A Novel Microfluidic Device Integrated with Chitosan-Modified Capillaries for Rapid ZIKV Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Sample Preparation
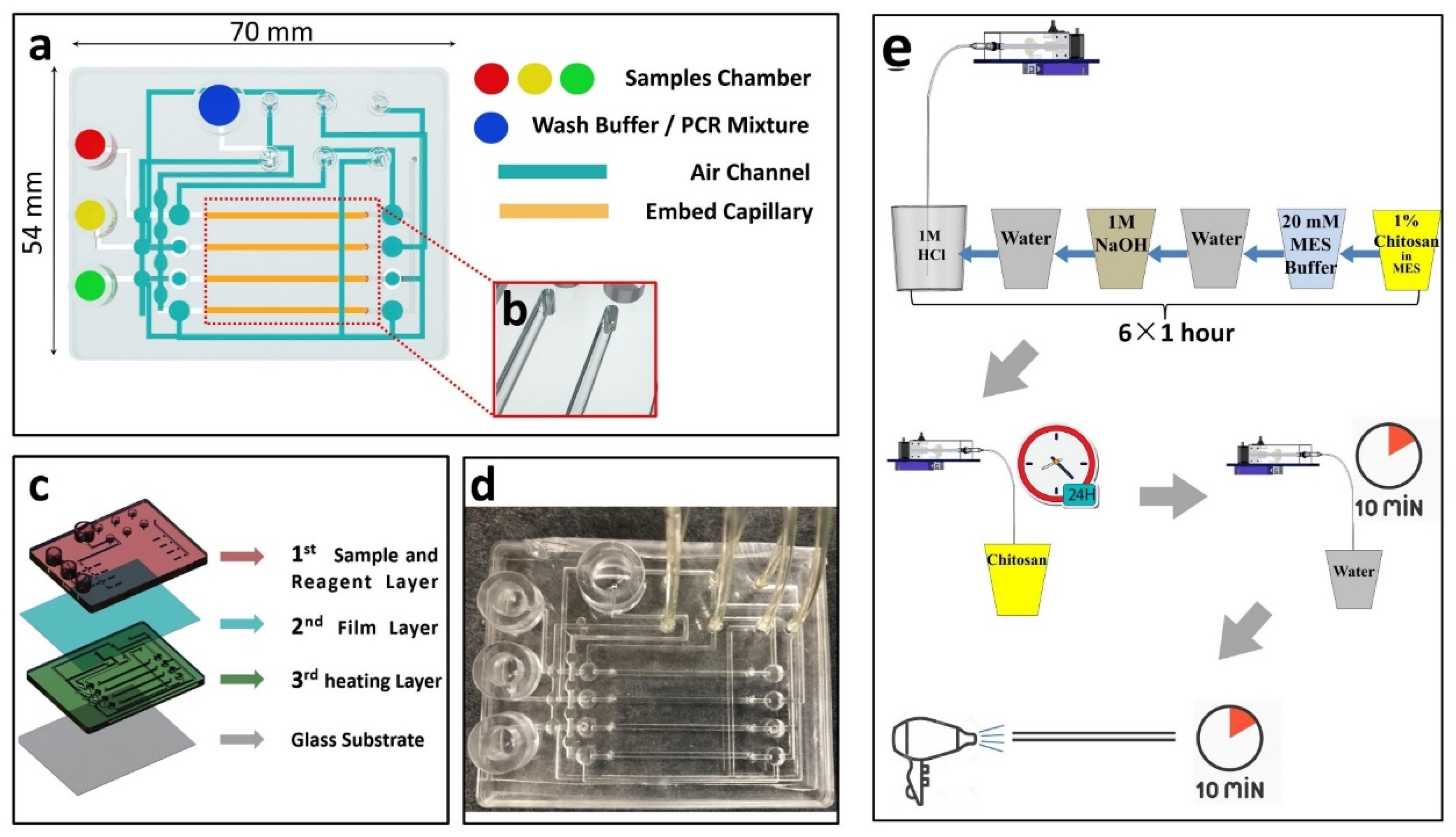
2.3. Design and Fabrication of the Capillary Microfluidic Chip
2.4. The Progress of Capillary Modified by Chitosan Solution
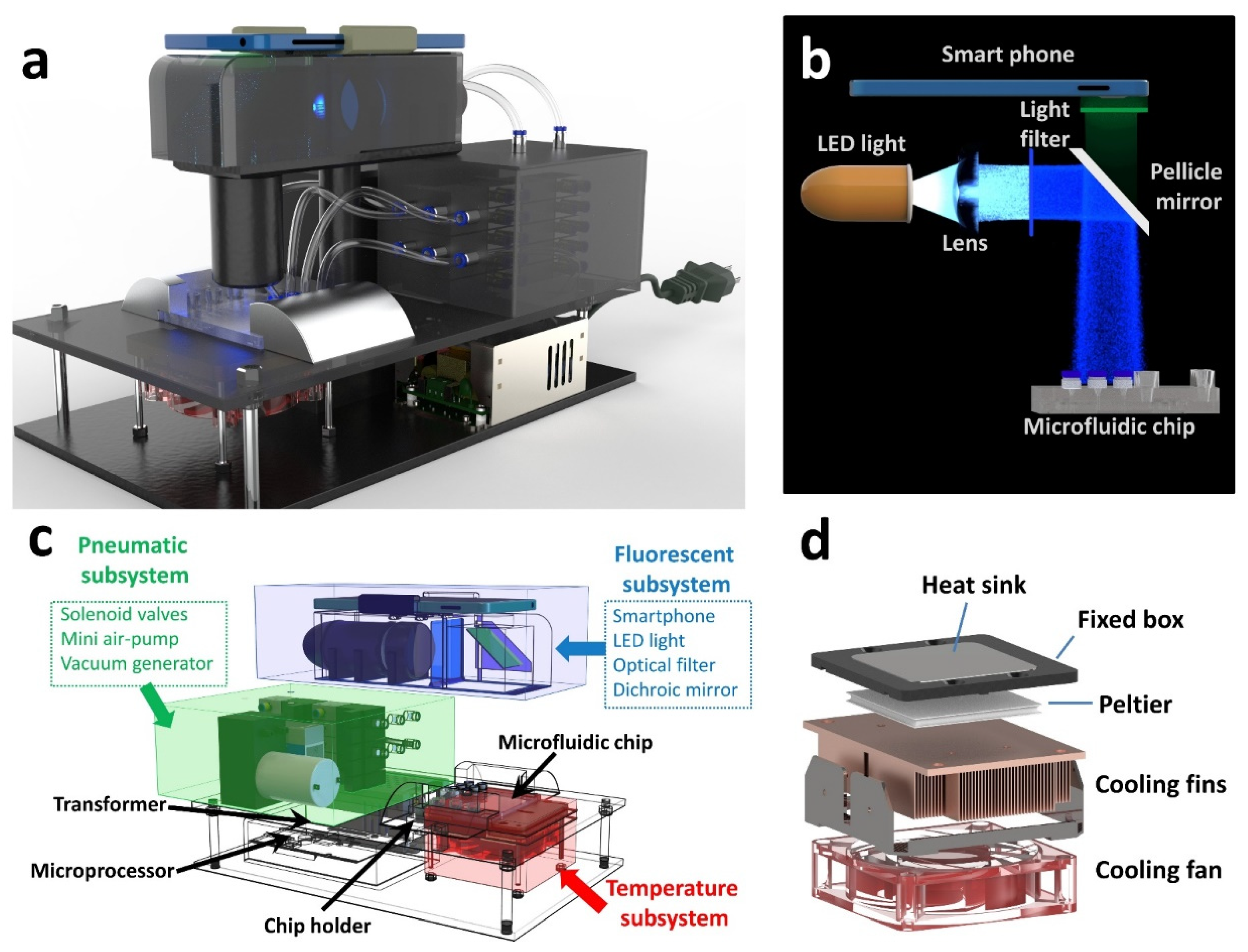
2.5. Custom-Made POC Device
2.6. Adsorption Kinetics of Nucleic Acid in Capillaries Modified by Chitosan
2.7. Preparation of in Situ PCR on the Chip
2.8. Fluorescence Signal Acquired and Data Analysis
3. Results
3.1. Nucleic Acids Extraction Strategy on the Capillary
3.2. Nucleic Acids Purification, Amplification, and Qualitative Detection on the C3-System
3.3. Characterization of Chitosan Modified Capillary and Confirmation of Adsorption Performance
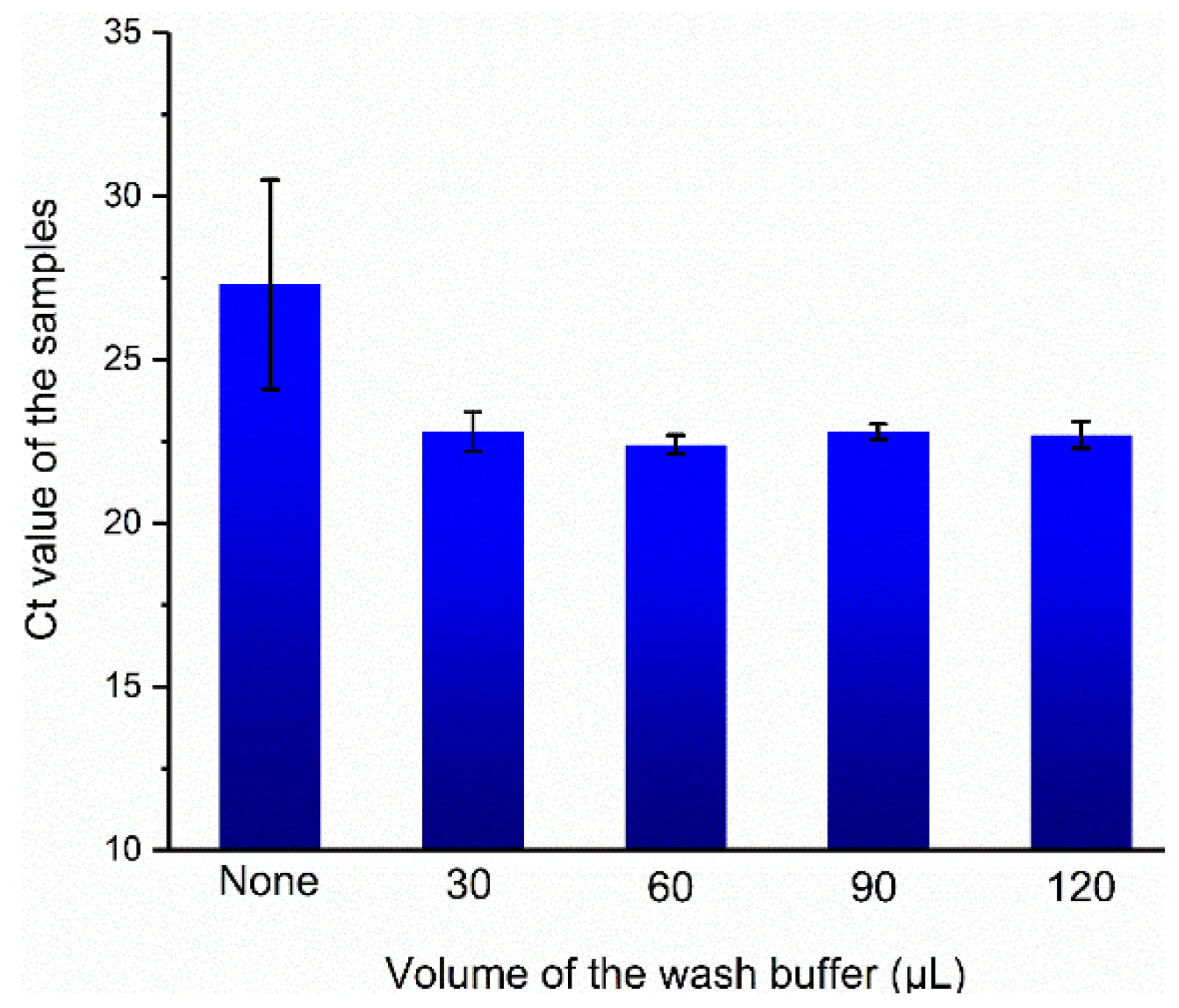
3.4. Optimization of Wash Conditions
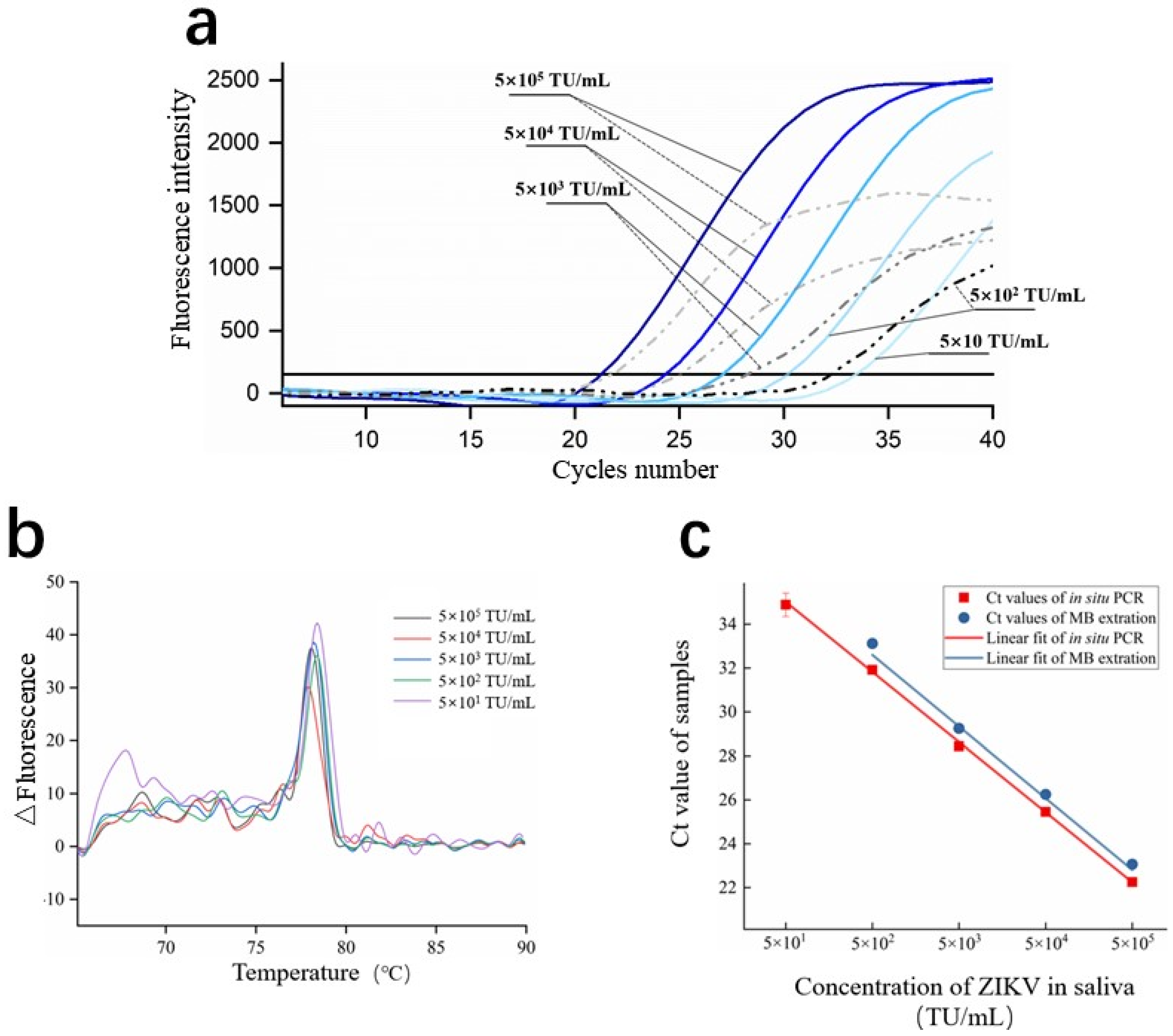
3.5. Real-Time PCR on C3-System
3.6. Amplification Efficiency of In Situ PCR
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wand, N.I.V.; Bonney, L.C.; Watson, R.J.; Graham, V.; Hewson, R. Point-of-care diagnostic assay for the detection of Zika virus using the recombinase polymerase amplification method. J. Gen. Virol. 2018, 99, 1012–1026. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Zika Virus. National Center for Emerging and Zoonotic Infectious Diseases (NCEZID). Available online: http://www.cdc.gov/zika/ (accessed on 5 February 2016).
- Sardi, S.I.; Somasekar, S.; Naccache, S.N.; Bandeira, A.C.; Tauro, L.B.; Campos, G.S.; Chiu, C.Y. Coinfections of Zika and Chikungunya Viruses in Bahia, Brazil, Identified by Metagenomic Next-Generation Sequencing. J. Clin. Microbiol. 2016, 54, 2348–2353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jin, X.; Zhu, Z.; Huang, L.; Liang, S.; Xu, Y.; Liao, R.; Zhou, L.; Zhang, Y.; Wilder-Smith, A. Early detection of Zika virus infection among travellers from areas of ongoing transmission in China. J. Travel Med. 2016, 23. [Google Scholar] [CrossRef] [PubMed]
- Paz-Bailey, G.; Rosenberg, E.S.; Doyle, K.; Munoz-Jordan, J.; Santiago, G.A.; Klein, L.; Perez-Padilla, J.; Medina, F.A.; Waterman, S.H.; Adams, L.E.; et al. Persistence of Zika Virus in Body Fluids-Final Report. N. Engl. J. Med. 2018, 379, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Bugallo, G.; Augusto Piedra, L.; Rodriguez, M.; Bisset, J.A.; Lourenco-de-Oliveira, R.; Weaver, S.C.; Vasilakis, N.; Vega-Rua, A. Vector-borne transmission and evolution of Zika virus. Nat. Ecol. Evol. 2019, 3, 561–569. [Google Scholar] [CrossRef]
- Haug, C.J.; Kieny, M.P.; Murgue, B. The Zika Challenge. N. Engl. J. Med. 2016, 374, 1801–1803. [Google Scholar] [CrossRef]
- Ward, M.J.; Alger, J.; Berrueta, M.; Bock, H.; Buekens, P.; Luisa Cafferata, M.; Ciganda, A.; Garcia, J.; Garcia, K.; Lopez, W.; et al. Zika Virus and the World Health Organization Criteria for Determining Recent Infection Using Plaque Reduction Neutralization Testing. Am. J. Trop Med. Hyg. 2018, 99, 780–782. [Google Scholar] [CrossRef]
- Zhu, C.-C.; Cui, J.-S.; Hu, A.-Z.; Yang, K.; Zhao, J.; Liu, Y.; Deng, G.-Q.; Zhu, L. Multiplex Nested Solid Phase PCR-Array Chip for Simultaneous Detection of Highly Pathogenic Microorganisms. Chin. J. Anal. Chem. 2019, 47, 1751–1758. [Google Scholar] [CrossRef]
- Zhu, C.; Hu, A.; Cui, J.; Yang, K.; Zhu, X.; Liu, Y.; Deng, G.; Zhu, L. A Lab-on-a-Chip Device Integrated DNA Extraction and Solid Phase PCR Array for the Genotyping of High-Risk HPV in Clinical Samples. Micromachines 2019, 10, 537. [Google Scholar] [CrossRef]
- Trieu, N.; Andreasen, S.Z.; Wolff, A.; Bang, D.D. From Lab on a Chip to Point of Care Devices: The Role of Open Source Microcontrollers. Micromachines 2018, 9, 403. [Google Scholar] [CrossRef]
- Mauk, M.G.; Song, J.; Liu, C.; Bau, H.H. Simple Approaches to Minimally-Instrumented, Microfluidic-Based Point-of-Care Nucleic Acid Amplification Tests. Biosensors 2018, 8, 17. [Google Scholar] [CrossRef]
- Hartman, M.R.; Ruiz, R.C.H.; Hamada, S.; Xu, C.Y.; Yancey, K.G.; Yu, Y.; Han, W.; Luo, D. Point-of-care nucleic acid detection using nanotechnology. Nanoscale 2013, 5, 10141–10154. [Google Scholar] [CrossRef]
- Zarei, M. Portable biosensing devices for point-of-care diagnostics: Recent developments and applications. TrAC Trends Anal. Chem. 2017, 91, 26–41. [Google Scholar] [CrossRef]
- Hui, J.; Gu, Y.; Zhu, Y.; Chen, Y.; Guo, S.-J.; Tao, S.-C.; Zhang, Y.; Liu, P. Multiplex sample-to-answer detection of bacteria using a pipette-actuated capillary array comb with integrated DNA extraction, isothermal amplification, and smartphone detection. Lab. Chip 2018. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Mason, M.G.; Wang, Y.; Wee, E.; Turni, C.; Blackall, P.J.; Trau, M.; Botella, J.R. Nucleic acid purification from plants, animals and microbes in under 30 seconds. PLoS Biol. 2017, 15, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Mauk, M.; Chen, D.; Qiu, X.; Kim, J.; Gale, B.; Bau, H.H. A PCR reactor with an integrated alumina membrane for nucleic acid isolation. Analyst 2010, 135, 2408–2414. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Geva, E.; Mauk, M.; Qiu, X.; Abrams, W.R.; Malamud, D.; Curtis, K.; Owen, S.M.; Bau, H.H. An isothermal amplification reactor with an integrated isolation membrane for point-of-care detection of infectious diseases. Analyst 2011, 136, 2069–2076. [Google Scholar] [CrossRef]
- Jangam, S.R.; Yamada, D.H.; McFall, S.M.; Kelso, D.M. Rapid, Point-of-Care Extraction of Human Immunodeficiency Virus Type 1 Proviral DNA from Whole Blood for Detection by Real-Time PCR. J. Clin. Microbiol. 2009, 47, 2363–2368. [Google Scholar] [CrossRef]
- Jangam, S.R.; Agarwal, A.K.; Sur, K.; Kelso, D.M. A point-of-care PCR test for HIV-1 detection in resource-limited settings. Biosens. Bioelectron. 2013, 42, 69–75. [Google Scholar] [CrossRef]
- Gan, W.; Gu, Y.; Han, J.; Li, C.X.; Sun, J.; Liu, P. Chitosan-Modified Filter Paper for Nucleic Acid Extraction and “in Situ PCR” on a Thermoplastic Microchip. Anal. Chem. 2017, 89, 3568–3575. [Google Scholar] [CrossRef]
- Schlappi, T.S.; McCalla, S.E.; Schoepp, N.G.; Ismagilov, R.F. Flow-through Capture and in Situ Amplification Can Enable Rapid Detection of a Few Single Molecules of Nucleic Acids from Several Milliliters of Solution. Anal. Chem. 2016, 88, 7647–7653. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Toley, B.J. Paper-based nucleic acid amplification tests for point-of-care diagnostics. Analyst 2018, 143, 2213–2234. [Google Scholar] [CrossRef] [PubMed]
- Trinh, K.T.L.; Zhang, Y.; Lee, N.Y. One-step DNA purification and amplification on an integrated plastic microdevice for on-site identification of foodborne pathogens. Anal. Chim. Acta 2018, 1040, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhou, X.; Xing, D. Lab-on-capillary: A rapid, simple and quantitative genetic analysis platform integrating nucleic acid extraction, amplification and detection. Lab. Chip 2017, 17, 4334–4341. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, J.; Wu, X.; Zhu, C.; Liu, Y.; Wang, A.; Deng, G.; Zhu, L.J.B.; Equipment, B. A rapid microfluidic platform with real-time fluorescence detection system for molecular diagnosis. Biotechnol. Biotechnol. Equip. 2019, 33, 223–230. [Google Scholar] [CrossRef]
- Li, B.; Li, L.; Guan, A.; Dong, Q.; Ruan, K.; Hu, R.; Li, Z. A smartphone controlled handheld microfluidic liquid handling system. Lab. Chip 2014, 14, 4085–4092. [Google Scholar] [CrossRef]
- Li, L.; Zhu, L.; Jiang, Q.; Liu, G. Optimizing of Thermal Cycler Control in Microfluidic PCR System. In Information Technology Applications in Industry; Zhang, J., Wang, Z.J., Zhu, S.R., Meng, X.M., Eds.; Trans Tech Publications Ltd.: Bäch SZ, Switzerland, 2013; Volume 263–266, p. 713. [Google Scholar]
- Zhu, L.; Zhu, C.; Deng, G.; Zhang, L.; Zhao, S.; Lin, J.; Li, L.; Jiao, P.; Liao, M.; Liu, Y. Rapid identification of H5 avian influenza virus in chicken throat swab specimens using microfluidic real-time RT-PCR. Anal. Methods 2014, 6, 2628–2632. [Google Scholar] [CrossRef]
- Petralia, S.; Sciuto, E.L.; Conoci, S. A novel miniaturized biofilter based on silicon micropillars for nucleic acid extraction. Analyst 2017, 142, 140–146. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Matsuda, K.; Uehara, M.; Honda, T.; Saito, Y. A novel automated device for rapid nucleic acid extraction utilizing a zigzag motion of magnetic silica beads. Anal. Chim. Acta 2016, 906, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Monica Bravo-Anaya, L.; Gricelda Fernandez-Solis, K.; Rosselgong, J.; Elena Nano-Rodriguez, J.L.; Carvajal, F.; Rinaudo, M. Chitosan-DNA polyelectrolyte complex: Influence of chitosan characteristics and mechanism of complex formation. Int. J. Biol. Macromol. 2019, 126, 1037–1049. [Google Scholar] [CrossRef]
- Oliveira, D.B.L.; Durigon, G.S.; Mendes, E.A.; Ladner, J.T.; Andreata-Santos, R.; Araujo, D.B.; Botosso, V.F.; Paola, N.D.; Neto, D.F.L.; Cunha, M.P.; et al. Persistence and Intra-Host Genetic Evolution of Zika Virus Infection in Symptomatic Adults: A Special View in the Male Reproductive System. Viruses 2018, 10, 615. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, Z.; Duan, Q.; Alsaedi, A.; Hayat, T.; Chen, C. Decoration of ZIF-8 on polypyrrole nanotubes for highly efficient and selective capture of U(VI). J. Clean. Prod. 2018, 204, 896–905. [Google Scholar] [CrossRef]
- Li, J.; Li, X.; Alsaedi, A.; Hayat, T.; Chen, C. Synthesis of highly porous inorganic adsorbents derived from metal-organic frameworks and their application in efficient elimination of mercury(II). J. Colloid Interface Sci. 2018, 517, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, J.D.A.B.; de Franca, C.A.; Gouveia, J.J.D.S.; Gouveia, G.V.; da Costa, M.M.; de Oliveira, H.P. Eudragit E100/poly(ethylene oxide) electrospun fibers for DNA removal from aqueous solution. J. Appl. Polym. Sci. 2019, 136. [Google Scholar] [CrossRef]
- Cai, P.; Huang, Q.; Zhang, X.; Chen, H. Adsorption of DNA on clay minerals and various colloidal particles from an Alfisol. Soil Biol. Biochem. 2006, 38, 471–476. [Google Scholar] [CrossRef]
- Zhang, X.; Servos, M.R.; Liu, J. Surface Science of DNA Adsorption onto Citrate-Capped Gold Nanoparticles. Langmuir 2012, 28, 3896–3902. [Google Scholar] [CrossRef]
- Pandit, K.R.; Nanayakkara, I.A.; Cao, W.; Raghavan, S.R.; White, I.M. Capture and Direct Amplification of DNA on Chitosan Microparticles in a Single PCR-Optimal Solution. Anal. Chem. 2015, 87, 11022–11029. [Google Scholar] [CrossRef]
- Nanayakkara, I.A.; Cao, W.; White, I.M. Simplifying Nucleic Acid Amplification from Whole Blood with Direct Polymerase Chain Reaction on Chitosan Microparticles. Anal. Chem. 2017, 89, 3773–3779. [Google Scholar] [CrossRef]



| Procedure on Chip | Opened Microvalves | Closed Microvalves | Energized Solenoid Valves |
|---|---|---|---|
| Samples loading/lysed samples removal | A B C H I J K L M | D E F G N O | 1 4 6 8 10 |
| Wash buffer loading/removal | D E F G H I J K L M | A B C N O | 2 3 6 8 10 |
| PCR mixture loading | D E F G H I J K L M N O P | A B C | 2 3 6 9 |
| Sample lysis/PCR amplification | D E F G | A B C H I J K L M N O | 2 3 5 7 8 10 |
| Parameter of the Langmuir Fit | Parameter of the Freundlich Fit | ||||
|---|---|---|---|---|---|
| R2 | N | R2 | |||
| 282.4 | 0.255 | 0.982 | 119.7 | 5.094 | 0.927 |
| Parameter of the Linear Fit of In Situ PCR | Parameter of the Linear Fit of MBs Extraction | ||||||
|---|---|---|---|---|---|---|---|
| R2 | Efficiency | R2 | Efficiency | ||||
| −3.192 | 8.47 | 0.998 | 98.3% | −3.189 | 10.17 | 0.994 | 98.1% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Zhao, J.; Hu, A.; Pan, J.; Deng, G.; Hua, C.; Zhu, C.; Liu, Y.; Yang, K.; Zhu, L. A Novel Microfluidic Device Integrated with Chitosan-Modified Capillaries for Rapid ZIKV Detection. Micromachines 2020, 11, 186. https://doi.org/10.3390/mi11020186
Zhu X, Zhao J, Hu A, Pan J, Deng G, Hua C, Zhu C, Liu Y, Yang K, Zhu L. A Novel Microfluidic Device Integrated with Chitosan-Modified Capillaries for Rapid ZIKV Detection. Micromachines. 2020; 11(2):186. https://doi.org/10.3390/mi11020186
Chicago/Turabian StyleZhu, Xinchao, Jun Zhao, Anzhong Hu, Jingyu Pan, Guoqing Deng, Changyi Hua, Cancan Zhu, Yong Liu, Ke Yang, and Ling Zhu. 2020. "A Novel Microfluidic Device Integrated with Chitosan-Modified Capillaries for Rapid ZIKV Detection" Micromachines 11, no. 2: 186. https://doi.org/10.3390/mi11020186
APA StyleZhu, X., Zhao, J., Hu, A., Pan, J., Deng, G., Hua, C., Zhu, C., Liu, Y., Yang, K., & Zhu, L. (2020). A Novel Microfluidic Device Integrated with Chitosan-Modified Capillaries for Rapid ZIKV Detection. Micromachines, 11(2), 186. https://doi.org/10.3390/mi11020186





