Nanosensors-Assisted Quantitative Analysis of Biochemical Processes in Droplets
Abstract
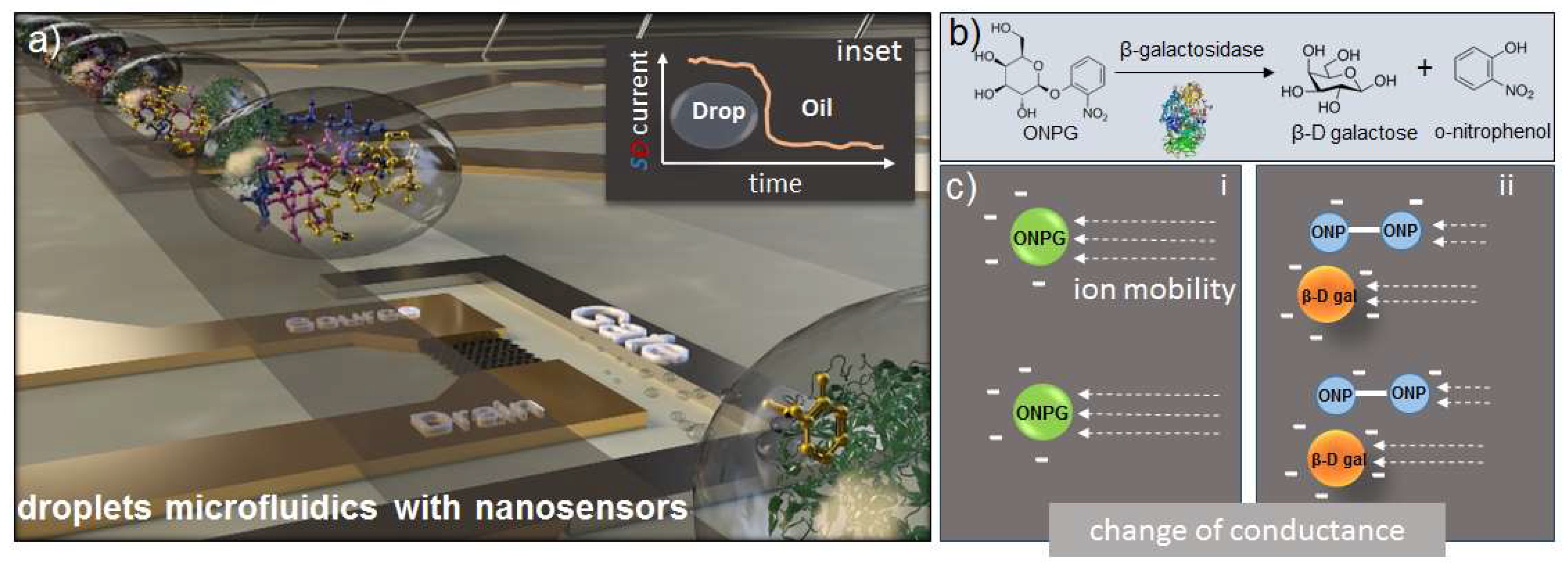
1. Introduction
2. Materials and Methods
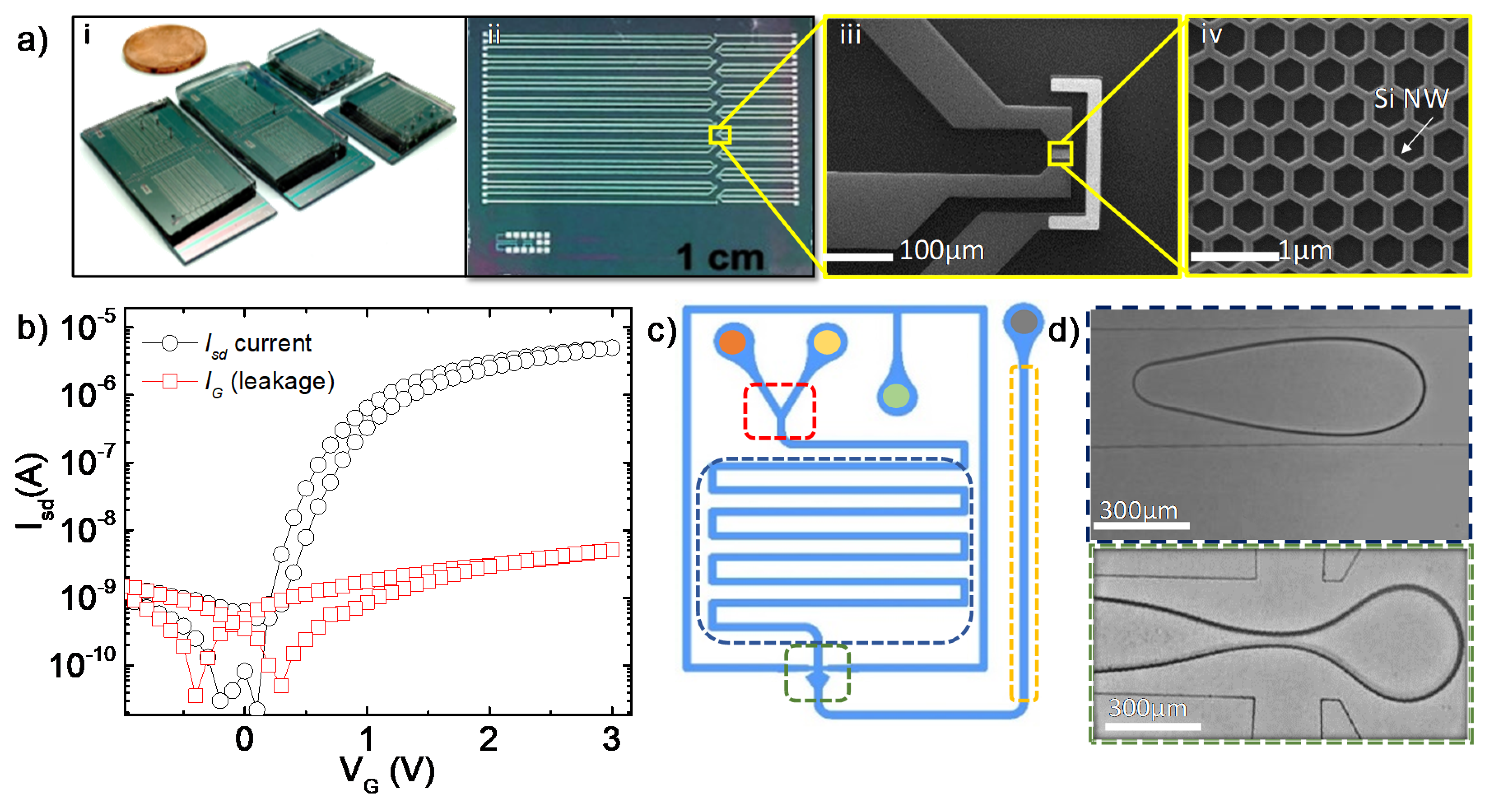
2.1. Fabrication and Characterization of the FET Devices
2.2. Microfluidic Chip
3. Results
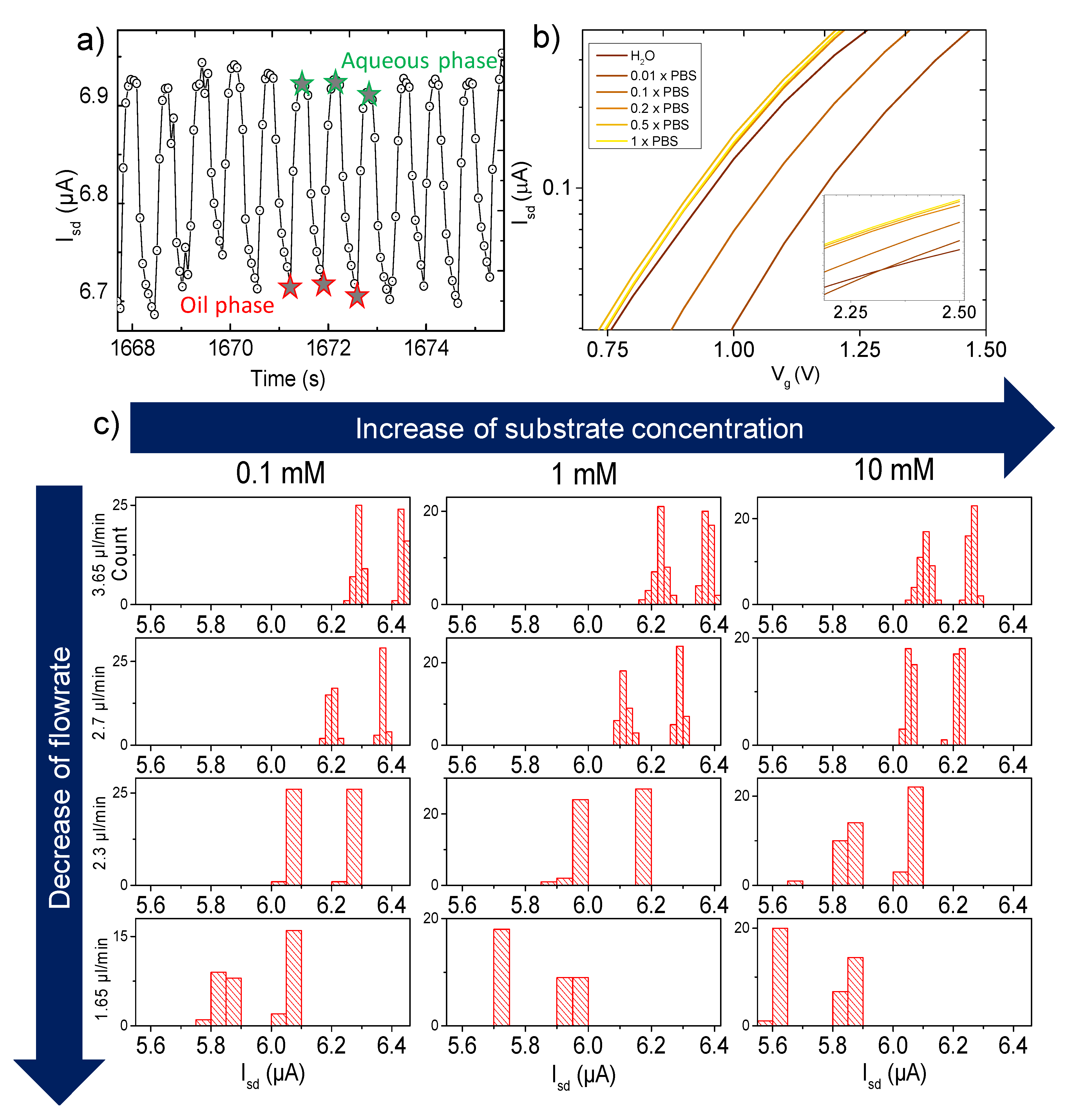
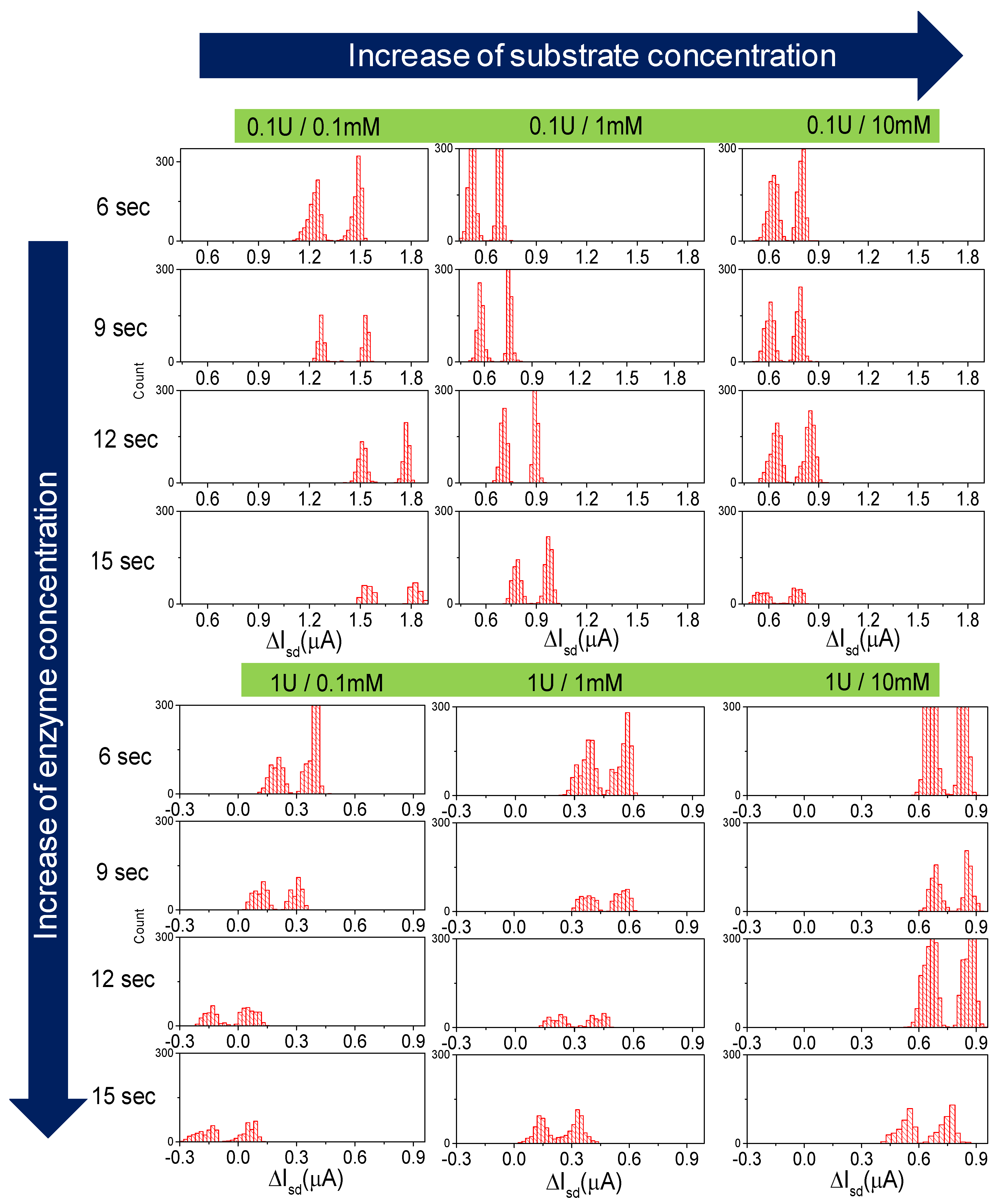
3.1. Measurements
3.2. Enzymatic Reaction
3.3. Spectrophotometry as Reference Test
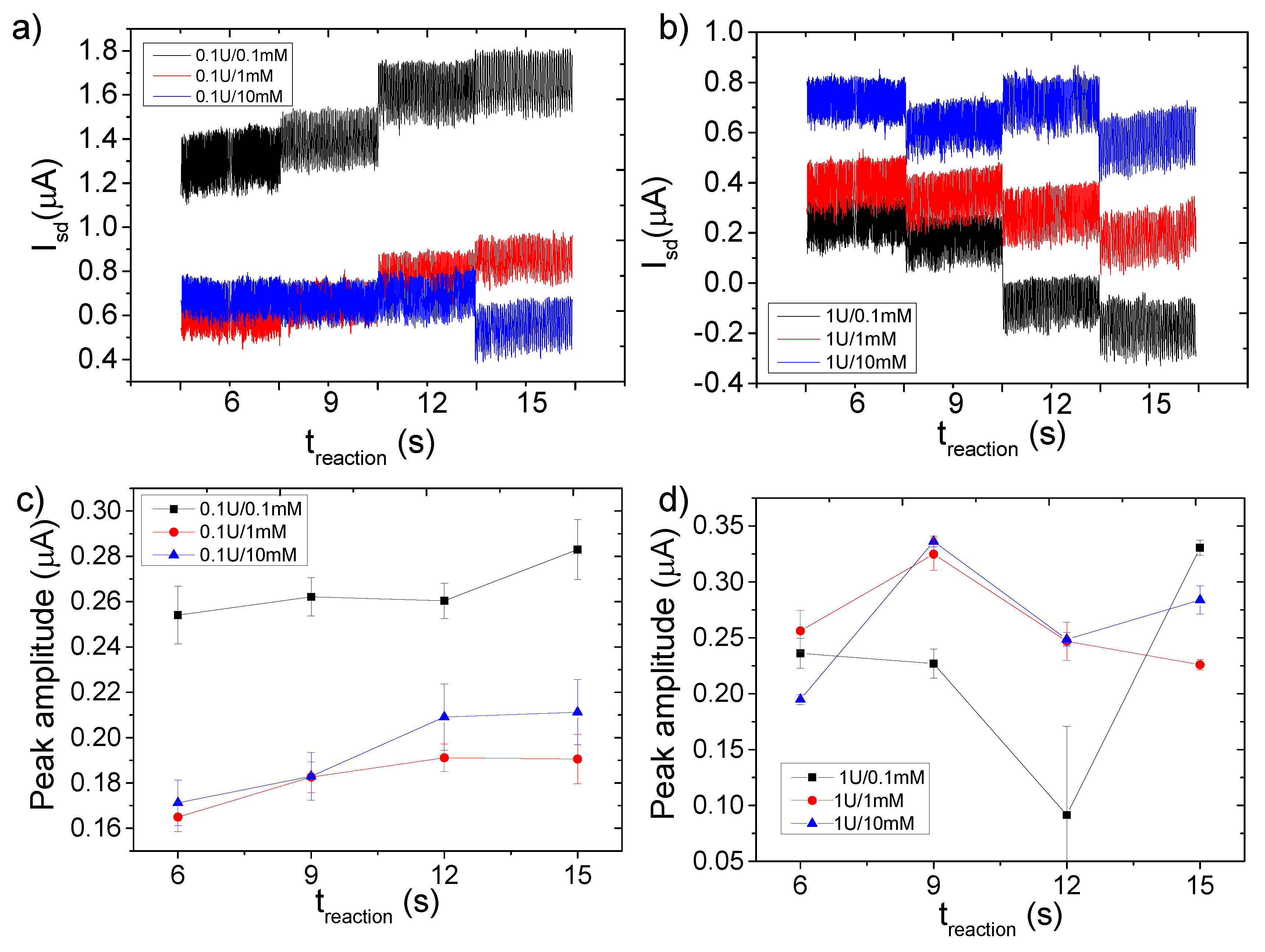
3.4. FET Monitoring of β-Galactosidase Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Abgrall, P.; Gué, A.-M. Lab-on-chip technologies: Making a microfluidic network and coupling it into a complete microsystem—A review. J. Micromech. Microeng. 2007, 17, R15–R49. [Google Scholar] [CrossRef]
- Hejazian, M.; Li, W.; Nguyen, N.-T. Lab on a chip for continuous-flow magnetic cell separation. Lab a Chip 2015, 15, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.J.; Agirregabiria, M.; Berganzo, J.; Mayora, K.; Elizalde, J.; Calle, A.; Domínguez, C.; Lechuga, L. Microfluidic-optical integrated CMOS compatible devices for label-free biochemical sensing. J. Micromech. Microeng. 2006, 16, 1006–1016. [Google Scholar] [CrossRef]
- Early detection: A long road ahead. Nat. Rev. Cancer 2018, 18, 401. [CrossRef]
- Riemann, D.; Cwikowski, M.; Turzer, S.; Giese, T.; Grallert, M.; Schütte, W.; Seliger, B. Blood immune cell biomarkers in lung cancer. Clin. Exp. Immunol. 2019, 195, 179–189. [Google Scholar] [CrossRef]
- Malamud, D. Saliva as a diagnostic fluid. Dent. Clin. N. Am. 2011, 55, 159–178. [Google Scholar] [CrossRef]
- Bidin, M.Z.; Shah, A.M.; Stanslas, J.; Seong, C.L.T.; Lim, C.T.S. Blood and urine biomarkers in chronic kidney disease: An update. Clin. Chim. Acta 2019, 495, 239–250. [Google Scholar] [CrossRef]
- Javaid, M.A.; Ahmed, A.S.; Durand, R.; Tran, S.D. Saliva as a diagnostic tool for oral and systemic diseases. J. Oral Biol. Craniofacial Res. 2016, 6, 67–76. [Google Scholar] [CrossRef]
- Agrawal, M.; Biswas, A. Molecular diagnostics of neurodegenerative disorders. Front. Mol. Biosci. 2015, 2, 360. [Google Scholar] [CrossRef]
- Singh, S.; Nagpal, M.; Singh, P.; Chauhan, P.; Zaidi, M.A. Tumor markers: A diagnostic tool. Natl. J. Maxillofac. Surg. 2016, 7, 17–20. [Google Scholar] [CrossRef]
- Aydin, S.; Ugur, K.; Aydin, S.; Sahin, I.; Yardim, M. Biomarkers in acute myocardial infarction: Current perspectives. Vasc. Heal. Risk Manag. 2019, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- McDowell, S.E.; Ferner, R.E. Biochemical monitoring of patients treated with antihypertensive therapy for adverse drug reactions. Drug Saf. 2011, 34, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Citartan, M.; Gopinath, S.C.B.; Tominaga, J.; Tang, T.-H. Label-free methods of reporting biomolecular interactions by optical biosensors. Analyst 2013, 138, 3576. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W. Poly(indole-5-carboxylic acid)-functionalized ZnO nanocomposite for electrochemical DNA hybridization detection. J. Solid State Electrochem. 2015, 20, 499–506. [Google Scholar] [CrossRef]
- Mazzaracchio, V.; Neagu, D.; Porchetta, A.; Marcoccio, E.; Pomponi, A.; Faggioni, G.; D’Amore, N.; Notargiacomo, A.; Pea, M.; Moscone, D.; et al. A label-free impedimetric aptasensor for the detection of Bacillus anthracis spore simulant. Biosens. Bioelectron. 2019, 126, 640–646. [Google Scholar] [CrossRef]
- Lin, G.; Baraban, L.; Han, L.; Karnaushenko, D.; Makarov, D.; Cuniberti, G.; Schmidt, O.G. Magnetoresistive emulsion analyzer. Sci. Rep. 2013, 3, 2548. [Google Scholar] [CrossRef]
- Ibarlucea, B.; Rim, T.; Baek, C.K.; De Visser, J.A.; Baraban, L.; Cuniberti, G. Nanowire sensors monitor bacterial growth kinetics and response to antibiotics. Lab Chip 2017, 17, 4283–4293. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, D.; Xu, L.; Li, Q.; Song, J.; Xu, S.; Xing, R.; Song, H. A sensitive label–free amperometric immunosensor for alpha-fetoprotein based on gold nanorods with different aspect ratio. Sci. Rep. 2015, 5, 9939. [Google Scholar] [CrossRef]
- Boisen, A.; Dohn, S.; Keller, S.S.; Schmid, S.; Tenje, M. Cantilever-like micromechanical sensors. Rep. Prog. Phys. 2011, 74, 36101. [Google Scholar] [CrossRef]
- Swierczewska, M.; Liu, G.; Lee, S.; Chen, X. ChemInform abstract: High-sensitivity nanosensors for biomarker detection. ChemInform 2012, 43, 2641–2655. [Google Scholar] [CrossRef]
- Rao, Y.; Zhang, G. Enhancing the sensitivity of SAW sensors with nanostructures. Curr. Nanosci. 2006, 2, 311–318. [Google Scholar] [CrossRef]
- Cui, Y.; Wei, Q.; Park, H.; Lieber, C.M. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science 2001, 293, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.G.; Wróbel, P.S.; Bachmatiuk, A.; Sun, J.; Gemming, T.; Liu, Z.; Rümmeli, M.H. Carbon nanostructures as a multi-functional platform for sensing applications. Chemosensors 2018, 6, 60. [Google Scholar] [CrossRef]
- Klinghammer, S.; Uhlig, T.; Patrovsky, F.; Böhm, M.; Schütt, J.; Pütz, N.; Baraban, L.; Eng, L.M.; Cuniberti, G.; Schuett, J. Plasmonic biosensor based on vertical arrays of gold nanoantennas. ACS Sens. 2018, 3, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Patolsky, F.; Cui, Y.; Wang, W.U.; Lieber, C.M. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat. Biotechnol. 2005, 23, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Sakata, T. Molecular-charge-contact-based ion-sensitive field-effect transistor sensor in microfluidic system for protein sensing. Sensors 2019, 19, 3393. [Google Scholar] [CrossRef] [PubMed]
- Schütt, J.; Ibarlucea, B.; Illing, R.; Zörgiebel, F.; Pregl, S.; Nozaki, D.; Weber, W.M.; Mikolajick, T.; Baraban, L.; Cuniberti, G.; et al. Compact nanowire sensors probe microdroplets. Nano Lett. 2016, 16, 4991–5000. [Google Scholar] [CrossRef]
- Late, D.J.; Huang, Y.-K.; Liu, B.; Acharya, J.; Shirodkar, S.N.; Luo, J.; Yan, A.; Charles, D.; Waghmare, U.V.; Dravid, V.P.; et al. Sensing behavior of atomically thin-layered MoS2 transistors. ACS Nano 2013, 7, 4879–4891. [Google Scholar] [CrossRef]
- Postek, W.; Gargulinski, P.; Scheler, O.; Kaminski, T.S.; Garstecki, P. Microfluidic screening of antibiotic susceptibility at a single-cell level shows the inoculum effect of cefotaxime on E. coli. Lab a Chip 2018, 18, 3668–3677. [Google Scholar] [CrossRef]
- Scheler, O.; Postek, W.; Garstecki, P. Recent developments of microfluidics as a tool for biotechnology and microbiology. Curr. Opin. Biotechnol. 2019, 55, 60–67. [Google Scholar] [CrossRef]
- Rambach, R.W.; Biswas, P.; Yadav, A.; Garstecki, P.; Franke, T. Fast selective trapping and release of picoliter droplets in a 3D microfluidic PDMS multi-trap system with bubbles. Analyst 2018, 143, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, T.S.; Garstecki, P.; Scheler, O. Droplet microfluidics for microbiology: Techniques, applications and challenges. Lab a Chip 2016, 16, 2168–2187. [Google Scholar] [CrossRef] [PubMed]
- Debski, P.R.; Sklodowska, K.; Michalski, J.A.; Korczyk, P.M.; Dolata, M.; Jakiela, S. Continuous recirculation of microdroplets in a closed loop tailored for screening of bacteria cultures. Micromachines 2018, 9, 469. [Google Scholar] [CrossRef] [PubMed]
- Yakdi, N.E.; Bricault, D.; Huet, F.; Ngo, K. Detection and sizing of single droplets flowing in a microfluidic device by impedance measurement. Procedia Eng. 2016, 168, 1466–1470. [Google Scholar] [CrossRef]
- Kong, T.F.; Shen, X.; Marcos; Yang, C. Lab-on-chip microfluidic impedance measurement for laminar flow ratio sensing and differential conductivity difference detection. Appl. Phys. Lett. 2017, 110, 233501. [Google Scholar] [CrossRef]
- Kiilerich-Pedersen, K.; Rozlosnik, N. Cell-based biosensors: Electrical sensing in microfluidic devices. Diagnostics 2012, 2, 83–96. [Google Scholar] [CrossRef]
- Lee, G.; Lee, J.; Kim, J.; Choi, H.S.; Kim, J.; Lee, S.; Lee, H. Single microfluidic electrochemical sensor system for simultaneous multi-pulmonary hypertension biomarker analyses. Sci. Rep. 2017, 7, 7545. [Google Scholar] [CrossRef]
- Illing, R.; Burkart, C.; Pfitzner, D.; Jungmann, D.; Baraban, L.; Cuniberti, G. Ecotoxicity assessment using ciliate cells in millifluidic droplets. Biomicrofluidics 2016, 10, 024115. [Google Scholar] [CrossRef]
- Karnaushenko, D.; Baraban, L.; Ye, D.; Uguz, I.; Mendes, R.G.; Rümmeli, M.H.; De Visser, J.A.G.M.; Schmidt, O.G.; Cuniberti, G.; Makarov, D. Monitoring microbial metabolites using an inductively coupled resonance circuit. Sci. Rep. 2015, 5, 12878. [Google Scholar] [CrossRef]
- Campos, P.P.; Moraes, M.L.; Volpati, D.; Miranda, P.B.; Oliveira, O.N.; Ferreira, M. Amperometric detection of lactose using β-Galactosidase immobilized in layer-by-layer films. ACS Appl. Mater. Interfaces 2014, 6, 11657–11664. [Google Scholar] [CrossRef]
- Suzuki, K.; Suzuki, Y. Globoid cell leucodystrophy (Krabbe’s disease): Deficiency of galactocerebroside β-galactosidase. Proc. Natl. Acad. Sci. USA 1970, 66, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Hahn, C.N.; Martin, M.D.P.; Schröder, M.; Vanier, M.T.; Suzuki, K.; Hara, Y.; D’Azzo, A. Generalized CNS disease and massive GM1-ganglioside accumulation in mice defective in lysosomal acid -galactosidase. Hum. Mol. Genet. 1997, 6, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.Y.; Han, J.A.; Im, J.S.; Morrone, A.; Johung, K.; Goodwin, E.C.; Kleijer, W.J.; DiMaio, D.; Hwang, E.S. Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell 2006, 5, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, C.; Stach, M.H.; Amtmann, A.; Young, M.E.; Reyes, J.G.R.; Huebner, H.; Buchholz, R. Measuring β-Galactosidase activity at pH 6 with a differential pH sensor. Electron. J. Biotechnol. 2009, 12, 2009. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Liu, J.; Ma, X.; Cui, C.; Deenik, P.R.; Henderson, P.K.P.; Sigler, A.L.; Cui, L. Real-time imaging of senescence in tumors with DNA damage. Sci. Rep. 2019, 9, 2102. [Google Scholar] [CrossRef]
- Rim, T.; Kim, K.; Kim, S.; Baek, C.-K.; Meyyappan, M.; Jeong, Y.-H.; Lee, J.-S. Improved electrical characteristics of honeycomb nanowire ISFETs. IEEE Electron Device Lett. 2013, 34, 1059–1061. [Google Scholar] [CrossRef]
- Shen, S.-H.; Wang, I.-S.; Cheng, H.; Lin, C.-T. An enhancement of high-k/oxide stacked dielectric structure for silicon-based multi-nanowire biosensor in cardiac troponin I detection. Sens. Actuators B Chem. 2015, 218, 303–309. [Google Scholar] [CrossRef]
- Kim, K.; Park, C.; Kwon, D.; Kim, D.; Meyyappan, M.; Jeon, S.; Lee, J.-S. Silicon nanowire biosensors for detection of cardiac troponin I (cTnI) with high sensitivity. Biosens. Bioelectron. 2016, 77, 695–701. [Google Scholar] [CrossRef]
- Castilho, P.C.; Crampton, M.R.; Yarwood, J. Infrared spectroscopic studies of hydrogen bonding in substituted nitrophenols: Substituent and solvent effects. Vib. Spectrosc. 1992, 3, 167–180. [Google Scholar] [CrossRef]
- Roskoski, R. Michaelis-menten kinetics. In xPharm: The Comprehensive Pharmacology Reference; Elsevier: Amsterdam, The Netherland, 2007; pp. 1–10. [Google Scholar]
- Lee, J.; Wipf, M.; Mu, L.; Adams, C.; Hannant, J.; Reed, M.A. Metal-coated microfluidic channels: An approach to eliminate streaming potential effects in nano biosensors. Biosens. Bioelectron. 2017, 87, 447–452. [Google Scholar] [CrossRef]
- Ward, K.; Fan, Z.H. Mixing in microfluidic devices and enhancement methods. J. Micromech. Microeng. 2015, 25, 94001. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belyaev, D.; Schütt, J.; Ibarlucea, B.; Rim, T.; Baraban, L.; Cuniberti, G. Nanosensors-Assisted Quantitative Analysis of Biochemical Processes in Droplets. Micromachines 2020, 11, 138. https://doi.org/10.3390/mi11020138
Belyaev D, Schütt J, Ibarlucea B, Rim T, Baraban L, Cuniberti G. Nanosensors-Assisted Quantitative Analysis of Biochemical Processes in Droplets. Micromachines. 2020; 11(2):138. https://doi.org/10.3390/mi11020138
Chicago/Turabian StyleBelyaev, Dmitry, Julian Schütt, Bergoi Ibarlucea, Taiuk Rim, Larysa Baraban, and Gianaurelio Cuniberti. 2020. "Nanosensors-Assisted Quantitative Analysis of Biochemical Processes in Droplets" Micromachines 11, no. 2: 138. https://doi.org/10.3390/mi11020138
APA StyleBelyaev, D., Schütt, J., Ibarlucea, B., Rim, T., Baraban, L., & Cuniberti, G. (2020). Nanosensors-Assisted Quantitative Analysis of Biochemical Processes in Droplets. Micromachines, 11(2), 138. https://doi.org/10.3390/mi11020138







