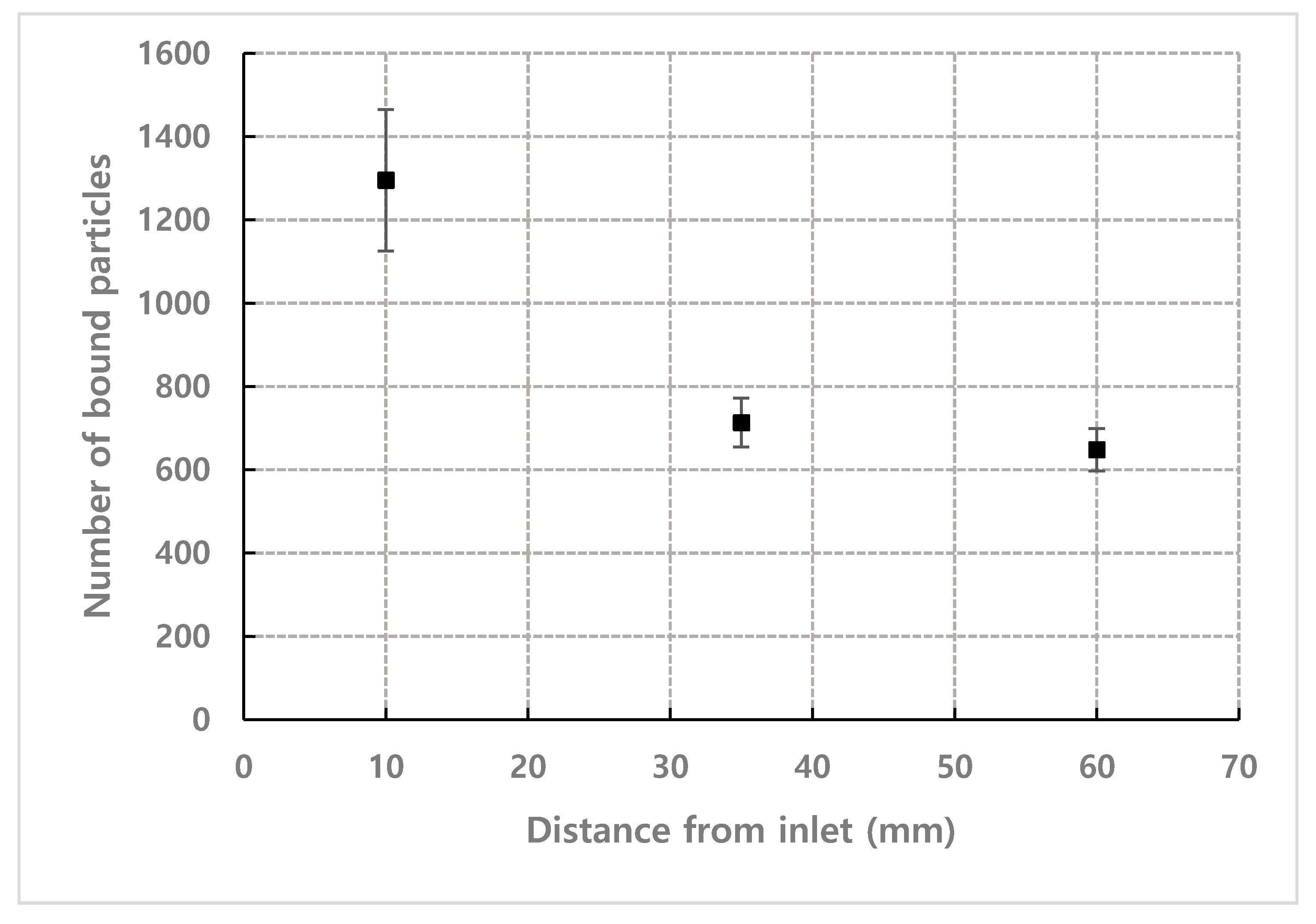

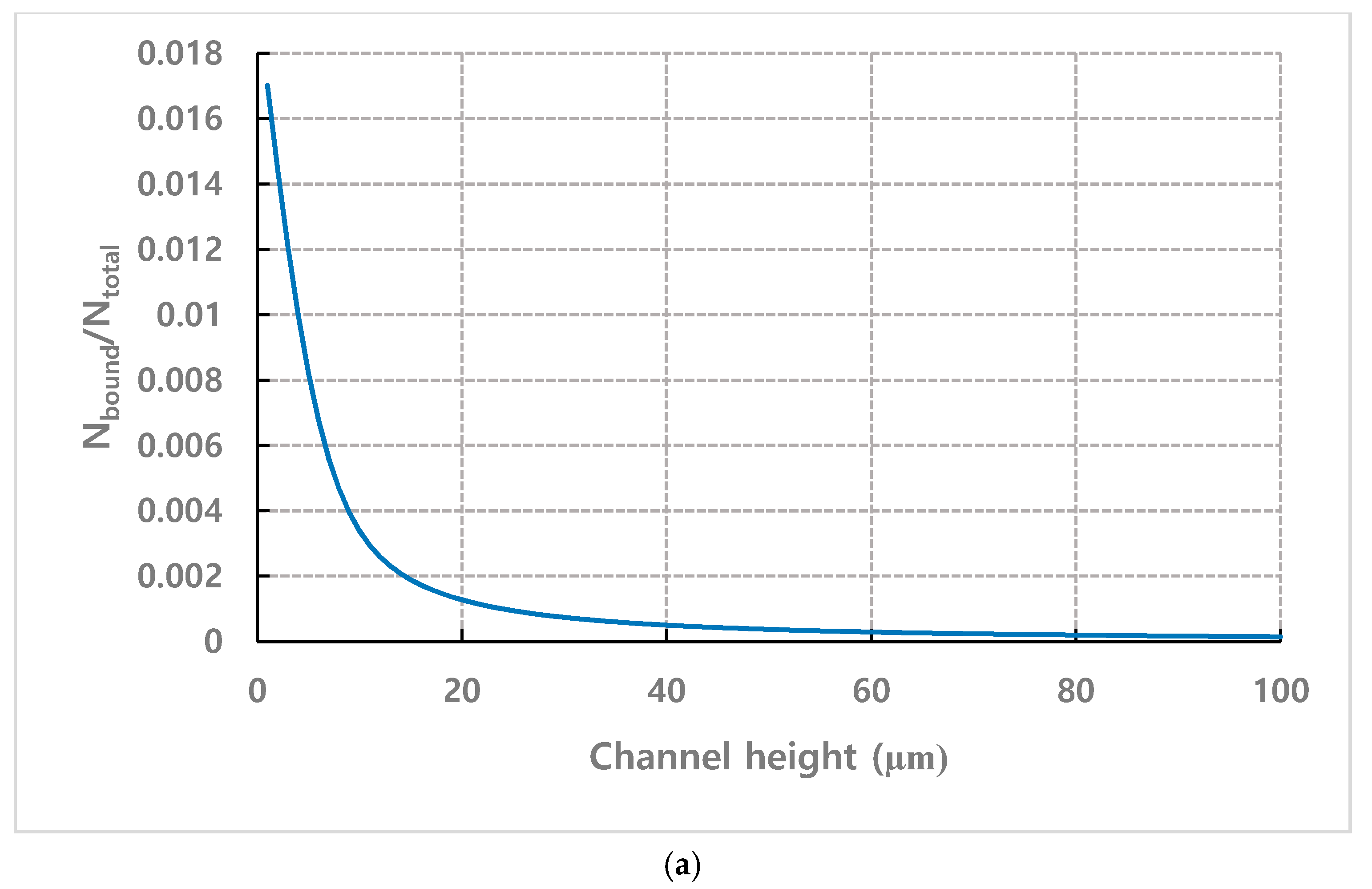
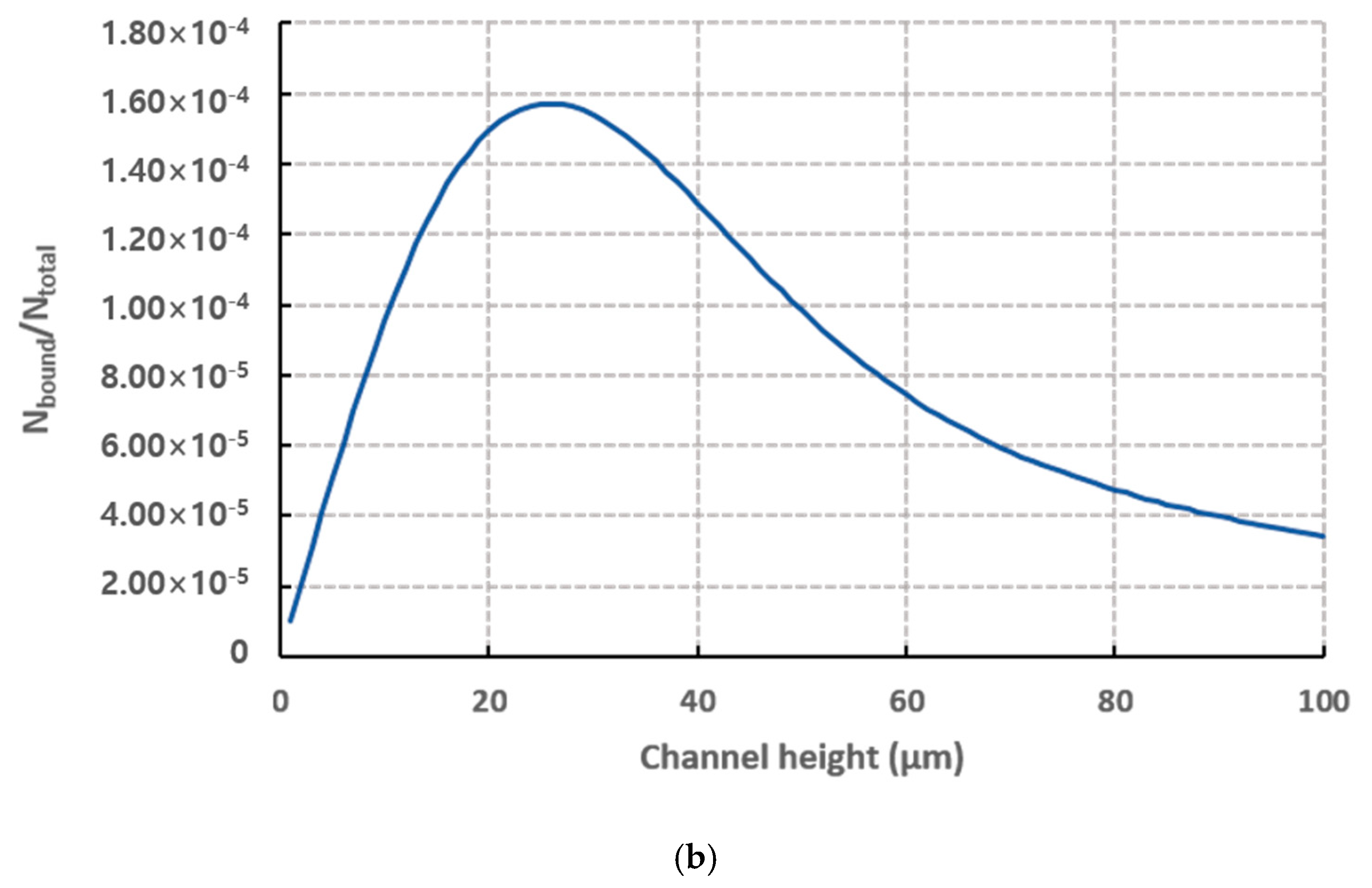
| Number Density (n) | Flow Rate | Observation Position (L) |
|---|---|---|
| 1.2 mL·h−1 | 35 mm | |
| 1.2 mL·h−1 | 35 mm | |
| 1.2 mL·h−1 | 35 mm | |
| 1.2 mL·h−1 | 35 mm | |
| 0.3 mL·h−1 | 35 mm | |
| 2.4 mL·h−1 | 35 mm | |
| 1.2 mL·h−1 | 10 mm | |
| 1.2 mL·h−1 | 60 mm |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, S.; Lee, W.I.; Lee, G.H.; Yoo, Y.-E. Analysis of the Binding of Analyte-Receptor in a Micro-Fluidic Channel for a Biosensor Based on Brownian Motion. Micromachines 2020, 11, 570. https://doi.org/10.3390/mi11060570
Choi S, Lee WI, Lee GH, Yoo Y-E. Analysis of the Binding of Analyte-Receptor in a Micro-Fluidic Channel for a Biosensor Based on Brownian Motion. Micromachines. 2020; 11(6):570. https://doi.org/10.3390/mi11060570
Chicago/Turabian StyleChoi, Sunghak, Woo Il Lee, Gyu Hee Lee, and Yeong-Eun Yoo. 2020. "Analysis of the Binding of Analyte-Receptor in a Micro-Fluidic Channel for a Biosensor Based on Brownian Motion" Micromachines 11, no. 6: 570. https://doi.org/10.3390/mi11060570
APA StyleChoi, S., Lee, W. I., Lee, G. H., & Yoo, Y.-E. (2020). Analysis of the Binding of Analyte-Receptor in a Micro-Fluidic Channel for a Biosensor Based on Brownian Motion. Micromachines, 11(6), 570. https://doi.org/10.3390/mi11060570





