An Immune–Magnetophoretic Device for the Selective and Precise Enrichment of Circulating Tumor Cells from Whole Blood
Abstract
1. Introduction
2. Materials and Methods
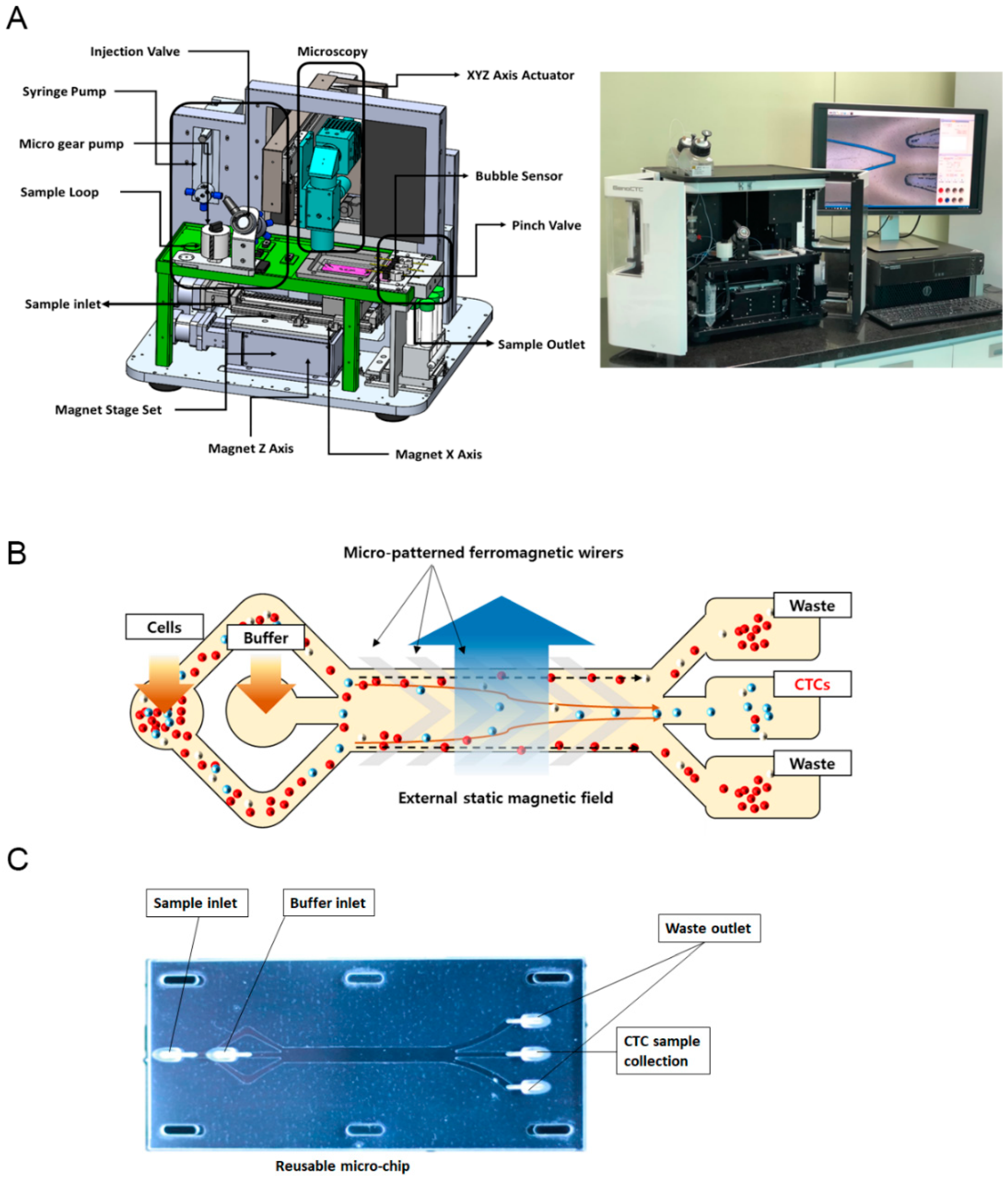
2.1. Device Design and Microchip Fabrication
2.2. Preparation of Antibody-Coated Magnetic Microbeads
2.3. Cell Culture and Blood Sample Collection
2.4. Sample Preparation
2.5. Immunocytochemistry
2.6. Flow Cytometry
3. Results
3.1. GenoCTC Working Principle
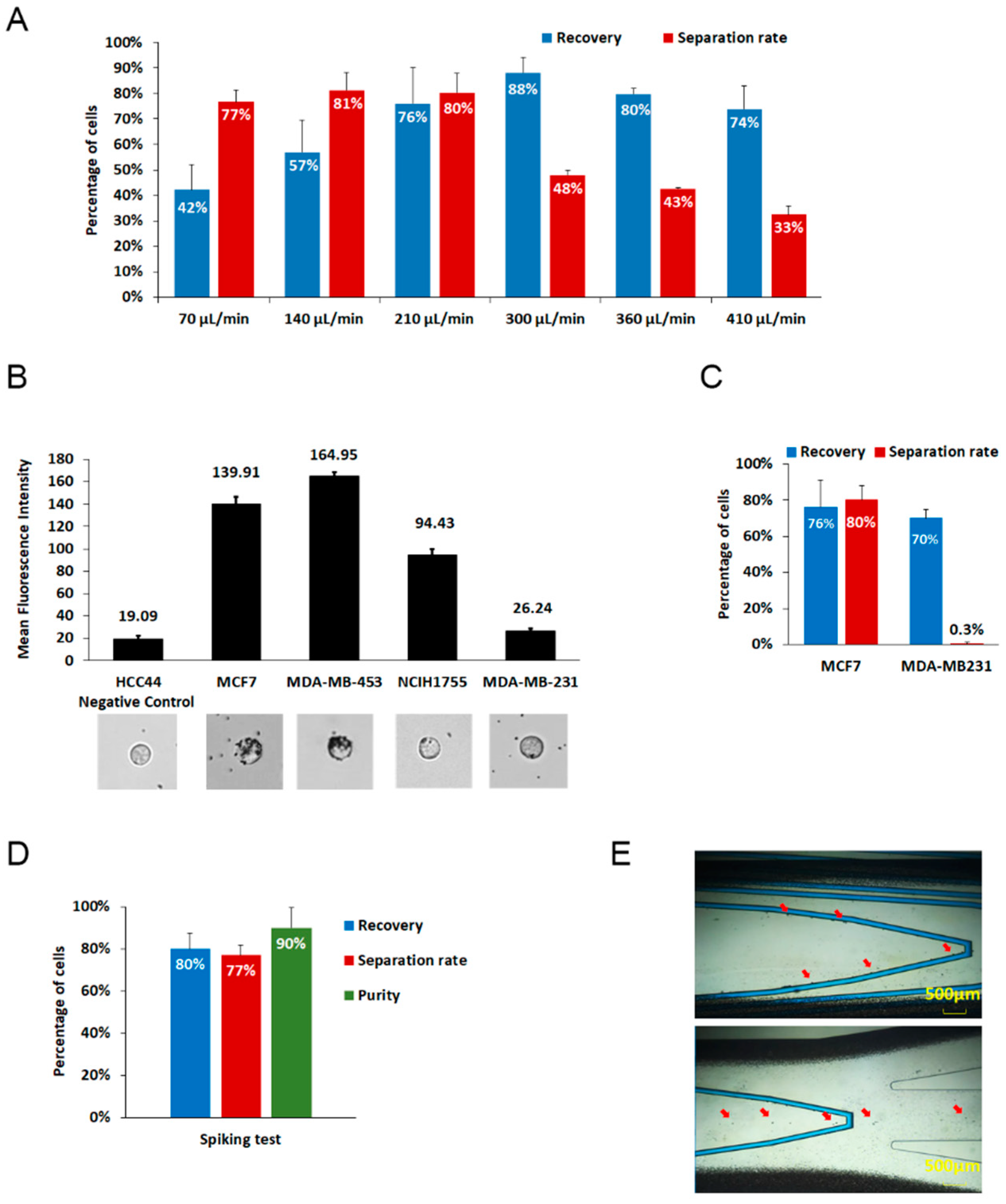
3.2. Analytical Performanc of GenoCTC
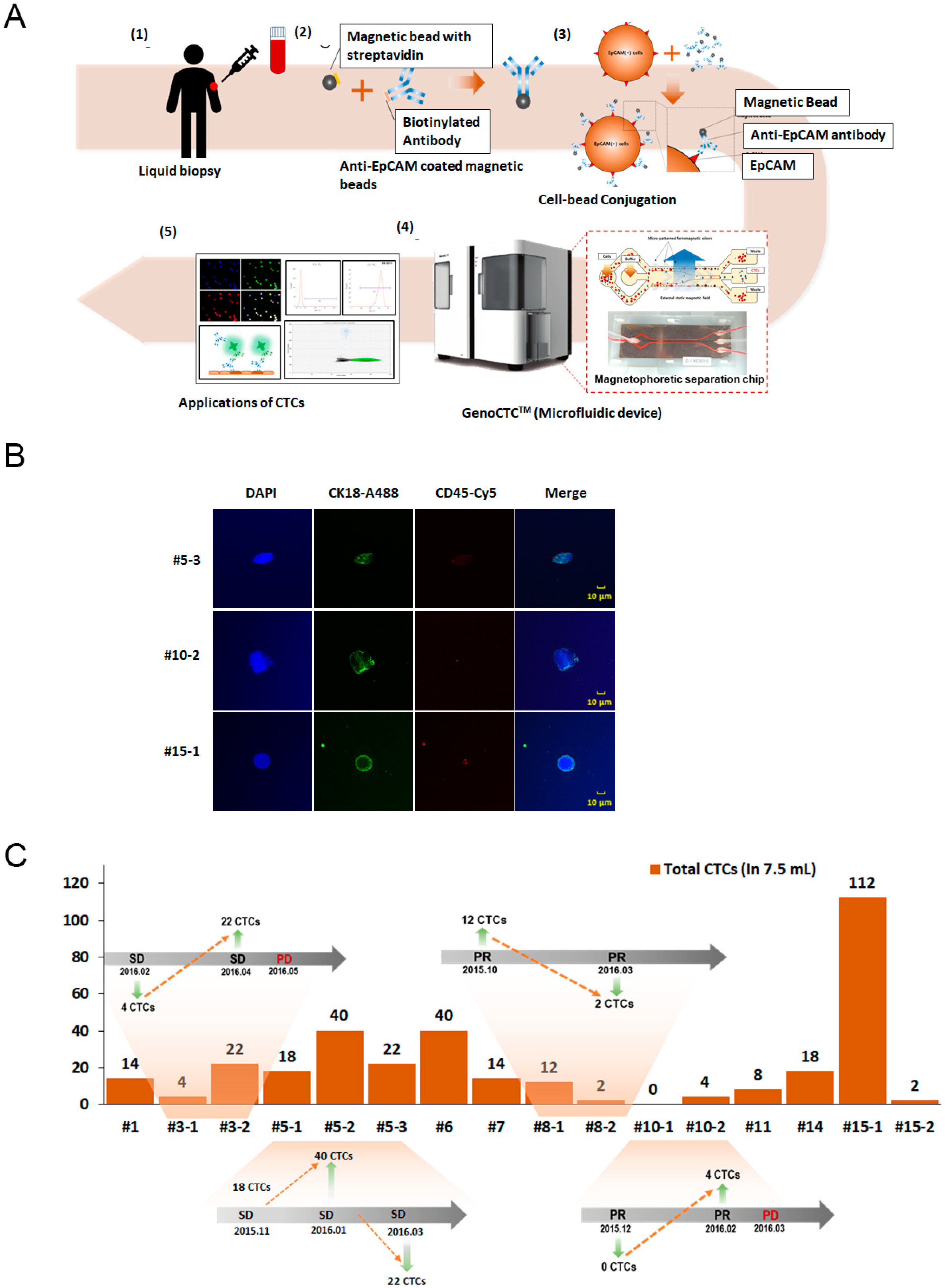
3.3. Isolation and Enumeration of CTCs from NSCLC Patients
3.4. Programmed Death Ligand 1(PDL1) and Tyrosine Protein Kinase (MET) Expression on CTCs from Non-Small Cell Lung Cancer (NSCLC) Patients
3.5. Enumeration and Analysis of CTCs from a Cholangiocarcinoma Patient Using Epithelial and Non-Epithelial Markers
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Karachaliou, N.; Mayo-De-Las-Casas, C.; Molina-Vila, M.A.; Rosell, R. Real-time liquid biopsies become a reality in cancer treatment. Ann. Transl. Med. 2015, 3, 36. [Google Scholar]
- Di Meo, A.; Bartlett, J.; Cheng, Y.; Pasic, M.D.; Yousef, G.M. Liquid biopsy: A step forward towards precision medicine in urologic malignancies. Mol. Cancer 2017, 16, 80. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Leveridge, M.J.; Finelli, A.; Kachura, J.R.; Evans, A.; Chung, H.; Shiff, D.A.; Fernandes, K.; Jewett, M.A.S. Outcomes of Small Renal Mass Needle Core Biopsy, Nondiagnostic Percutaneous Biopsy, and the Role of Repeat Biopsy. Eur. Urol. 2011, 60, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed]
- Lianidou, E. Circulating Tumor Cell Isolation: A Marathon Race Worth Running. Clin. Chem. 2014, 60, 287–289. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Song, Y.; Tian, T.; Shi, Y.; Liu, W.; Zou, Y.; Khajvand, T.; Wang, S.; Zhu, Z.; Yang, C. Enrichment and single-cell analysis of circulating tumor cells. Chem. Sci. 2016, 8, 1736–1751. [Google Scholar] [CrossRef] [PubMed]
- Danova, M.; Torchio, M.; Mazzini, G. Isolation of rare circulating tumor cells in cancer patients: Technical aspects and clinical implications. Expert Rev. Mol. Diagn. 2011, 11, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but not in Healthy Subjects or Patients with Nonmalignant Diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Xu, J.Y.; Handy, B.; Michaelis, C.L.; Waguespack, S.; Hu, M.I.; Busaidy, N.; Jimenez, C.; Cabanillas, M.E.; Fritsche, H.A.; Cote, G.J.; et al. Detection and Prognostic Significance of Circulating Tumor Cells in Patients With Metastatic Thyroid Cancer. J. Clin. Endocrinol. Metab. 2016, 101, 4461–4467. [Google Scholar] [CrossRef]
- Earl, J.; Garcia-Nieto, S.; Martinez-Avila, J.C.; Montans, J.; Dehesa, A.S.; Rodriguez-Garrote, M.; Lisa, E.; Mendía, E.; Lobo, E.; Malats, N.; et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer 2015, 15, 797. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Adalsteinsson, V.A.; Cibulskis, K.; Choudhury, A.D.; Rosenberg, M.; Cruz-Gordillo, P.; Francis, J.M.; Zhang, C.-Z.; Shalek, A.K.; Satija, R.; et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 2014, 32, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Stott, S.L.; Hsu, C.-H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [PubMed]
- Murlidhar, V.; Zeinali, M.; Grabauskiene, S.; Ghannad-Rezaie, M.; Wicha, M.S.; Simeone, D.M.; Ramnath, N.; Reddy, R.M.; Nagrath, S. A radial flow microfluidic device for ultra-high-throughput affinity-based isolation of circulating tumor cells. Small 2014, 10, 4895–4904. [Google Scholar] [CrossRef]
- Vona, G.; Sabile, A.; Louha, M.; Sitruk, V.; Romana, S.P.; Schütze, K.; Capron, F.; Franco, M.; Pazzagli, M.; Vekemans, M.; et al. Isolation by Size of Epithelial Tumor Cells. Am. J. Pathol. 2000, 156, 57–63. [Google Scholar] [CrossRef]
- Hosokawa, M.; Hayata, T.; Fukuda, Y.; Arakaki, A.; Yoshino, T.; Tanaka, T.; Matsunaga, T. Size-Selective Microcavity Array for Rapid and Efficient Detection of Circulating Tumor Cells. Anal. Chem. 2010, 82, 6629–6635. [Google Scholar] [CrossRef]
- Coumans, F.A.W.; Van Dalum, G.; Beck, M.; Terstappen, L.W.M.M. Filter Characteristics Influencing Circulating Tumor Cell Enrichment from Whole Blood. PLoS ONE 2013, 8, e61770. [Google Scholar] [CrossRef]
- Ligthart, S.T.; Coumans, F.A.W.; Bidard, F.-C.; Simkens, L.H.J.; Punt, C.J.A.; De Groot, M.R.; Attard, G.; De Bono, J.S.; Pierga, J.-Y.; Terstappen, L.W.M.M. Circulating Tumor Cells Count and Morphological Features in Breast, Colorectal and Prostate Cancer. PLoS ONE 2013, 8, e67148. [Google Scholar] [CrossRef]
- Park, S.Y.; Ang, R.R.; Duffy, S.P.; Bazov, J.; Chi, K.N.; Black, P.C.; Ma, H. Morphological differences between circulating tumor cells from prostate cancer patients and cultured prostate cancer cells. PLoS ONE 2014, 9, e85264. [Google Scholar] [CrossRef]
- Shishido, S.N.; Carlsson, A.; Nieva, J.J.; Bethel, K.; Hicks, J.; Bazhenova, L.; Kuhn, P. Circulating tumor cells as a response monitor in stage IV non-small cell lung cancer. J. Transl. Med. 2019, 17, 294. [Google Scholar] [CrossRef]
- Park, J.W.; Cho, S.M.; Jung, M.Y.; Ihm, C.; Lee, N. Microdevice for Separation of Circulating Tumor Cells using Embedded Magnetophoresis with V-shaped Ni-Co Nanowires and Immuno-nanomagnetic Beads. ETRI J. 2015, 37, 233–240. [Google Scholar] [CrossRef]
- Jin, C.; Mcfaul, S.M.; Duffy, S.P.; Deng, X.; Tavassoli, P.; Black, P.C.; Ma, H. Technologies for label-free separation of circulating tumor cells: From historical foundations to recent developments. Lab Chip 2014, 14, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [PubMed]
- Coumans, F.; Doggen, C.J.M.; Attard, G.; De Bono, J.S.; Terstappen, L.W.M.M. All circulating EpCAM+CK+CD45- objects predict overall survival in castration-resistant prostate cancer. Ann. Oncol. 2010, 21, 1851–1857. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients With Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Liu, M.C.; Shields, P.G.; Warren, R.D.; Cohen, P.; Wilkinson, M.; Ottaviano, Y.L.; Rao, S.B.; Eng-Wong, J.; Seillier-Moiseiwitsch, F.; Noone, A.-M.; et al. Circulating Tumor Cells: A Useful Predictor of Treatment Efficacy in Metastatic Breast Cancer. J. Clin. Oncol. 2009, 27, 5153–5159. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Q.; Wang, T.; Bian, L.; Zhang, S.; Hu, H.; Li, S.; Hu, Z.; Wu, S.; Liu, B.; et al. Circulating tumor cells in HER2-positive metastatic breast cancer patients: A valuable prognostic and predictive biomarker. BMC Cancer 2013, 13, 202. [Google Scholar] [CrossRef]
- Che, J.; Yu, V.; Dhar, M.; Renier, C.; Matsumoto, M.; Heirich, K.; Garon, E.B.; Goldman, J.; Rao, J.; Sledge, G.W.; et al. Classification of large circulating tumor cells isolated with ultra-high throughput microfluidic Vortex technology. Oncotarget 2016, 7, 12748–12760. [Google Scholar] [CrossRef]
- Hattori, M.; Nakanishi, H.; Yoshimura, M.; Iwase, M.; Yoshimura, A.; Adachi, Y.; Gondo, N.; Kotani, H.; Sawaki, M.; Fujita, N.; et al. Circulating tumor cell detection in tumor draining veins of breast cancer patients. Sci Rep. 2019, 9, 18195. [Google Scholar] [CrossRef]
- Toss, A.; Mu, Z.; Fernandez, S.; Cristofanilli, M. CTC enumeration and characterization: Moving toward personalized medicine. Ann. Transl. Med. 2014, 2, 108. [Google Scholar]
- Pantel, K.; Speicher, M.R. The biology of circulating tumor cells. Oncogene 2015, 35, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Jiang, Y.; Li, P.; Wang, Y.; Xue, J.; Li, N.; Li, D.; Wang, R.; Dang, Y.; Hu, Z.; et al. Dynamic change of PD-L1 expression on circulating tumor cells in advanced solid tumor patients undergoing PD-1 blockade therapy. OncoImmunology 2018, 7, e1438111. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Long, E.; Bence, C.; Butori, C.; Lassalle, S.; Bouhlel, L.; Fazzalari, L.; Zahaf, K.; Lalvee, S.; Washetine, K.; et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: A potential issue for anti-PD-L1 therapeutic strategies. Ann. Oncol. 2016, 27, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Fehm, T.; Becker, S.; Duerr-Stoerzer, S.; Sotlar, K.; Mueller, V.; Wallwiener, D.; Lane, N.; Solomayer, E.; Uhr, J. Determination of HER2 status using both serum HER2 levels and circulating tumor cells in patients with recurrent breast cancer whose primary tumor was HER2 negative or of unknown HER2 status. Breast Cancer Res. 2007, 9, R74. [Google Scholar] [CrossRef]
- Pestrin, M.; Bessi, S.; Galardi, F.; Truglia, M.; Biggeri, A.; Biagioni, C.; Cappadona, S.; Biganzoli, L.; Giannini, A.; Di Leo, A. Correlation of HER2 status between primary tumors and corresponding circulating tumor cells in advanced breast cancer patients. Breast Cancer Res. Treat. 2009, 118, 523–530. [Google Scholar] [CrossRef]
- Ilie, M.; Szafer-Glusman, E.; Hofman, V.; Long-Mira, E.; Suttmann, R.; Darbonne, W.; Butori, C.; Lalvée, S.; Fayada, J.; Selva, E.; et al. Expression of MET in circulating tumor cells correlates with expression in tumor tissue from advanced-stage lung cancer patients. Oncotarget 2017, 8, 26112–26121. [Google Scholar] [CrossRef]
- Satelli, A.; Batth, I.S.; Brownlee, Z.; Rojas, C.; Meng, Q.H.; Kopetz, S.; Li, S. Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Sci. Rep. 2016, 6, 28910. [Google Scholar] [CrossRef]
- Mazel, M.; Jacot, W.; Pantel, K.; Bartkowiak, K.; Topart, D.; Cayrefourcq, L.; Rossille, D.; Maudelonde, T.; Fest, T.; Alix-Panabières, C. Frequent expression of PD-L1 on circulating breast cancer cells. Mol. Oncol. 2015, 9, 1773–1782. [Google Scholar] [CrossRef]
- Nicolazzo, C.; Raimondi, C.; Mancini, M.; Caponnetto, S.; Gradilone, A.; Gandini, O.; Mastromartino, M.; Del Bene, G.; Prete, A.A.; Longo, F.; et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci. Rep. 2016, 6, 31726. [Google Scholar] [CrossRef]
- Kammula, U.S.; Kuntz, E.J.; Francone, T.D.; Zeng, Z.; Shia, J.; Landmann, R.; Paty, P.B.; Weiser, M.R. Molecular co-expression of the c-Met oncogene and hepatocyte growth factor in primary colon cancer predicts tumor stage and clinical outcome. Cancer Lett. 2007, 248, 219–228. [Google Scholar] [CrossRef]
- Shoji, H.; Yamada, Y.; Taniguchi, H.; Nagashima, K.; Okita, N.; Takashima, A.; Honma, Y.; Iwasa, S.; Kato, K.; Hamaguchi, T.; et al. Clinical impact of c-MET expression and genetic mutational status in colorectal cancer patients after liver resection. Cancer Sci. 2014, 105, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, A.; Wagner, J.; Gorges, T.M.; Taenzer, A.; Uzunoglu, F.G.; Driemel, C.; Stoecklein, N.H.; Knoefel, W.T.; Angenendt, S.; Hauch, S.; et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci. Rep. 2016, 6, 28010. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Zhang, L.; Kim, D.-W.; Liu, X.; Lee, D.H.; Yang, J.C.-H.; Ahn, M.-J.; Vansteenkiste, J.F.; Su, W.-C.; Felip, E.; et al. Phase Ib/II Study of Capmatinib (INC280) Plus Gefitinib After Failure of Epidermal Growth Factor Receptor (EGFR) Inhibitor Therapy in Patients With EGFR-Mutated, MET Factor–Dysregulated Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2018, 36, 3101–3109. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Lorusso, P.M.; Hamid, O.; Janku, F.; Kittaneh, M.; Catenacci, D.V.T.; Chan, E.; Bekaii-Saab, T.; Gadgeel, S.M.; Loberg, R.D.; et al. Phase I Study of AMG 337, a Highly Selective Small-molecule MET Inhibitor, in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2018, 25, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Jardim, D.L.; Johnson, F.M.; Subbiah, V.; Piha-Paul, S.A.; Tsimberidou, A.M.; Falchook, G.S.; Karp, D.; Zinner, R.; Wheler, J.; et al. Phase I study of the combination of crizotinib (as a MET inhibitor) and dasatinib (as a c-SRC inhibitor) in patients with advanced cancer. Investig. New Drugs 2017, 36, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Roohullah, A.; Cooper, A.; Lomax, A.J.; Aung, J.; Barge, A.; Chow, L.; McHale, M.; Desai, J.; Whittle, J.R.; Tran, B.; et al. A phase I trial to determine safety and pharmacokinetics of ASLAN002, an oral MET superfamily kinase inhibitor, in patients with advanced or metastatic solid cancers. Investig. New Drugs 2018, 36, 886–894. [Google Scholar] [CrossRef]
- Rizvi, S.; Khan, S.A.; Hallemeier, C.L.; Kelley, R.K.; Gores, G.J. Cholangiocarcinoma—Evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2017, 15, 95–111. [Google Scholar] [CrossRef]
- Yang, J.D.; Campion, M.B.; Liu, M.C.; Chaiteerakij, R.; Giama, N.H.; Ahmed Mohammed, H.; Zhang, X.; Hu, C.; Campion, V.L.; Jen, J.; et al. Circulating tumor cells are associated with poor overall survival in patients with cholangiocarcinoma. Hepatology 2016, 63, 148–158. [Google Scholar] [CrossRef]
- Kim, S.; Han, S.-I.; Park, M.-J.; Jeon, C.-W.; Joo, Y.-D.; Choi, I.-H.; Han, K.-H. Circulating Tumor Cell Microseparator Based on Lateral Magnetophoresis and Immunomagnetic Nanobeads. Anal. Chem. 2013, 85, 2779–2786. [Google Scholar] [CrossRef]
- Forbes, T.P.; Forry, S.P. Microfluidic magnetophoretic separations of immunomagnetically labeled rare mammalian cells. Lab Chip 2012, 12, 1471. [Google Scholar] [CrossRef]
- Adams, J.D.; Kim, U.; Soh, H.T. Multitarget magnetic activated cell sorter. Proc. Natl. Acad. Sci. USA 2008, 105, 18165–18170. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Yamato, M.; Nara, A. Particle Trapping and Undulation of a Liquid Surface Using a Microscopically Modulated Magnetic Field. Langmuir 2004, 20, 572–574. [Google Scholar] [CrossRef] [PubMed]
- Königsberg, R.; Obermayr, E.; Bises, G.; Pfeiler, G.; Gneist, M.; Wrba, F.; De Santis, M.; Zeillinger, R.; Hudec, M.; Dittrich, C. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol. 2011, 50, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Schneck, H.; Gierke, B.; Uppenkamp, F.; Behrens, B.; Niederacher, D.; Stoecklein, N.H.; Templin, M.F.; Pawlak, M.; Fehm, T.; Neubauer, H. Correction: EpCAM-Independent Enrichment of Circulating Tumor Cells in Metastatic Breast Cancer. PLoS ONE 2016, 11, e0149315. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Roese, L.; Zollner, T.M.; Krahn, T.; Von Ahsen, O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 178. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Gires, O. EpCAM (CD326) finding its role in cancer. Br. J. Cancer 2007, 96, 417–423. [Google Scholar] [CrossRef]
- Patriarca, C.; Macchi, R.M.; Marschner, A.K.; Mellstedt, H. Epithelial cell adhesion molecule expression (CD326) in cancer: A short review. Cancer Treat. Rev. 2012, 38, 68–75. [Google Scholar] [CrossRef]
- Munz, M.; Baeuerle, P.A.; Gires, O. The Emerging Role of EpCAM in Cancer and Stem Cell Signaling. Cancer Res. 2009, 69, 5627–5629. [Google Scholar] [CrossRef]
- De Wit, S.; Van Dalum, G.; Lenferink, A.T.M.; Tibbe, A.G.J.; Hiltermann, T.J.N.; Groen, H.J.M.; Van Rijn, C.J.M.; Terstappen, L.W.M.M. The detection of EpCAM+ and EpCAM− circulating tumor cells. Sci. Rep. 2015, 5, 12270. [Google Scholar] [CrossRef]
- Schulze, K.; Gasch, C.; Staufer, K.; Nashan, B.; Lohse, A.W.; Pantel, K.; Riethdorf, S.; Wege, H. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int. J. Cancer 2013, 133, 2165–2171. [Google Scholar] [CrossRef]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumor cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Park, M.-H.; Reátegui, E.; Li, W.; Tessier, S.N.; Wong, K.H.K.; Jensen, A.E.; Thapar, V.; Ting, D.; Toner, M.; Stott, S.L.; et al. Enhanced Isolation and Release of Circulating Tumor Cells Using Nanoparticle Binding and Ligand Exchange in a Microfluidic Chip. J. Am. Chem. Soc. 2017, 139, 2741–2749. [Google Scholar] [CrossRef] [PubMed]
- Renier, C.; Pao, E.; Che, J.; Liu, H.E.; Lemaire, C.A.; Matsumoto, M.; Triboulet, M.; Srivinas, S.; Jeffrey, S.S.; Rettig, M.; et al. Label-free isolation of prostate circulating tumor cells using Vortex microfluidic technology. NPJ Precis. Oncol. 2017, 1, 15. [Google Scholar] [CrossRef] [PubMed]
- Warkiani, M.E.; Guan, G.; Luan, K.B.; Lee, W.C.; Bhagat, A.A.; Chaudhuri, P.K.; Tan, D.S.-W.; Lim, D.W.-T.; Lee, S.C.; Chen, P.C.Y.; et al. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip 2014, 14, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Pratt, E.D.; Huang, C.; Hawkins, B.G.; Gleghorn, J.P.; Kirby, B. Rare cell capture in microfluidic devices. Chem. Eng. Sci. 2011, 66, 1508–1522. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chelakkot, C.; Ryu, J.; Kim, M.Y.; Kim, J.-S.; Kim, D.; Hwang, J.; Park, S.H.; Ko, S.B.; Park, J.W.; Jung, M.Y.; et al. An Immune–Magnetophoretic Device for the Selective and Precise Enrichment of Circulating Tumor Cells from Whole Blood. Micromachines 2020, 11, 560. https://doi.org/10.3390/mi11060560
Chelakkot C, Ryu J, Kim MY, Kim J-S, Kim D, Hwang J, Park SH, Ko SB, Park JW, Jung MY, et al. An Immune–Magnetophoretic Device for the Selective and Precise Enrichment of Circulating Tumor Cells from Whole Blood. Micromachines. 2020; 11(6):560. https://doi.org/10.3390/mi11060560
Chicago/Turabian StyleChelakkot, Chaithanya, Jiyeon Ryu, Mi Young Kim, Jin-Soo Kim, Dohyeong Kim, Juhyun Hwang, Sung Hoon Park, Seok Bum Ko, Jeong Won Park, Moon Youn Jung, and et al. 2020. "An Immune–Magnetophoretic Device for the Selective and Precise Enrichment of Circulating Tumor Cells from Whole Blood" Micromachines 11, no. 6: 560. https://doi.org/10.3390/mi11060560
APA StyleChelakkot, C., Ryu, J., Kim, M. Y., Kim, J.-S., Kim, D., Hwang, J., Park, S. H., Ko, S. B., Park, J. W., Jung, M. Y., Kim, R. N., Song, K., Kim, Y. J., Choi, Y.-L., Lee, H. S., & Shin, Y. K. (2020). An Immune–Magnetophoretic Device for the Selective and Precise Enrichment of Circulating Tumor Cells from Whole Blood. Micromachines, 11(6), 560. https://doi.org/10.3390/mi11060560





