Lab-on-Chip Platform for Culturing and Dynamic Evaluation of Cells Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Elements of the LOC Platform
2.2. Cultured Microobjects
3. Results
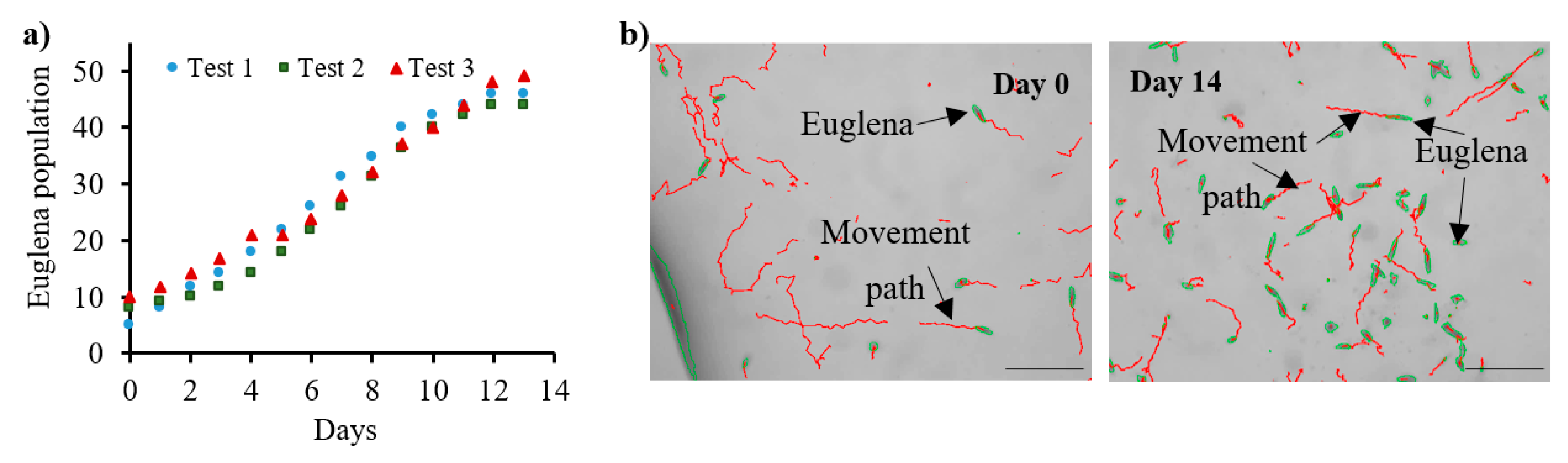
3.1. Euglena Gracilis Investigation
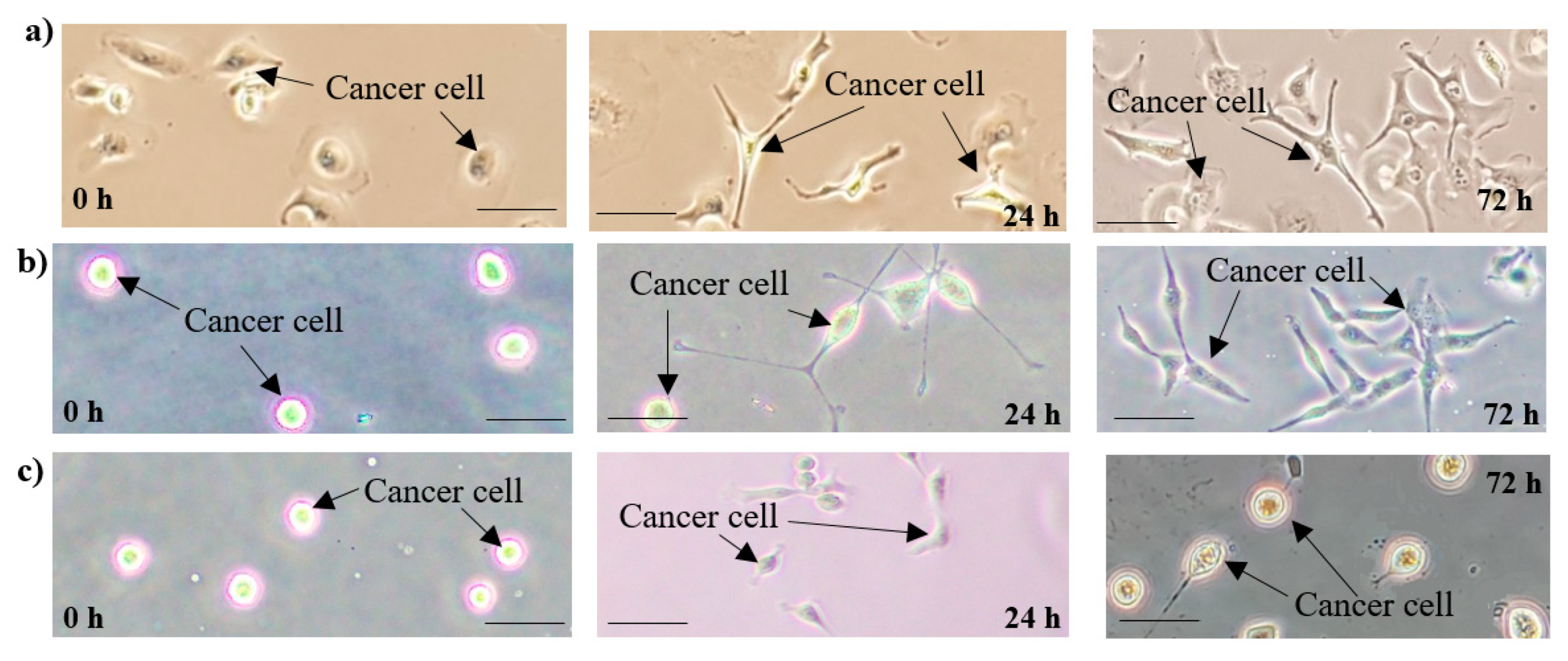
3.2. Ovarian Cancer Cells Development (Cell Line SKOV-3)
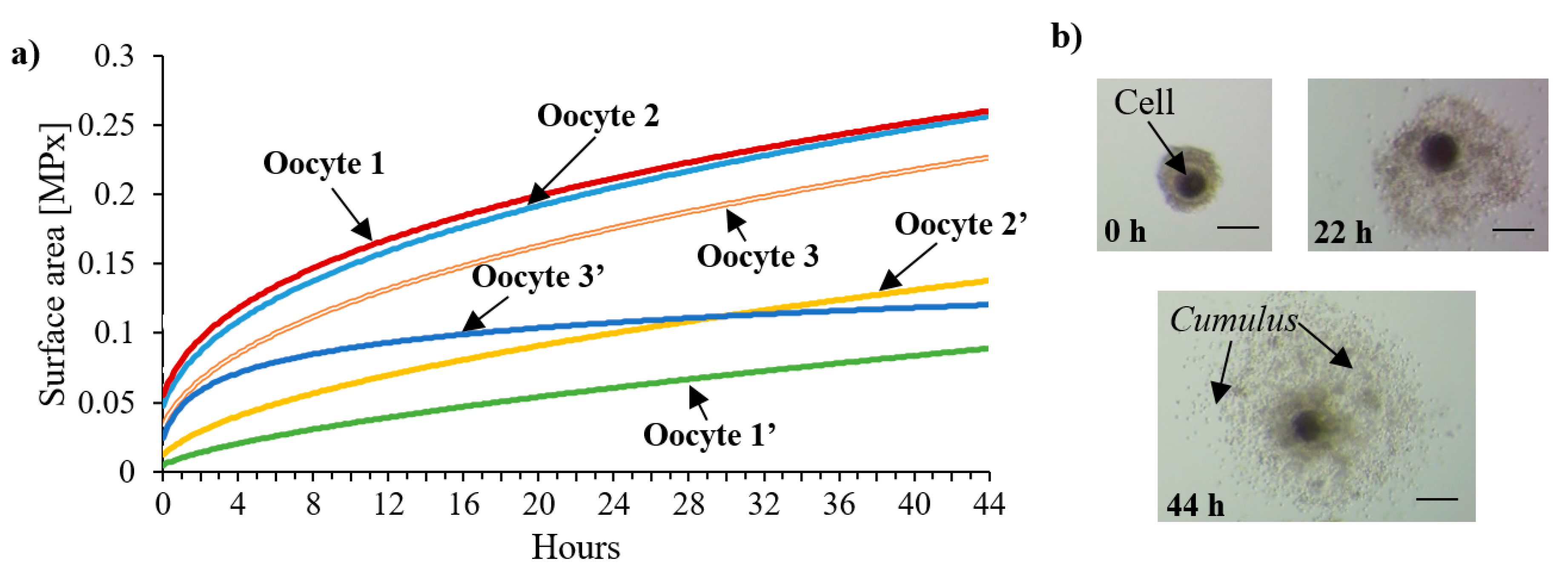
3.3. In Vitro Maturation (IVM) of Porcine Oocytes
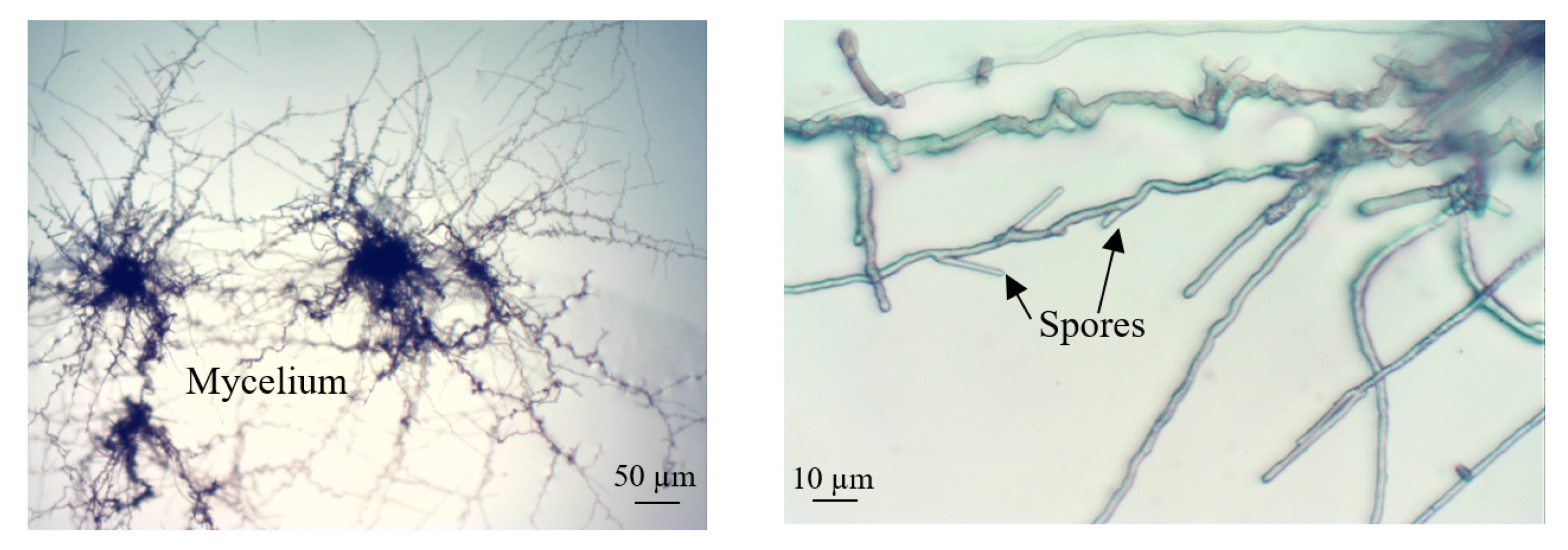
3.4. Culture of Cladosporium Macrocarpum
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sosa-Hernández, J.E.; Villalba-Rodríguez, A.M.; Romero-Castillo, K.D.; Aguilar-Aguila-Isaías, M.A.; García-Reyes, I.E.; Hernández-Antonio, A.; Ahmed, I.; Sharma, A.; Parra-Saldívar, R.; Iqbal, H. Organs-on-a-Chip Module: A Review from the Development and Applications Perspective. Micromachines 2018, 9, 536. [Google Scholar]
- Van Duinen, V.; Trietsch, S.J.; Joore, J.; Vulto, P.; Hankemeier, T. Microfluidic 3D cell culture: From tools to tissue models. Curr. Opin. Biotechnol. 2015, 35, 118–126. [Google Scholar] [CrossRef]
- Young, E.W.K.; Beebe, D.J. Fundamentals of microfluidic cell culture in controlled microenvironments. Chem. Soc. Rev. 2010, 39, 1036–1048. [Google Scholar] [CrossRef] [PubMed]
- Riedl, A.; Schlederer, M.; Pudelko, K.; Stadler, M.; Walter, S.; Unterleuthner, D.; Unger, C.; Kramer, N.; Hengstschläger, M.; Kenner, L.; et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J. Cell Sci. 2017, 130, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Cevenini, L.; Calabretta, M.M.; Lopreside, A.; Branchini, B.R.; Southworth, T.L.; Michelini, E.; Roda, A. Bioluminescence Imaging of Spheroids for High-throughput Longitudinal Studies on 3D Cell Culture Models. Photochem. Photobiol. 2017, 93, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Sart, S.; Tomasi, R.F.-X.; Amselem, G.; Baroud, C.N. Multiscale cytometry and regulation of 3D cell cultures on a chip. Nat. Commun. 2017, 8, 469. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. Slas Discov. Adv. Life Sci. R D 2017, 22, 456–472. [Google Scholar]
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Paul Solomon, F.D. 3D Cell Culture Systems: Advantages and Applications. J. Cell Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Gupta, N.; Liu, J.R.; Patel, B.; Solomon, D.E.; Vaidya, B.; Gupta, V. Microfluidics-based 3D cell culture models: Utility in novel drug discovery and delivery research. Bioeng. Transl. Med. 2016, 1, 63–81. [Google Scholar] [CrossRef]
- Bürgel, S.C.; Escobedo, C.; Haandbæk, N.; Hierlemann, A. On-chip electroporation and impedance spectroscopy of single-cells. Sens. Actuators B Chem. 2015, 210, 82–90. [Google Scholar] [CrossRef]
- Madison, A.C.; Royal, M.W.; Vigneault, F.; Chen, L.; Griffin, P.B.; Horowitz, M. Scalable Device for Automated Microbial Electroporation in a Digital Microfluidic Platform. ACS Synth. Biol. 2017, 6, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Bithi, S.S.; Vanapalli, S.A. Microfluidic cell isolation technology for drug testing of single tumor cells and their clusters. Sci. Rep. 2017, 7, 41707. [Google Scholar] [CrossRef] [PubMed]
- Valente, K.P.; Khetani, S.; Kolahchi, A.R.; Sanati-Nezhad, A.; Suleman, A.; Akbari, M. Microfluidic technologies for anticancer drug studies. Drug Discov. Today 2017, 22, 1654–1670. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, N.; Singha, S.; Panda, T.; Das, S.K. A diffusion based long-range and steady chemical gradient generator on a microfluidic device for studying bacterial chemotaxis. J. Micromech. Microeng. 2016, 26, 035011. [Google Scholar] [CrossRef]
- Walczak, R.; Śniadek, P.; Dziuban, J.A.; Kluger, J.; Chełmońska-Soyta, A. Supravital fluorometric apoptosis detection in a single mouse embryo using lab-on-a-chip. Lab Chip 2011, 11, 3263–3268. [Google Scholar] [CrossRef]
- Walczak, R.; Śniadek, P.; Dziuban, J.A.; Kempisty, B.; Jackowska, M.; Antosik, P.; Jaśkowski, J.M. Lab-on-a-chip spectrophotometric characterization of porcine oocytes, Sensor. Actuat. B Chem. 2012, 165, 38–43. [Google Scholar] [CrossRef]
- Berthier, E.; Young, E.W.; Beebe, D. Engineers are from PDMS-land, Biologists are from Polystyrenia. Lab Chip 2012, 12, 1224–1237. [Google Scholar] [CrossRef]
- Ren, K.; Zhou, J.; Wu, H. Materials for Microfluidic Chip Fabrication. Acc. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef]
- Regehr, K.J.; Domenech, M.; Koepsel, J.T.; Carver, K.C.; Ellison-Zelski, S.J.; Murphy, W.L.; Schuler, L.A.; Alarid, E.T.; Beebe, D.J. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip 2009, 9, 132–139. [Google Scholar] [CrossRef]
- Su, X.G.; Young, E.W.K.; Underkofler, H.A.S.; Kamp, T.J.; January, C.T.; Beebe, D.J. Microfluidic cell culture and its application in high-throughput drug screening Cardiotoxicity assay for hERG channels. J. Biomol. Screen. 2011, 16, 101–111. [Google Scholar] [CrossRef]
- Piruska, A.; Nikcevic, I.; Lee, S.H.; Ahn, C.; Heineman, W.R.; Limbach, P.A.; Seliskar, C.J. The autofluorescence of plastic materials and chips measured under laser irradiation. Lab Chip 2005, 5, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Domansky, K.; Sliz, J.D.; Wen, N.; Hinojosa, C.; Thompson, G.; Fraser, J.P.; Hamkins-Indik, T.; Hamilton, G.A.; Levner, D.; Ingber, D.E. SEBS elastomers for fabrication of microfluidic devices with reduced drug absorption by injection molding and extrusion. Microfluid. Nanofluid. 2017, 21, 107. [Google Scholar] [CrossRef]
- Kim, B.J.; Meng, E. Micromachining of Parylene C for bioMEMS. Polym. Adv. Technol. 2016, 27, 564–576. [Google Scholar] [CrossRef]
- Aymerich, M.; Gómez-Varela, A.I.; Álvarez, E.; Flores-Arias, M.T. Study of Different Sol-Gel Coatings to Enhance the Lifetime of PDMS Devices: Evaluation of Their Biocompatibility. Materials 2016, 9, 728. [Google Scholar] [CrossRef]
- Lee, S.H.; Shim, K.Y.; Kim, B.; Sung, J.H. Hydrogel-based three-dimensional cell culture for organ-on-a-chip applications. Biotechnol. Prog. 2017, 33, 580–589. [Google Scholar] [CrossRef]
- Xu, J.; Kawano, H.; Liu, W.; Hanada, Y.; Lu, P.; Miyawaki, A.; Midorikawa, K.; Sugioka, K. Controllable alignment of elongated microorganisms in 3D microspace using electrofluidic devices manufactured by hybrid femtosecond laser microfabrication. Microsyst. Nanoeng. 2017, 3, 16078. [Google Scholar] [CrossRef]
- Yalikun, Y.; Hosokawa, Y.; Iino, T.; Tanaka, Y. An all-glass 12 μm ultra-thin and flexible micro-fluidic chip fabricated by femtosecond laser processing. Lab Chip 2016, 16, 2427–2433. [Google Scholar] [CrossRef]
- Podwin, A.; Kubicki, W.; Dziuban, J.A. Study of the behavior of Euglena viridis, Euglena gracilis and Lepadella patella cultured in all-glass microaquarium. Biomed. Microdevices 2017, 19, 63. [Google Scholar] [CrossRef]
- Podwin, A.; Walczak, R.; Dziuban, J.A. A 3D Printed Membrane-Based Gas Microflow Regulator for On-Chip Cell Culture. Appl. Sci. 2018, 8, 579. [Google Scholar] [CrossRef]
- Lizanets, D.; Walczak, R. Cell detection and tracking in lab-on-a-chip devices by image processing. Opt. Appl. 2018, 48, 15–24. [Google Scholar]
- Pokrzywnicka, A.; Śniadek, P.; Małyszka, N.; Lizanets, D.; Kubicki, W.; Pawlak, P.; Walczak, R. MEMS cytometer for porcine oocyte deformation measurement. J. Micromech. Microeng. 2019, 29, 095004. [Google Scholar] [CrossRef]
- Przystupski, D.; Michel, O.; Rossowska, J.; Kwiatkowski, S.; Saczko, J.; Kulbacka, J. The modulatory effect of green tea catechin on drug resistance in human ovarian cancer cells. Med. Chem. Res. 2019, 28, 657–667. [Google Scholar] [CrossRef]
- Moor, R.M.; Dai, Y.; Lee, C.; Fulka, J., Jr. Oocyte maturation and embryonic failure. Hum. Reprod. Update 1998, 4, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Cramer, M.; Myers, J. Growth and Photosynthetic Characteristics of Euglena gracilis. Arch. Fur Mikrobiol. 1952, 17, 384–402. [Google Scholar] [CrossRef]
- Ozasa, K.; Lee, J.; Song, S.; Hara, M.; Maeda, M. Gas/liquid sensing via chemotaxis of euglena cells confined in an isolated micro-aquarium. Lab Chip 2013, 13, 4033–4039. [Google Scholar] [CrossRef]
- Ozasa, K.; Lee, J.; Song, S.; Hara, M.; Maeda, M. Two-dimensional optical feedback control of Euglena confined in closed-type microfluidic channels. Lab Chip 2011, 11, 1933–1940. [Google Scholar] [CrossRef]
- Ozasa, K.; Won, J.; Song, S.; Tamaki, S.; Ishikawa, T.; Maeda, M. Temporal change of photophobic step-up responses of Euglena gracilis investigated through motion analysis. PLoS ONE 2017, 12, e0172813. [Google Scholar] [CrossRef]
- Abeydeera, L.R.; Day, B.N. Fertilization and subsequent development in vitro of pig oocytes inseminated in a modified tris-buffered medium with frozen–thawed ejaculated spermatozoa. Biol. Reprod. 1997, 57, 729–734. [Google Scholar] [CrossRef]
- Provencher, D.M.; Lounis, H.; Champoux, L.; Tetrault, M.; Manderson, E.N.; Wang, J.C.; Eydoux, P.; Savoie, R.; Tonin, P.N.; Mes-Masson, A.M. Characterization of four novel epithelial ovarian cancer cell lines. Vitro Cell. Dev. Biol. Anim. 2000, 36, 357–361. [Google Scholar] [CrossRef]
- Wheeler, M.B.; Rubessa, M. Integration of microfluidics in animal in vitro embryo production. Mol. Hum. Reprod. 2017, 23, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Ferraz, M.A.M.M. Improved bovine embryo production in an oviduct-on-a-chip system: Prevention of poly-spermic fertilization and parthenogenic activation. Lab Chip 2017, 17, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.E.; Grossmann, G.; i Solvasa, X.C.; de Mello, A.J. Soil-on-a-Chip: Microfluidic platforms for environmental organismal studies. Lab Chip 2016, 16, 228–241. [Google Scholar] [CrossRef] [PubMed]





| Internal Structures Geometry | ||||||
|---|---|---|---|---|---|---|
| Overall Dimensions [mm] | Microchannel(s) | Microchamber(s) | Via Hole Diameter [mm] | |||
| Depth | Width | Depth | Width | |||
| LOC 1 | 76 × 26 | 5 μm | 500 μm | 100 μm | 4 mm | 1 |
| LOC 2 | 35 × 17 | 400 μm | 3.5 mm | - | - | 2 |
| LOC 3 | 76 × 26 | 150 μm | 1 mm | 150 μm | 7 mm | 1 |
| LOC 4 | 50 × 25 | 80/400 μm | 150/500 μm | - | - | 1/1.5 |
| Microchamber 1 | Microchamber 2 | |||
|---|---|---|---|---|
| Time [min] | Illumination [nm] | Mobility 1 | Illumination [nm] | Mobility 1 |
| 0 | Ambient | 2.17 | Ambient | 2.17 |
| 5 | 615 | ↑2.23 | 470 | ↓1.54 |
| 10 | 615 | ↑2.33 | 615 | ↑2.07 |
| 15 | 615 | ↓2.22 | 470 | ↓1.65 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Podwin, A.; Lizanets, D.; Przystupski, D.; Kubicki, W.; Śniadek, P.; Kulbacka, J.; Wymysłowski, A.; Walczak, R.; Dziuban, J.A. Lab-on-Chip Platform for Culturing and Dynamic Evaluation of Cells Development. Micromachines 2020, 11, 196. https://doi.org/10.3390/mi11020196
Podwin A, Lizanets D, Przystupski D, Kubicki W, Śniadek P, Kulbacka J, Wymysłowski A, Walczak R, Dziuban JA. Lab-on-Chip Platform for Culturing and Dynamic Evaluation of Cells Development. Micromachines. 2020; 11(2):196. https://doi.org/10.3390/mi11020196
Chicago/Turabian StylePodwin, Agnieszka, Danylo Lizanets, Dawid Przystupski, Wojciech Kubicki, Patrycja Śniadek, Julita Kulbacka, Artur Wymysłowski, Rafał Walczak, and Jan A. Dziuban. 2020. "Lab-on-Chip Platform for Culturing and Dynamic Evaluation of Cells Development" Micromachines 11, no. 2: 196. https://doi.org/10.3390/mi11020196
APA StylePodwin, A., Lizanets, D., Przystupski, D., Kubicki, W., Śniadek, P., Kulbacka, J., Wymysłowski, A., Walczak, R., & Dziuban, J. A. (2020). Lab-on-Chip Platform for Culturing and Dynamic Evaluation of Cells Development. Micromachines, 11(2), 196. https://doi.org/10.3390/mi11020196






