Single-Molecule Counting of Nucleotide by Electrophoresis with Nanochannel-Integrated Nano-Gap Devices
Abstract
:1. Introduction
2. Materials and Methods
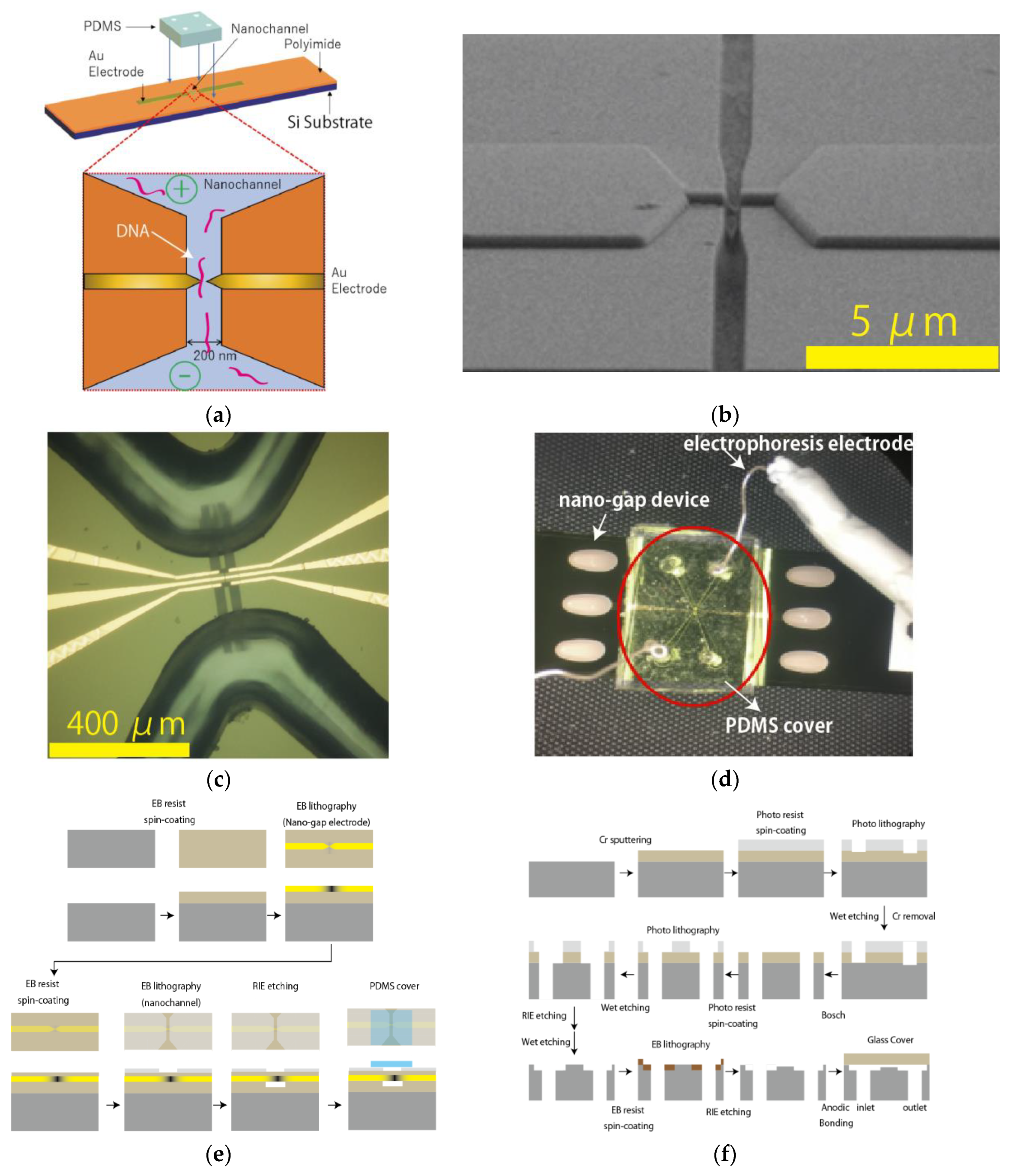
2.1. Fabrication of Nanochannel-Integrated Mechanically Controllable Break Junction (MCBJ) Devices for Single-Molecule Detection
2.2. Fabrication of Nanochannel Device for Single-Sample Observation by Optical Imaging
2.3. Imaging of Fluorescence-Stained λDNA Molecules and Fluorescence Particle Image by Fluorescence Optical Microscope
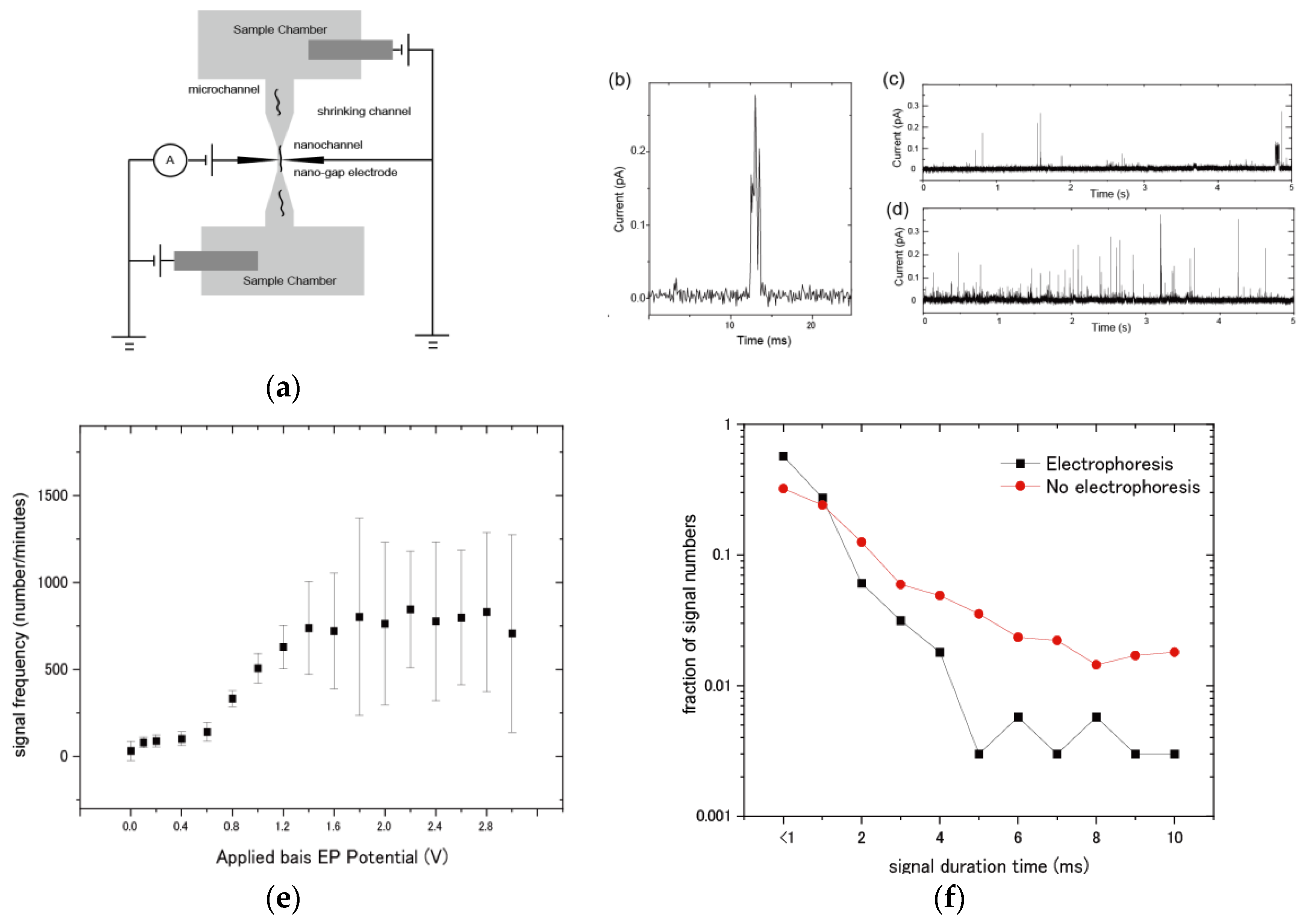
2.4. Procedure for Current Measurements
3. Results
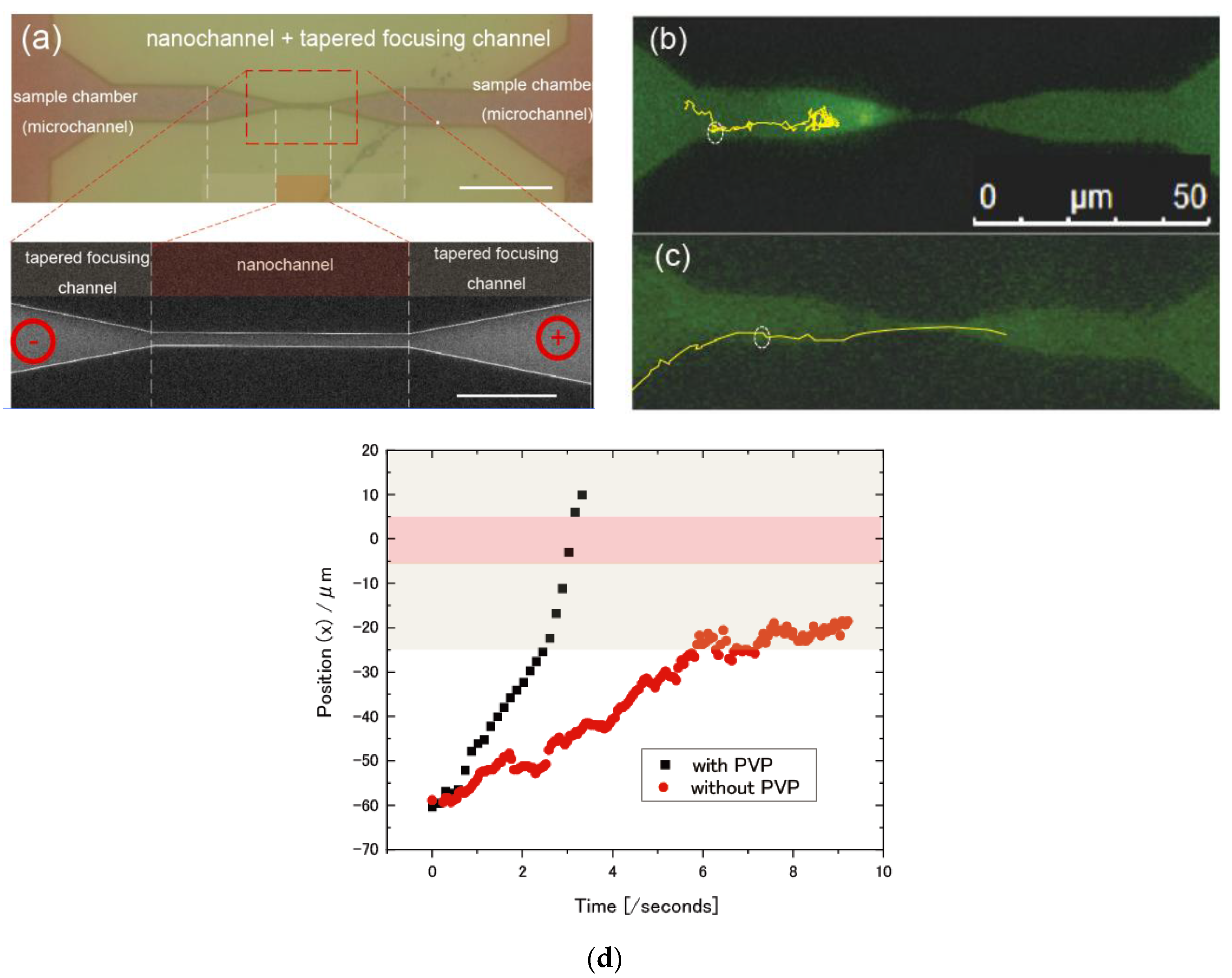
3.1. Direct Observation of Sample Behaviors in Nanochannel Device
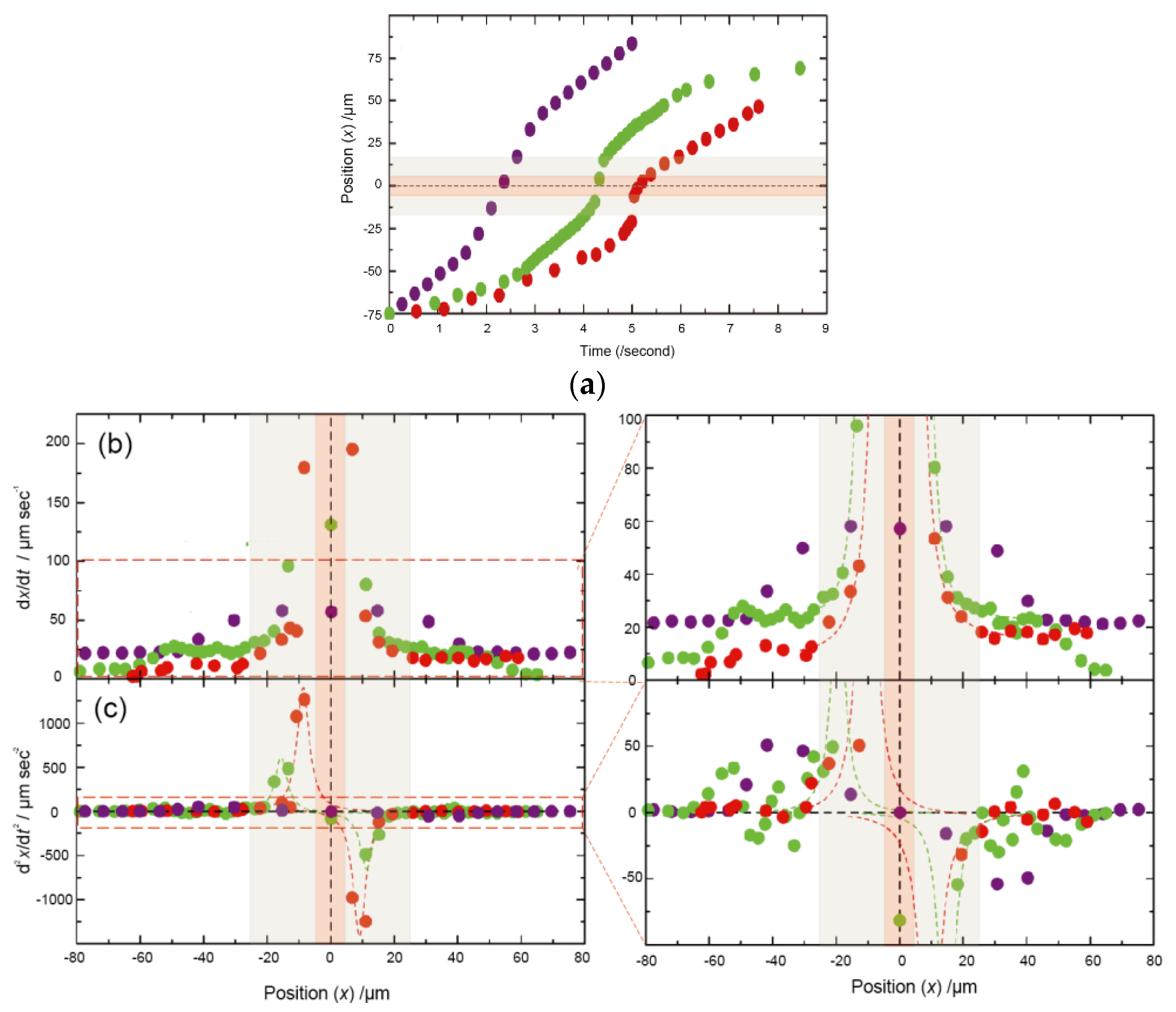
3.2. Dependency of Sample Electrophoretic Behaviors on the Shape of Nanochannel Devices
3.3. Evaluation of Molecular Detection and Signal Behaviors of DNA Samples by Electrophoresis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Schoch, R.B.; Han, J.Y.; Renaud, P. Transport phenomena in nanofluidics. Rev. Mod. Phys. 2007, 80, 839–883. [Google Scholar] [CrossRef] [Green Version]
- Abgrall, P.; Nguyen, N.T. Nanofluidic devices and their applications. Anal. Chem. 2008, 80, 2326–2341. [Google Scholar] [CrossRef] [PubMed]
- De la Escosura-Muniz, A.; Merkoci, A. Nanochannels Preparation and Application in Biosensing. ACS Nano 2012, 6, 7556–7583. [Google Scholar] [CrossRef]
- Rivet, C.; Lee, H.; Hirsch, A.; Hamilton, S.; Lu, H. Microfluidics for medical diagnostics and biosensors. Chem. Eng. Sci. 2011, 66, 1490–1507. [Google Scholar] [CrossRef]
- Liu, R.H.; Bonanno, J.; Yang, J.N.; Lenigk, R.; Grodzinski, P. Single-use, thermally actuated paraffin valves for microfluidic applications. Sens. Act. B 2004, 98, 328–336. [Google Scholar] [CrossRef]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Zwolak, M.; Di Ventra, M. Electronic signature of DNA nucleotides via transverse transport. Nano Lett. 2005, 5, 421–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagerqvist, J.; Zwolak, M.; Di Ventra, M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 2006, 6, 779–782. [Google Scholar] [CrossRef] [Green Version]
- Ohshiro, T.; Tsustui, M.; Matsubara, K.; Furuhashi, M.; Taniguchi, M.; Kawai, T. Single-Molecule Electrical Random Resequencing of DNA and RNA. Sci. Rep. 2012, 2, 501–507. [Google Scholar] [CrossRef]
- Ohshiro, T.; Tsutsui, M.; Yokota, K.; Taniguchi, M. Quantitative analysis of DNA with single-molecule sequencing. Sci. Rep. 2018, 8, 8517. [Google Scholar] [CrossRef] [Green Version]
- Ohshiro, T.; Tsutsui, T.; Yokota, K.; Furuhashi, M.; Taniguchi, M.; Kawai, T. Single-Molecule Proteomic Analysis of Post-translational Modification. Nat. Nanotech. 2014, 9, 835–840. [Google Scholar] [CrossRef]
- Ohshiro, T.; Komoto, Y.; Konno, M.; Koseki, J.; Asai, A.; Ishii, H.; Taniguchi, M. Direct Analysis of Incorporation of an Anticancer Drug into DNA at Single-Molecule Resolution. Sci. Rep. 2019, 9, 3886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuhata, T.; Ohshiro, T.; Akimoto, G.; Ueki, R.; Taniguchi, M.; Sando, S. Highly Conductive Nucleotide Analogue Facilitates Base-Calling in Quantum-Tunneling-Based DNA Sequencing. ACS Nano 2019, 135, 5028–5035. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Ohshiro, T.; Komoto, Y.; Takaai, T.; Yoshida, T.; Washio, T. High-Precision Single-Molecule Identification Based on Single-Molecule Information within a Noisy Matrix. J. Phys. Chem. C 2019, 123, 15867–15873. [Google Scholar] [CrossRef]
- Furuhata, T.; Ohshiro, T.; Akimoto, G.; Ueki, R.; Taniguchi, M.; Sando, S. Chemical Labeling-Assisted Detection of Nucleobase Modifications by Quantum Tunneling-Based Single Molecule Sensing. ChemBioChem 2020, 21, 335–339. [Google Scholar] [CrossRef]
- Komoto, Y.; Ohshiro, T.; Yoshida, T.; Tarusawa, E.; Yagi, T.; Washio, T.; Taniguchi, M. Time-resolved neurotransmitter detection in mouse brain tissue using an artificial intelligence-nanogap. Sci. Rep. 2020, 10, 11244. [Google Scholar] [CrossRef] [PubMed]
- Jyh-Ping, H.; Li-Hsien, Y.; Ming-Hong, K. Evaluation of the electric force in electrophoresis. J. Colloid. Int. Sci. 2007, 305, 324–329. [Google Scholar]
- Chang, H.; Yeo, L.Y. Electrokinetically-Driven Microfluidics and Nanofluidics; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Bocquet, L.; Barrat, J.L. Flow boundary conditions from nano- to micro-scales. Soft Matter 2007, 3, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Koga, Y.; Kuriyama, R.; Sato, Y. Effects of Micromachining Processes on Electro-Osmotic Flow Mobility of Glass Surfaces. Micromachines 2013, 4, 67. [Google Scholar] [CrossRef] [Green Version]
- Tsutsui, M.; Shoji, K.; Taniguchi, M.; Kawai, T. Formation and self-breaking mechanism of stable atom-sized junctions. Nano Lett. 2008, 8, 345–349. [Google Scholar] [CrossRef]
- Agrait, N.; Yeyati, A.L.; van Ruitenbeek, J.M. Quantum properties of atomic sized conductors. Phys. Rep. 2003, 377, 81–279. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.M.H.; Lee, D.H.Y.; Liaw, C.Y.N.; Lao, A.I.K.; Hsing, I.M. Detailed characterization of anodic bonding process between glass and thin-film coated silicon substrates. Sens. Actuators 2000, 86, 103–107. [Google Scholar] [CrossRef]
- Kaniansky, D.; Masár, M.; Bielčíková, J. Electroosmotic flow suppressing additives for capillary zone electrophoresis in a hydrodynamically closed separation system. J. Chromatogr. A 1997, 792, 483–494. [Google Scholar] [CrossRef]
- Kaneta, T.; Ueda, T.; Hata, K.; Imasaka, T. Suppression of electroosmotic flow and its application to determination of electrophoretic mobilities in a poly(vinylpyrrolidone)-coated capillary. J. Chromatogr. A 2006, 1106, 52–55. [Google Scholar] [CrossRef]
- Castillo-Fernandez, O.; Salieb-Beugelaar, G.B.; van Nieuwkasteele, J.W.; Bomer, J.G.; Arundell, M.; Samitier, J.; van den Berg, A.; Eijkel, J.C.T. Electrokinetic DNA transport in 20 nm-high nanoslits: Evidence for movement through a wall-adsorbed polymer nanogel. Electrophoresis 2011, 32, 2402–2409. [Google Scholar] [CrossRef]
- Menard, L.D.; Ramsey, J.M. Electrokinetically-Driven Transport of DNA through Focused Ion Beam Milled Nanofluidic Channels. Anal. Chem. 2013, 85, 1146–1153. [Google Scholar] [CrossRef] [Green Version]
- Milanova, D.; Chambers, R.D.; Bahga, S.S.; Santiago, J.G. Effect of PVP on the electroosmotic mobility of wet-etched glass microchannels. Electrophoresis 2012, 33, 3259–3262. [Google Scholar] [CrossRef]
- Thacker, V.V.; Ghosal, S.; Hernández-Ainsa, S.; Bell, N.A.W.; Keyser, U.F. Studying DNA translocation in nanocapillaries using single molecule fluorescence. Appl. Phys. Lett. 2012, 101, 223704. [Google Scholar] [CrossRef] [Green Version]
- Laohakunakorn, N.; Thacker, V.; Muthukumar, M.; Keyser, U.K. Electroosmotic Flow Reversal Outside Glass Nanopores. Nano Lett. 2015, 15, 695–702. [Google Scholar] [CrossRef]
- Reisner, W.; Beech, J.P.; Larsen, N.B.; Flyvbjerg, H.; Kristensen, A.; Tegenfeldt, J.O. Nanoconfinement-Enhanced Conformational Response of Single DNA Molecules to Changes in Ionic Environment. Phys. Rev. Lett. 2007, 99, 058302. [Google Scholar] [CrossRef] [Green Version]
- Perssona, F.; Tegenfeldt, J.O. DNA in nanochannels—Directly visualizing genomic information. Chem. Soc. Rev. 2010, 39, 985. [Google Scholar] [CrossRef] [PubMed]
- Reisner, W.; Pedersen, J.N.; Austin, R.H. DNA confinement in nanochannels: Physics and biological applications. Rep. Prog. Phys. 2012, 75, 106601. [Google Scholar] [CrossRef] [PubMed]
- Khandurina, J.; Guttman, A. Bioanalysis in microfluidic devices. J. Chromatogr. A 2002, 943, 275–285. [Google Scholar] [CrossRef]
- Hsieh, C.; Balducci, A.; Patrick, S. Doyle, Ionic Effects on the Equilibrium Dynamics of DNA Confined in Nanoslits. Nano Lett. 2008, 8, 1683–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mannion, J.; Reccius, C.H.; Cross, J.D.; Craighead, H.G. Conformational analysis of single DNA molecules undergoing entropically induced motion in nanochannels. Biophys. J. 2006, 90, 4538–4545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pires, L.R.; Vinayakumar, K.B.; Turos, M.; Miguel, V.; Gaspar, J. A Perspective on Microneedle-Based Drug Delivery and Diagnostics in Paediatrics. J. Pers. Med. 2019, 9, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azmana, M.; Mahmood, S.; Hiles, A.R.; Mandal, U.K.; Japairai, K.A.; Raman, S. Transdermal drug delivery system through polymeric microneedle. J. Drug Deliv. Sci. Technol. 2020, 60, 101877. [Google Scholar] [CrossRef]
- Ceze, L.; Nivala, J.; Strauss, K. Molecular digital data storage using DNA. Nat. Rev. Genet. 2019, 20, 456–466. [Google Scholar] [CrossRef]
- Akram, F.; Haq, I.U.; Ali, H.; Laghari, A.T. Trends to store digital data in DNA: An overview. Mol. Biol. Rep. 2018, 45, 1479–1490. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohshiro, T.; Komoto, Y.; Taniguchi, M. Single-Molecule Counting of Nucleotide by Electrophoresis with Nanochannel-Integrated Nano-Gap Devices. Micromachines 2020, 11, 982. https://doi.org/10.3390/mi11110982
Ohshiro T, Komoto Y, Taniguchi M. Single-Molecule Counting of Nucleotide by Electrophoresis with Nanochannel-Integrated Nano-Gap Devices. Micromachines. 2020; 11(11):982. https://doi.org/10.3390/mi11110982
Chicago/Turabian StyleOhshiro, Takahito, Yuki Komoto, and Masateru Taniguchi. 2020. "Single-Molecule Counting of Nucleotide by Electrophoresis with Nanochannel-Integrated Nano-Gap Devices" Micromachines 11, no. 11: 982. https://doi.org/10.3390/mi11110982
APA StyleOhshiro, T., Komoto, Y., & Taniguchi, M. (2020). Single-Molecule Counting of Nucleotide by Electrophoresis with Nanochannel-Integrated Nano-Gap Devices. Micromachines, 11(11), 982. https://doi.org/10.3390/mi11110982






