A Novel Microfluidic Method Utilizing a Hydrofoil Structure to Improve Circulating Tumor Cell Enrichment: Design and Analytical Validation
Abstract
:1. Introduction
- (i)
- non-invasive tumor sampling which significantly reduces the patients’ discomfort,
- (ii)
- frequent sampling to monitor therapy efficacy and evolution of resistance mutations, paving the way for a more precise disease management (through the selection of more appropriate targeted therapies, which avoids the side-effects and costs associated with inappropriate chemotherapy),
- (iii)
- better representation of the spatial and temporal heterogeneity of the solid tumors.
1.1. Principle of Particle Focusing in Spiral Microfluidic Channels
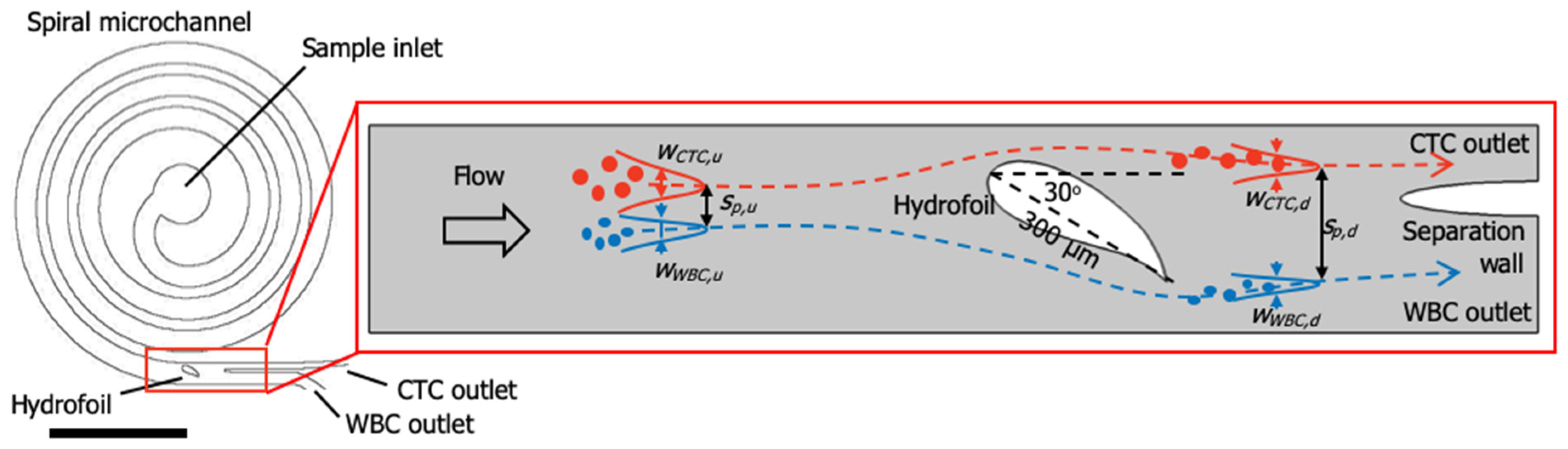
1.2. Device Design and Simulations
2. Materials and Methods
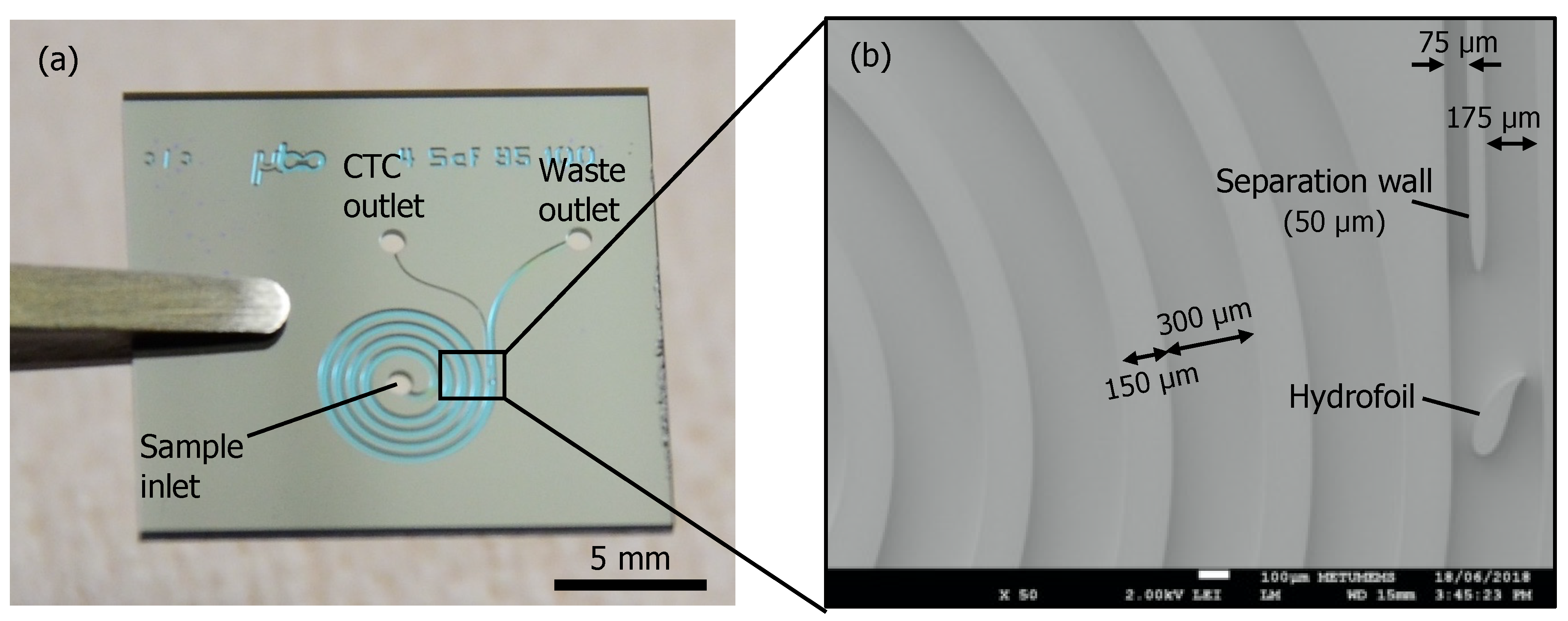
2.1. Device Fabrication
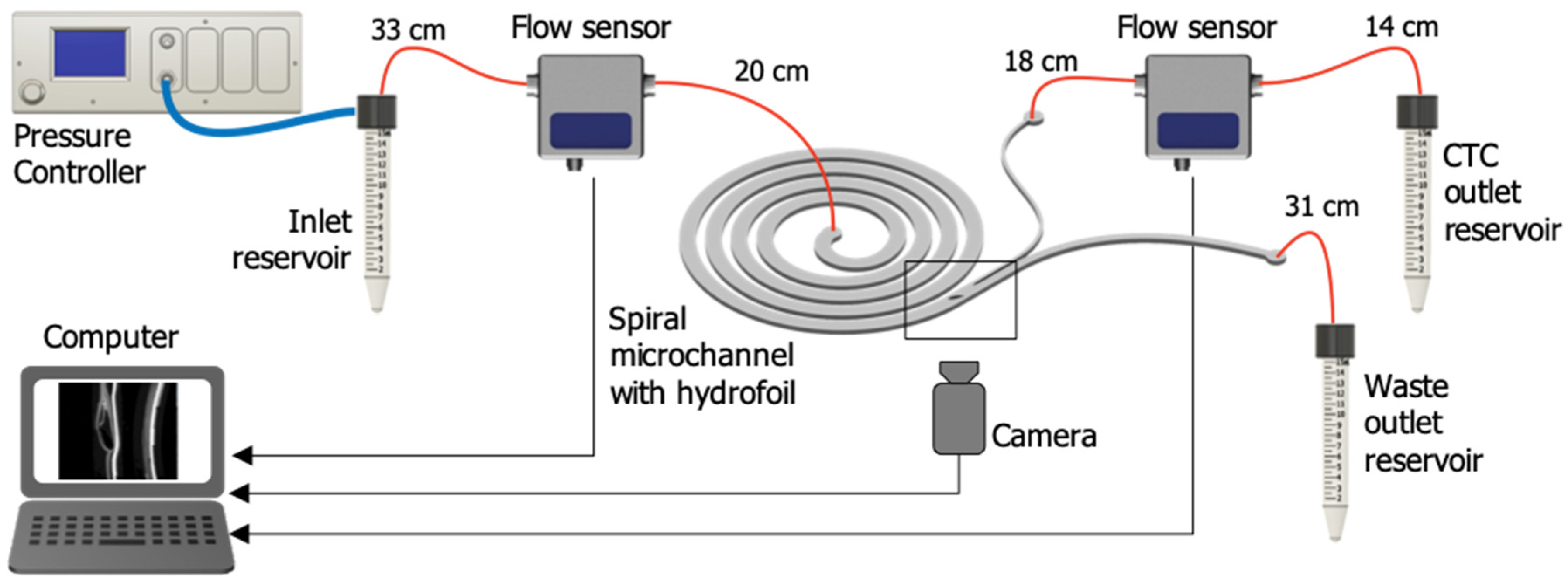
2.2. Microfluidic Setup
2.3. Chip Conditioning
2.4. Flow Rate Selection
2.5. Cell Culture, Blood Collection, and Sample Preparation
2.6. Analytical Performance Characterization
2.7. Analysis of WBC Depletion and CTC Recovery Rates
2.8. Viability Analysis
3. Results and Discussion
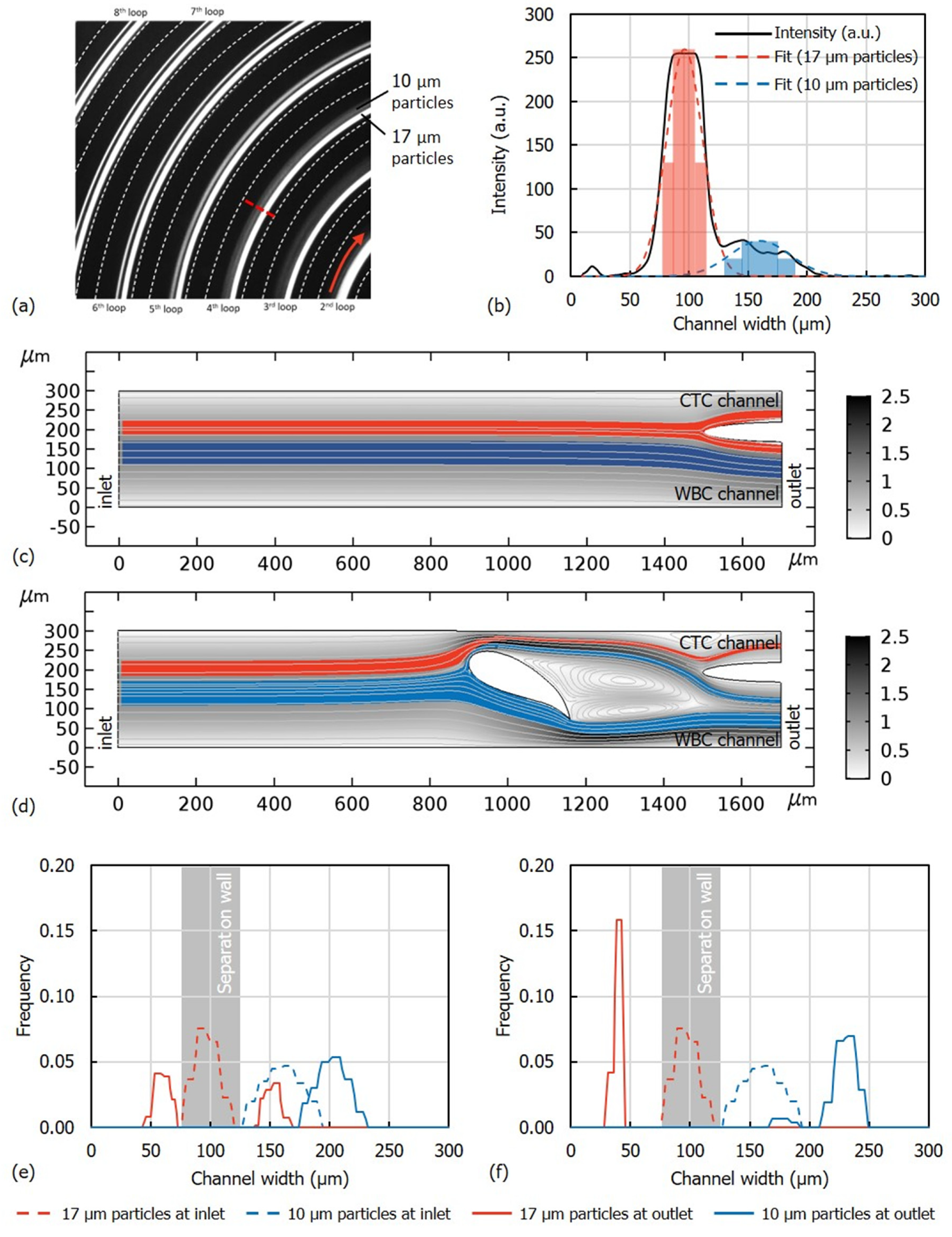
3.1. Flow Rate Optimization
3.2. WBC Depletion and CTC Recovery
3.3. Effect on Cell Viability
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization Press Release, International Agency for Research on Cancer: Latest Global Cancer Data. Available online: https://www.who.int/cancer/PRGlobocanFinal.pdf (accessed on 23 September 2020).
- Bardelli, A.; Pantel, K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017, 31, 172–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gold, B.; Cankovic, M.; Furtado, L.V.; Meier, F.; Gocke, C.D. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J. Mol. Diagn. 2015, 17, 209–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Adv. Med. Oncol. 2018, 10, 1758835918794630. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Lorente, D.; Omlin, A.; Olmos, D.; De Bono, J.S. Circulating tumor cells: A multifunctional biomarker. Clin. Cancer Res. 2014, 20, 2553–2558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maheswaran, S.; Haber, D.A. Circulating tumor cells: A window into cancer biology and metastasis. Curr. Opin. Genet. Dev. 2010, 20, 96–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, T.; Hayashi, N.; Iguchi, T.; Ito, S.; Eguchi, H.; Mimori, K. Clinical and biological significance of circulating tumor cells in cancer. Mol. Oncol. 2016, 10, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Tayoun, T.; Faugeroux, V.; Oulhen, M.; Aberlenc, A. Farace CTC-Derived Models: A Window into the Seeding Capacity of Circulating Tumor Cells (CTCs). Cells 2019, 8, 1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Chu, G.C.Y.; Mrdenovic, S.; Annamalai, A.A.; Hendifar, A.E.; Nissen, N.N.; Tomlinson, J.S.; Lewis, M.; Palanisamy, N.; Tseng, H.R.; et al. Cultured circulating tumor cells and their derived xenografts for personalized oncology. Asian J. Urol. 2016, 3, 240–253. [Google Scholar] [CrossRef] [Green Version]
- Alix-Panabières, C.; Bartkowiak, K.; Pantel, K. Functional studies on circulating and disseminated tumor cells in carcinoma patients. Mol. Oncol. 2016, 10, 443–449. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.C.; Doyle, G.V.; Terstappen, L.W.M.M. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J. Oncol. 2010, 2010, 1–8. [Google Scholar] [CrossRef]
- Agnoletto, C.; Corrà, F.; Minotti, L.; Baldassari, F.; Crudele, F.; Cook, W.J.J.; Di Leva, G.; D’Adamo, A.P.; Gasparini, P.; Volinia, S. Heterogeneity in circulating tumor cells: The relevance of the stem-cell subset. Cancers (Basel) 2019, 11, 483. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.B.; Lim, C.T.; Lim, W.T. Single-cell analysis of circulating tumor cells: Why heterogeneity matters. Cancers (Basel) 2019, 11, 1595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro-Giner, F.; Aceto, N. Tracking cancer progression: From circulating tumor cells to metastasis. Genome Med. 2020, 12, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micalizzi, D.S.; Maheswaran, S.; Haber, D.A. A conduit to metastasis: Circulating tumor cell biology. Genes Dev. 2017, 31, 1827–1840. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, R.; Soares, J.; Peixoto, A.; Cotton, S.; Lima, L.; Santos, L.L.; Ferreira, J.A. Circulating tumor cells in bladder cancer: Emerging technologies and clinical implications foreseeing precision oncology. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Bankó, P.; Lee, S.Y.; Nagygyörgy, V.; Zrínyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Hao, S.J.; Wan, Y.; Xia, Y.Q.; Zou, X.; Zheng, S.Y. Size-based separation methods of circulating tumor cells. Adv. Drug Deliv. Rev. 2018, 125, 3–20. [Google Scholar] [CrossRef]
- Jackson, J.M.; Witek, M.A.; Kamande, J.W.; Soper, S.A. Materials and microfluidics: Enabling the efficient isolation and analysis of circulating tumour cells. Chem. Soc. Rev. 2017, 46, 4245–4280. [Google Scholar] [CrossRef]
- Pødenphant, M.; Ashley, N.; Koprowska, K.; Mir, K.U.; Zalkovskij, M.; Bilenberg, B.; Bodmer, W.; Kristensen, A.; Marie, R. Separation of cancer cells from white blood cells by pinched flow fractionation. Lab. Chip 2015, 15, 4598–4606. [Google Scholar] [CrossRef] [Green Version]
- Karabacak, N.M.; Spuhler, P.S.; Fachin, F.; Lim, E.J.; Pai, V.; Ozkumur, E.; Martel, J.M.; Kojic, N.; Smith, K.; Chen, P.; et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protoc. 2014, 9, 694–710. [Google Scholar] [CrossRef] [Green Version]
- Fachin, F.; Spuhler, P.; Martel-Foley, J.M.; Edd, J.F.; Barber, T.A.; Walsh, J.; Karabacak, M.; Pai, V.; Yu, M.; Smith, K.; et al. Monolithic Chip for High-throughput Blood Cell Depletion to Sort Rare Circulating Tumor Cells. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Warkiani, M.E.; Khoo, B.L.; Wu, L.; Tay, A.K.P.; Bhagat, A.A.S.; Han, J.; Lim, C.T. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat. Protoc. 2016, 11, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Martel, J.M.; Smith, K.C.; Dlamini, M.; Pletcher, K.; Yang, J.; Karabacak, M.; Haber, D.A.; Kapur, R.; Toner, M. Continuous Flow Microfluidic Bioparticle Concentrator. Sci. Rep. 2015, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Liu, C.; Li, M.; Wang, J.; Xianyu, Y.; Hu, G.; Jiang, X. Size-based hydrodynamic rare tumor cell separation in curved microfluidic channels. Biomicrofluidics 2013, 7, 011802. [Google Scholar] [CrossRef] [Green Version]
- Bhagat, A.A.S.; Bow, H.; Hou, H.W.; Tan, S.J.; Han, J.; Lim, C.T. Microfluidics for cell separation. Med. Biol. Eng. Comput. 2010, 48, 999–1014. [Google Scholar] [CrossRef]
- Di Carlo, D.; Edd, J.F.; Humphry, K.J.; Stone, H.A.; Toner, M. Particle segregation and dynamics in confined flows. Phys. Rev. Lett. 2009, 102, 094503. [Google Scholar] [CrossRef] [Green Version]
- Nivedita, N.; Papautsky, I. Continuous separation of blood cells in spiral microfluidic devices. Biomicrofluidics 2013, 7, 54101. [Google Scholar] [CrossRef] [Green Version]
- Hanai, T.T. Column Eficiency. Hplc A Prac. Guid. 1999, 96–108. [Google Scholar]
- Hardelauf, H.; Sisnaiske, J.; Taghipour-Anvari, A.A.; Jacob, P.; Drabiniok, E.; Marggraf, U.; Frimat, J.P.; Hengstler, J.G.; Neyer, A.; Van Thriel, C.; et al. High fidelity neuronal networks formed by plasma masking with a bilayer membrane: Analysis of neurodegenerative and neuroprotective processes. Lab. Chip 2011, 11, 2763–2771. [Google Scholar] [CrossRef]
- Warkiani, M.E.; Khoo, B.L.; Tan, D.S.-W.; Bhagat, A.A.S.; Lim, W.-T.; Yap, Y.S.; Lee, S.C.; Soo, R.A.; Han, J.; Lim, C.T. An ultra-high-throughput spiral microfluidic biochip for the enrichment of circulating tumor cells. Analyst 2014, 139, 3245–3255. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.W.; Warkiani, M.E.; Khoo, B.L.; Li, Z.R.; Soo, R.A.; Tan, D.S.-W.; Lim, W.-T.; Han, J.; Bhagat, A.A.S.; Lim, C.T. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci. Rep. 2013, 3, 1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhagat, A.A.S.; Kuntaegowdanahalli, S.S.; Papautsky, I. Continuous particle separation in spiral microchannels using dean flows and differential migration. Lab. Chip 2008, 8, 1906–1914. [Google Scholar] [CrossRef] [PubMed]
- Warkiani, M.E.; Guan, G.; Luan, K.B.; Lee, W.C.; Bhagat, A.A.S.; Kant Chaudhuri, P.; Tan, D.S.W.; Lim, W.T.; Lee, S.C.; Chen, P.C.Y.; et al. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab. Chip 2014, 14, 128–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Guan, G.; Hou, H.W.; Bhagat, A.A.S.; Han, J. Separation of Leukocytes from Blood Using Spiral Channel with Trapezoid Cross-Section. Anal. Chem. 2012, 84, 9324–9331. [Google Scholar] [CrossRef]
- Kretzmer, G.; Schügerl, K. Response of mammalian cells to shear stress. Appl. Microbiol. Biotechnol. 1991, 34, 613–616. [Google Scholar] [CrossRef]
- Ketpun, D.; Sailasuta, A.; Suwannaphan, T.; Bhanpattanakul, S.; Pimpin, A.; Srituravanich, W.; Sripumkhai, W.; Jeamsaksiri, W.; Piyaviriyakul, P. The viability of single cancer cells after exposure to hydrodynamic shear stresses in a spiral microchannel: A canine cutaneous mast cell tumor model. Micromachines 2017, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Armistead, F.J.; Gala De Pablo, J.; Gadêlha, H.; Peyman, S.A.; Evans, S.D. Cells Under Stress: An Inertial-Shear Microfluidic Determination of Cell Behavior. Biophys. J. 2019, 116, 1127–1135. [Google Scholar] [CrossRef] [Green Version]
- Landwehr, G.M.; Kristof, A.J.; Rahman, S.M.; Pettigrew, J.H.; Coates, R.; Balhoff, J.B.; Triantafillu, U.L.; Kim, Y.; Melvin, A.T. Biophysical analysis of fluid shear stress induced cellular deformation in a microfluidic device. Biomicrofluidics 2018, 12, 054109. [Google Scholar] [CrossRef]
- Suwannaphan, T.; Srituravanich, W.; Sailasuta, A.; Piyaviriyakul, P.; Bhanpattanakul, S.; Jeamsaksiri, W.; Sripumkhai, W.; Pimpin, A. Investigation of leukocyte viability and damage in spiral microchannel and contraction-expansion array. Micromachines 2019, 10, 772. [Google Scholar] [CrossRef] [Green Version]
- Barnes, J.M.; Nauseef, J.T.; Henry, M.D. Resistance to Fluid Shear Stress Is a Conserved Biophysical Property of Malignant Cells. PLoS ONE 2012, 7, e50973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, M.J.; King, M.R. Computational and experimental models of cancer cell response to fluid shear stress. Front. Oncol. 2013, 3, 44. [Google Scholar] [CrossRef] [Green Version]
- Martel, J.M.; Toner, M. Particle Focusing in Curved Microfluidic Channels. Sci. Rep. 2013, 3, 3340. [Google Scholar] [CrossRef] [Green Version]
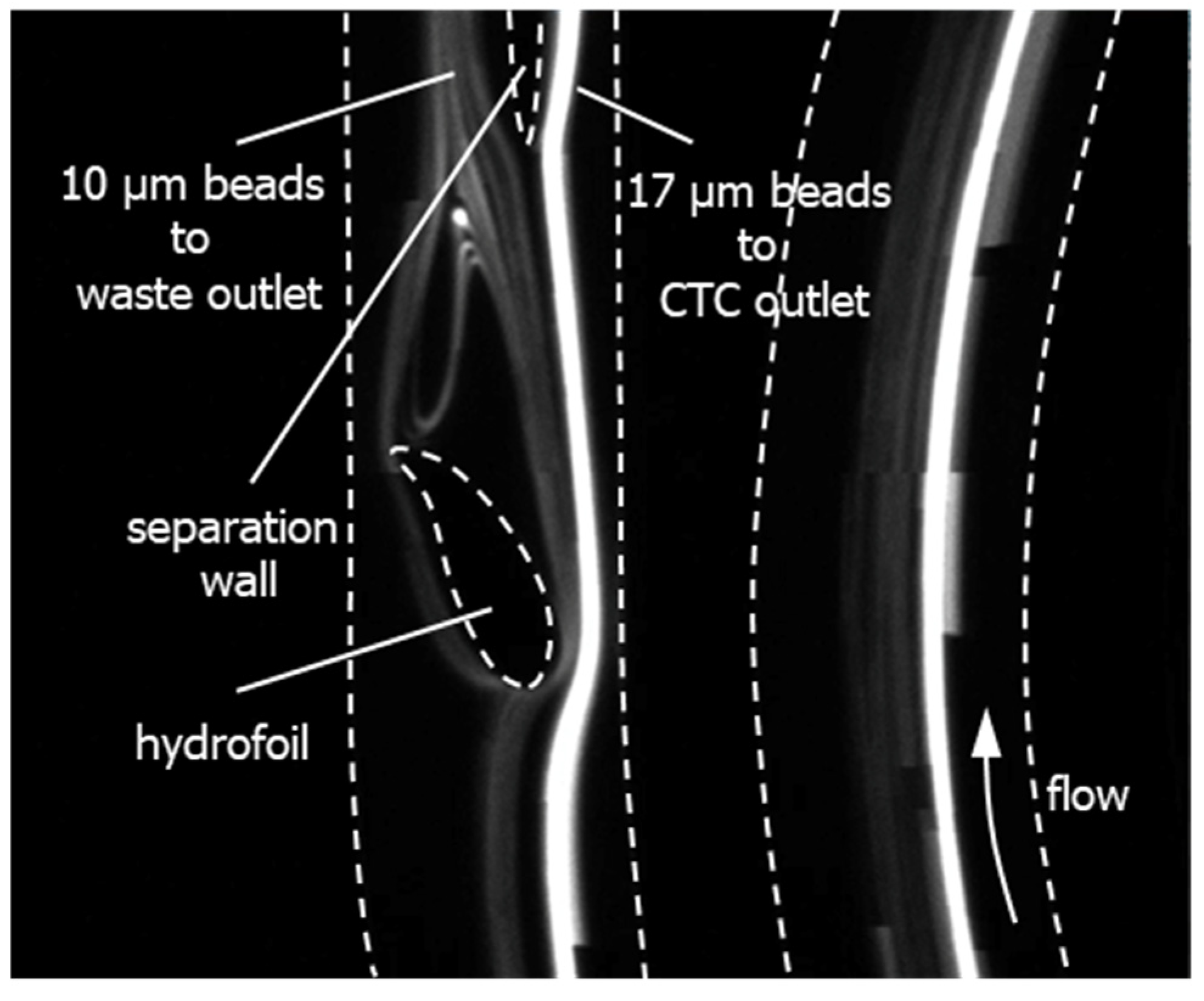
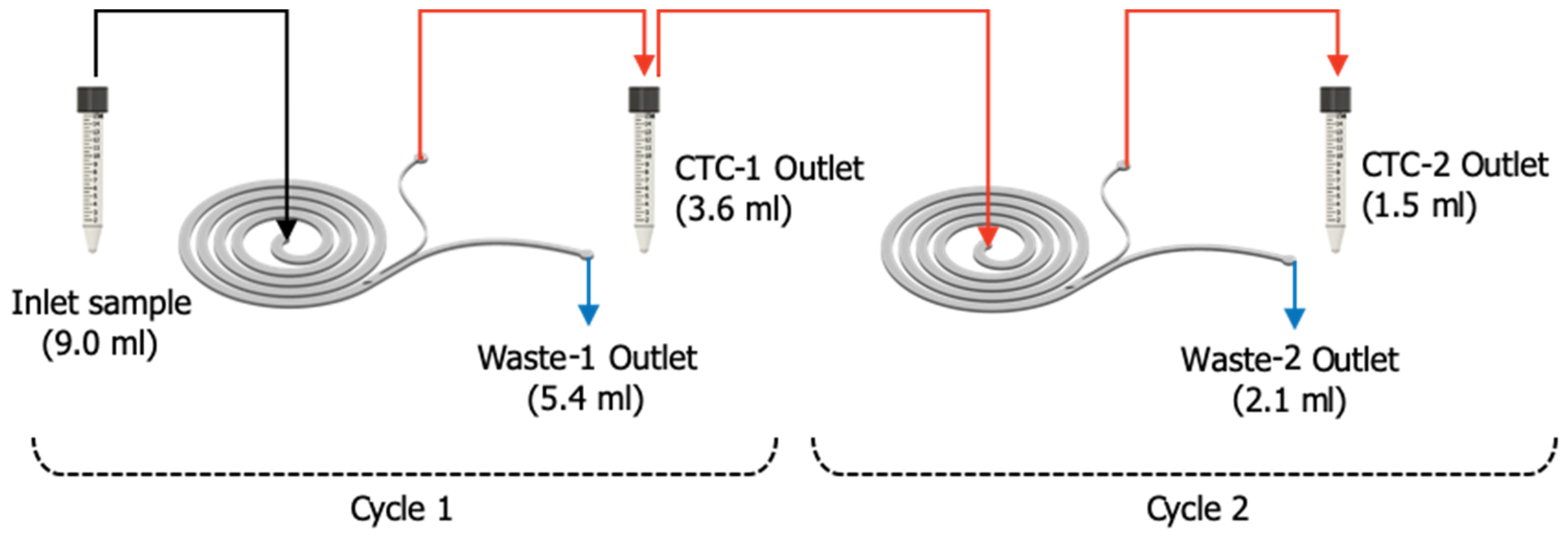
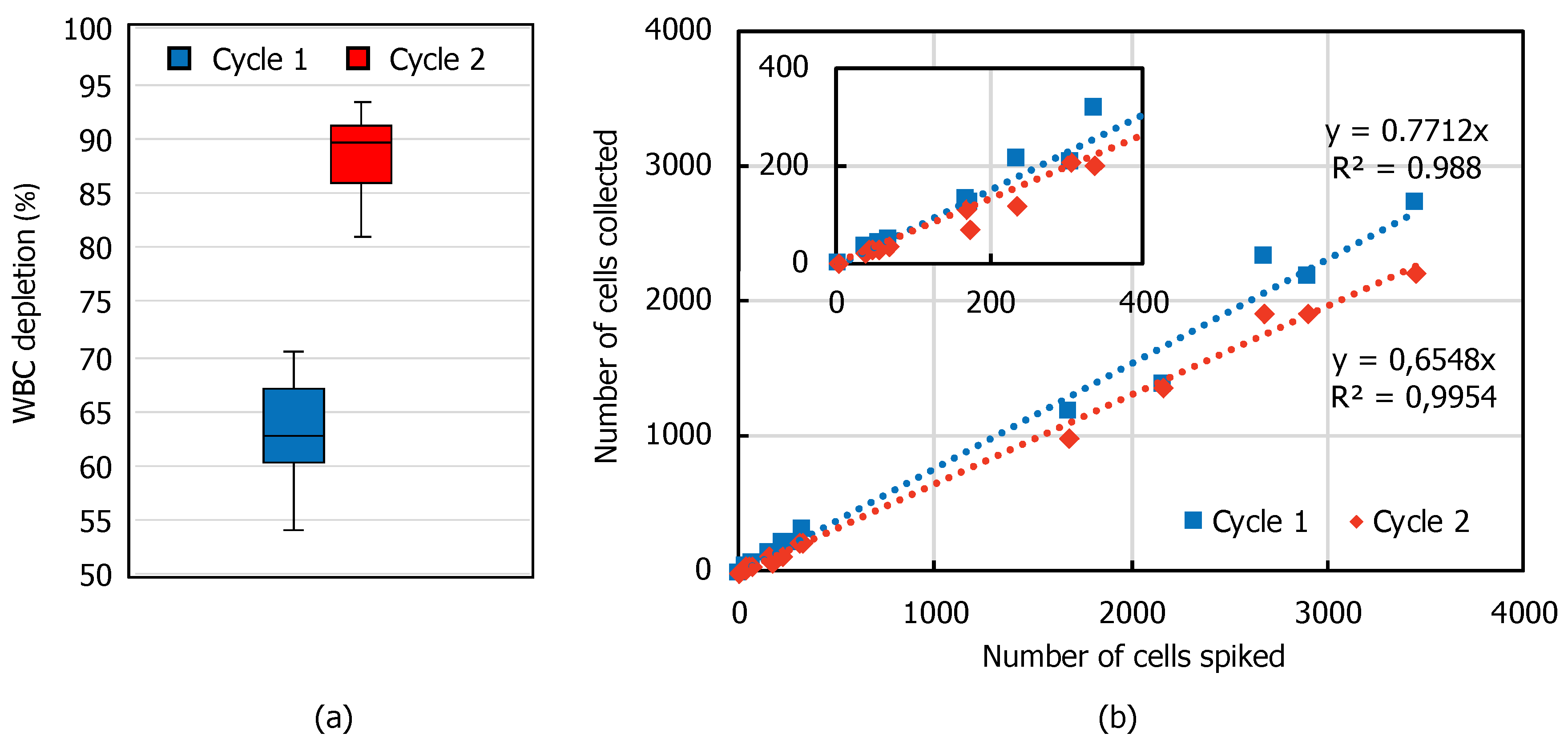
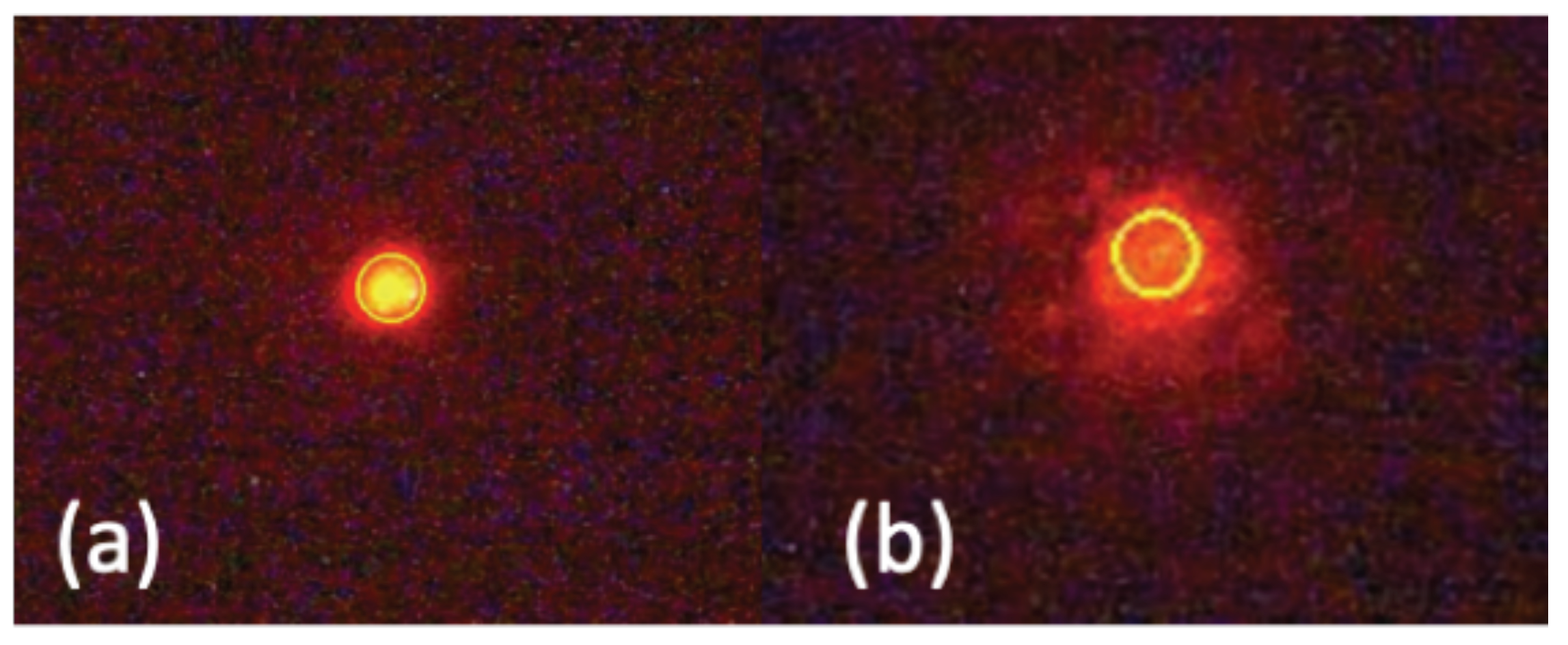
| Flow Rate (mL/min) | 1.0 | 1.2 | 1.4 |
|---|---|---|---|
| Recovery rate ± STD (%) | 99.0 ± 1.7 | 99.1 ± 1.6 | 98.1 ± 0.2 |
| Purity ± STD (%) | 79.2 ± 5.8 | 97.7 ± 0.3 | 96.2 ± 1.7 |
| Viable Cell Loss ± SEM (%) | Total Cell Loss ± SEM (%) | |
|---|---|---|
| Cycle 1 | 15.7 ± 8.5 | 15.3 ± 7.5 |
| Cycle 2 | 32.8 ± 10.2 | 29.7 ± 9.3 |
| Control | 3.3 ± 2.3 | 4.7 ± 2.3 |
| Inlet | Cycle 1 | Cycle 2 | |||
|---|---|---|---|---|---|
| CTC outlet | Waste outlet | CTC outlet | Waste outlet | ||
| Viability ± SEM (%) | 81.7 ± 2.9 | 84.7 ± 2.3 | 62.5 ± 6.9 | 85.2 ± 1.6 | 73.5 ± 3.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Özkayar, G.; Mutlu, E.; Şahin, Ş.; Demircan Yalçın, Y.; Töral, T.; Külah, H.; Yildirim, E.; Zorlu, Ö.; Özgür, E. A Novel Microfluidic Method Utilizing a Hydrofoil Structure to Improve Circulating Tumor Cell Enrichment: Design and Analytical Validation. Micromachines 2020, 11, 981. https://doi.org/10.3390/mi11110981
Özkayar G, Mutlu E, Şahin Ş, Demircan Yalçın Y, Töral T, Külah H, Yildirim E, Zorlu Ö, Özgür E. A Novel Microfluidic Method Utilizing a Hydrofoil Structure to Improve Circulating Tumor Cell Enrichment: Design and Analytical Validation. Micromachines. 2020; 11(11):981. https://doi.org/10.3390/mi11110981
Chicago/Turabian StyleÖzkayar, Gürhan, Ege Mutlu, Şebnem Şahin, Yağmur Demircan Yalçın, Taylan Töral, Haluk Külah, Ender Yildirim, Özge Zorlu, and Ebru Özgür. 2020. "A Novel Microfluidic Method Utilizing a Hydrofoil Structure to Improve Circulating Tumor Cell Enrichment: Design and Analytical Validation" Micromachines 11, no. 11: 981. https://doi.org/10.3390/mi11110981
APA StyleÖzkayar, G., Mutlu, E., Şahin, Ş., Demircan Yalçın, Y., Töral, T., Külah, H., Yildirim, E., Zorlu, Ö., & Özgür, E. (2020). A Novel Microfluidic Method Utilizing a Hydrofoil Structure to Improve Circulating Tumor Cell Enrichment: Design and Analytical Validation. Micromachines, 11(11), 981. https://doi.org/10.3390/mi11110981





