A Toolbox for Organelle Mechanobiology Research—Current Needs and Challenges
Abstract
1. Introduction
2. Mechanobiology of Mitochondria
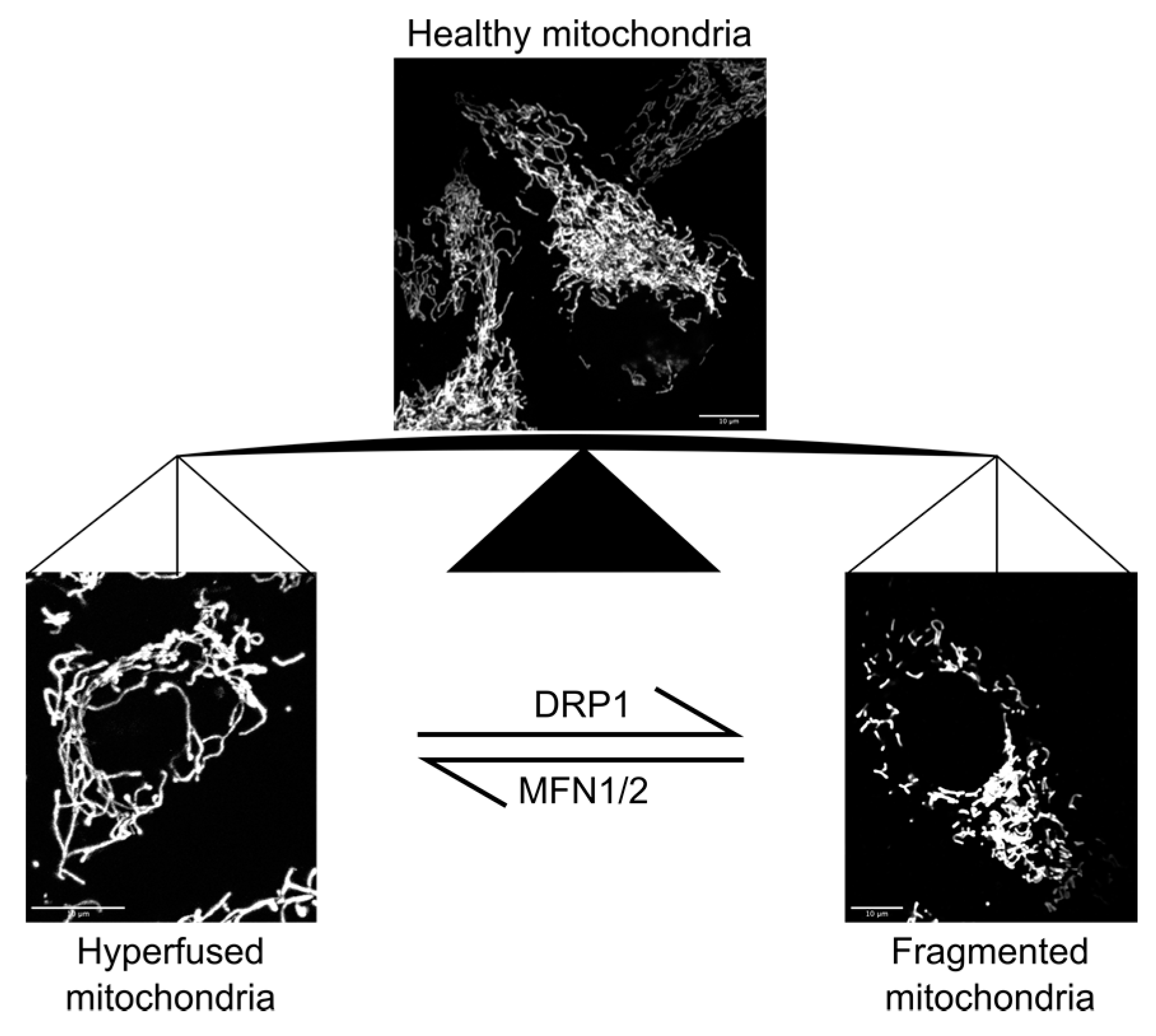
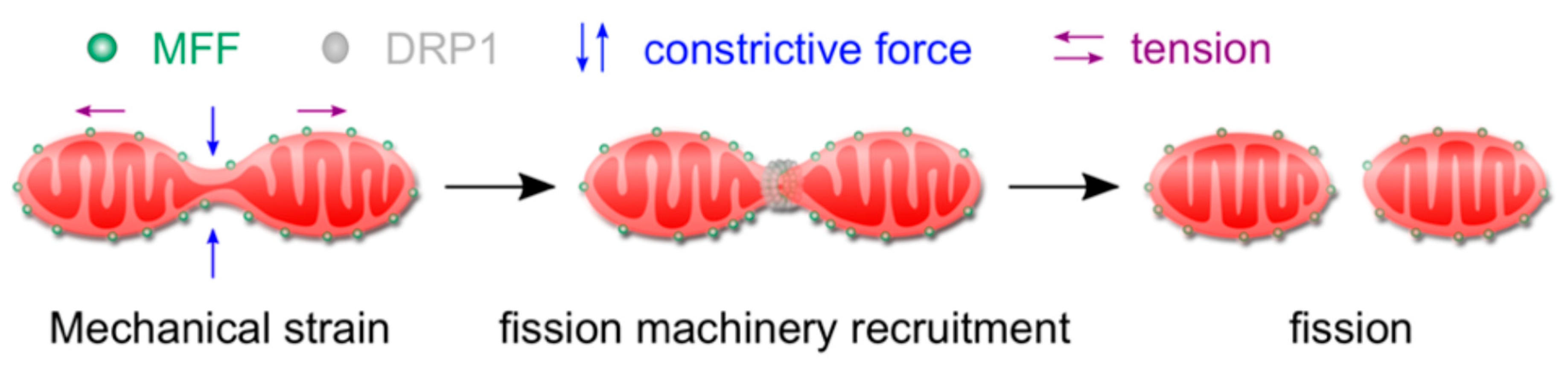
2.1. Mitochondrial Function and Response to Mechanical Stimulation
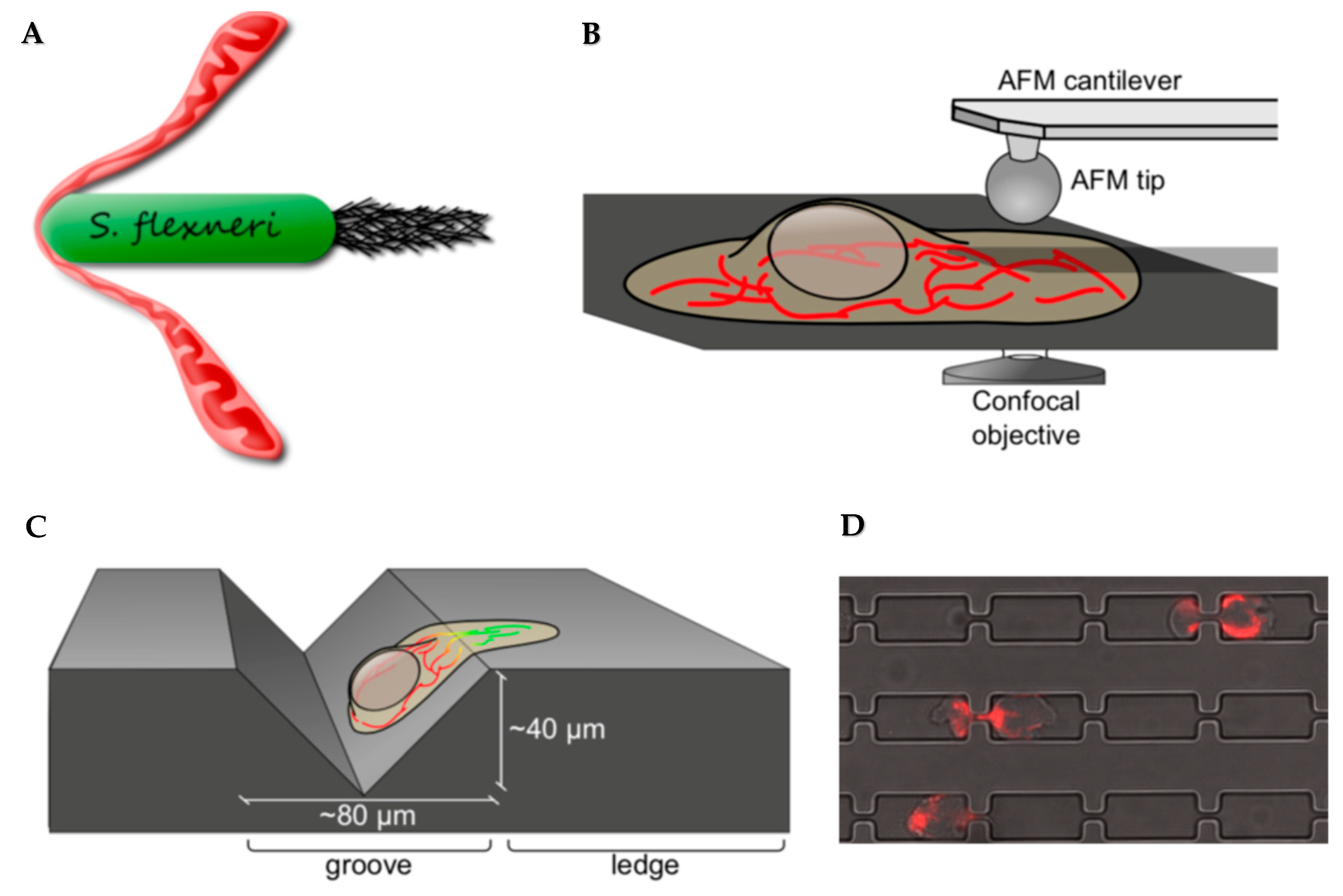
2.2. Current Challenges in Studying Mechanobiology of Mitochondria and Other Intracellular Organelles
2.3. Exciting Questions to Answer in Mitochondria Mechanobiology, and the Tools We Will Need
3. Future Engineering Tools for Studying Mechanobiology of Mitochondria and Other Intracellular Organelles
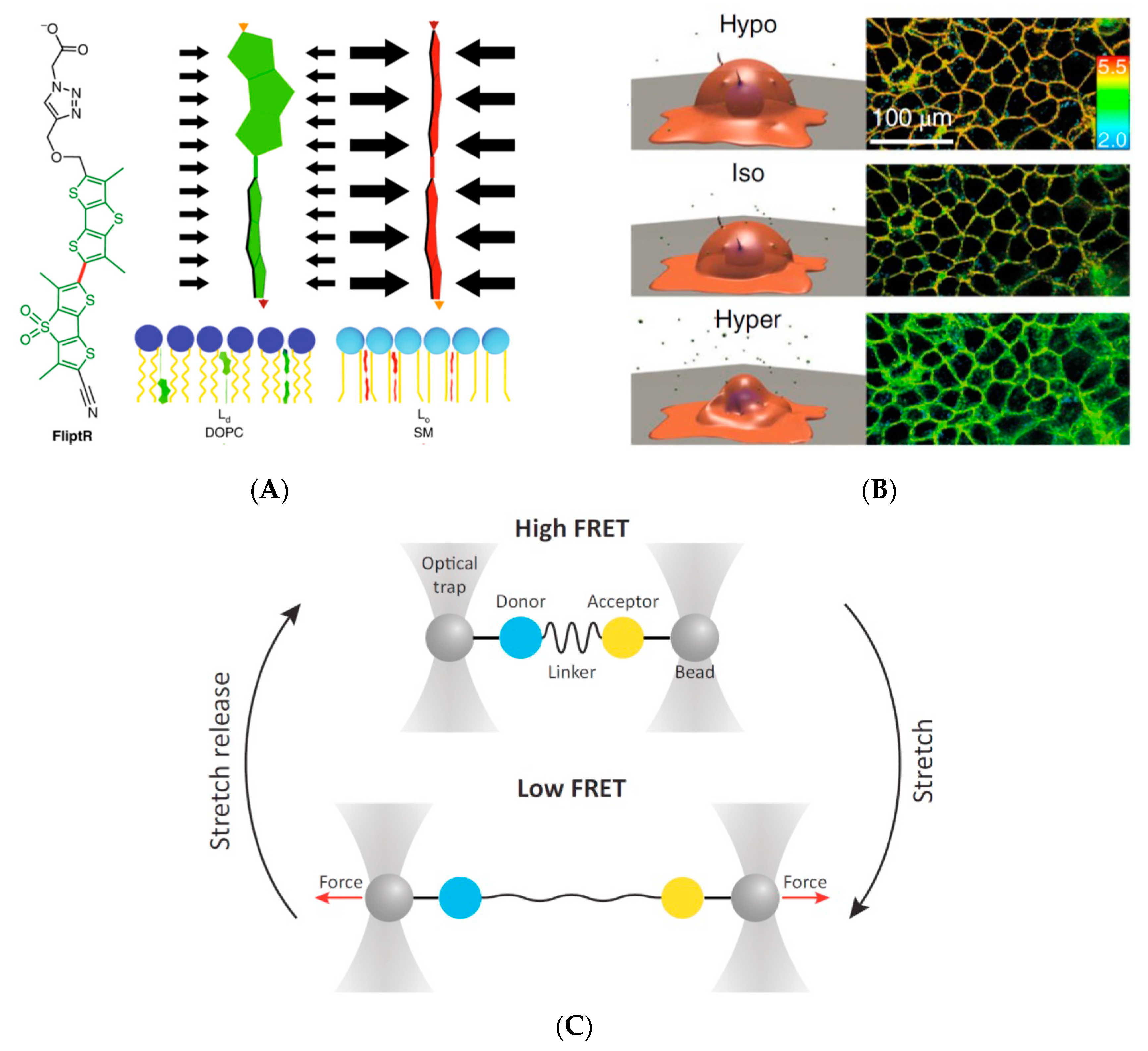
3.1. External Field Control for Intracellular Mechano-Stimulation
3.2. Detection of Forces/Tension on a Biological Membrane
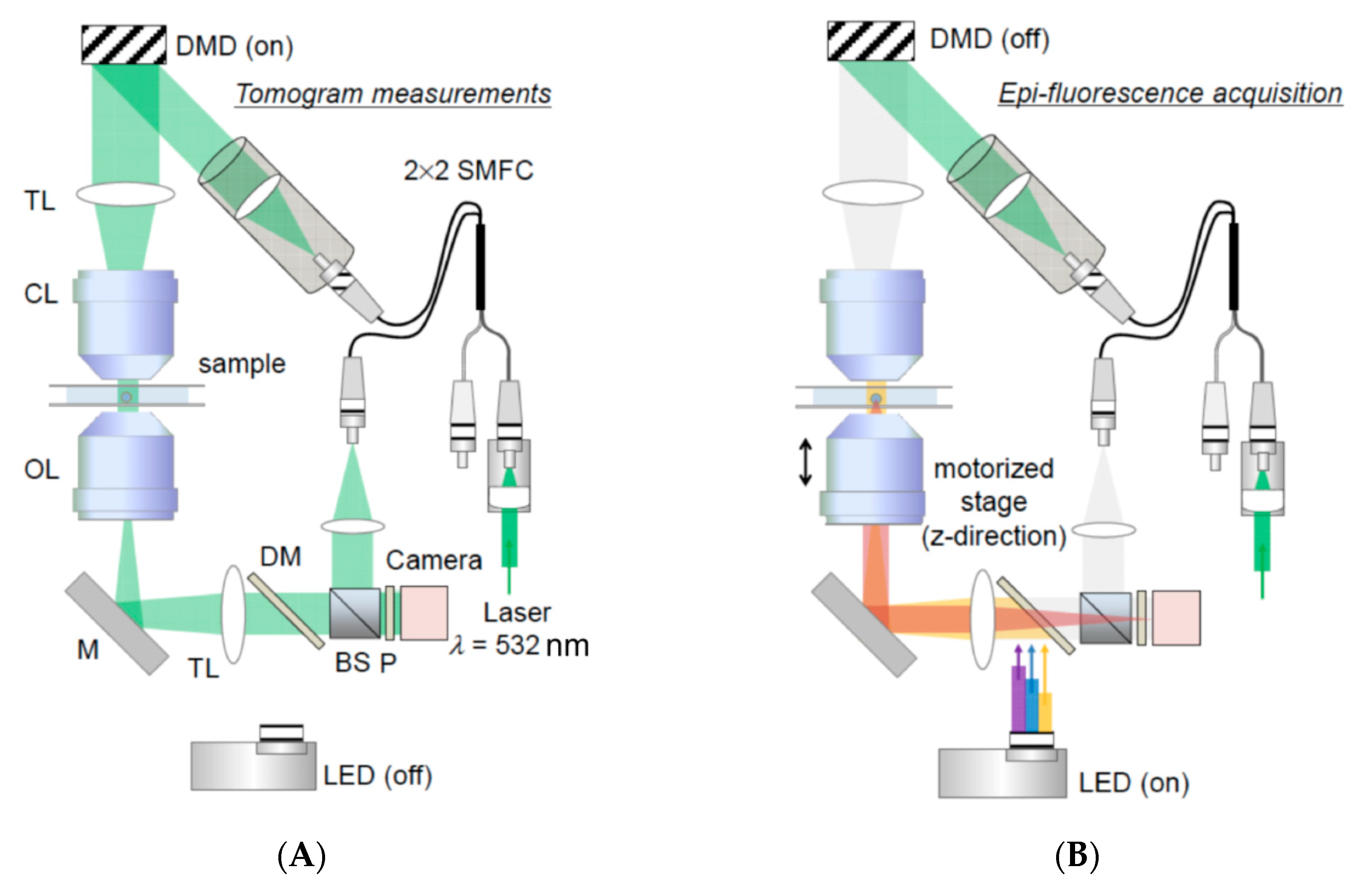
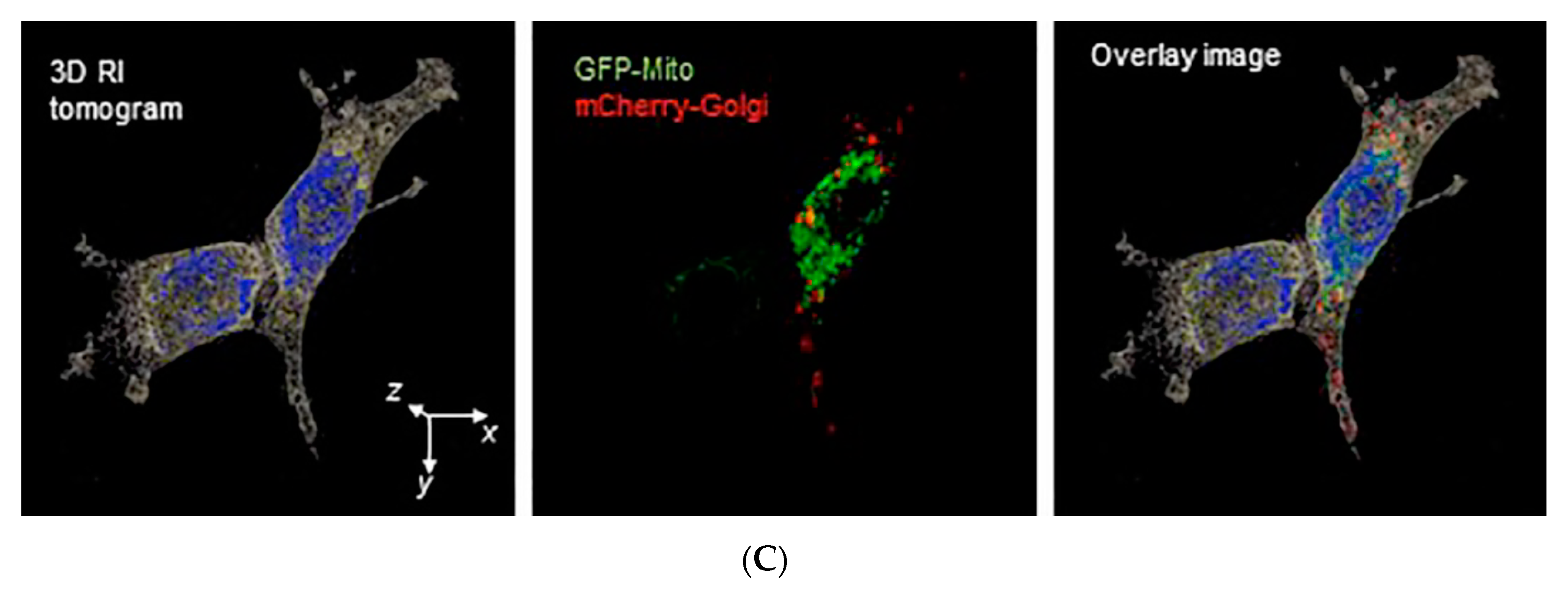
3.3. Assessing Biophysical Properties of Intracellular Organelles by Quantitative Phase Imaging (QPI)
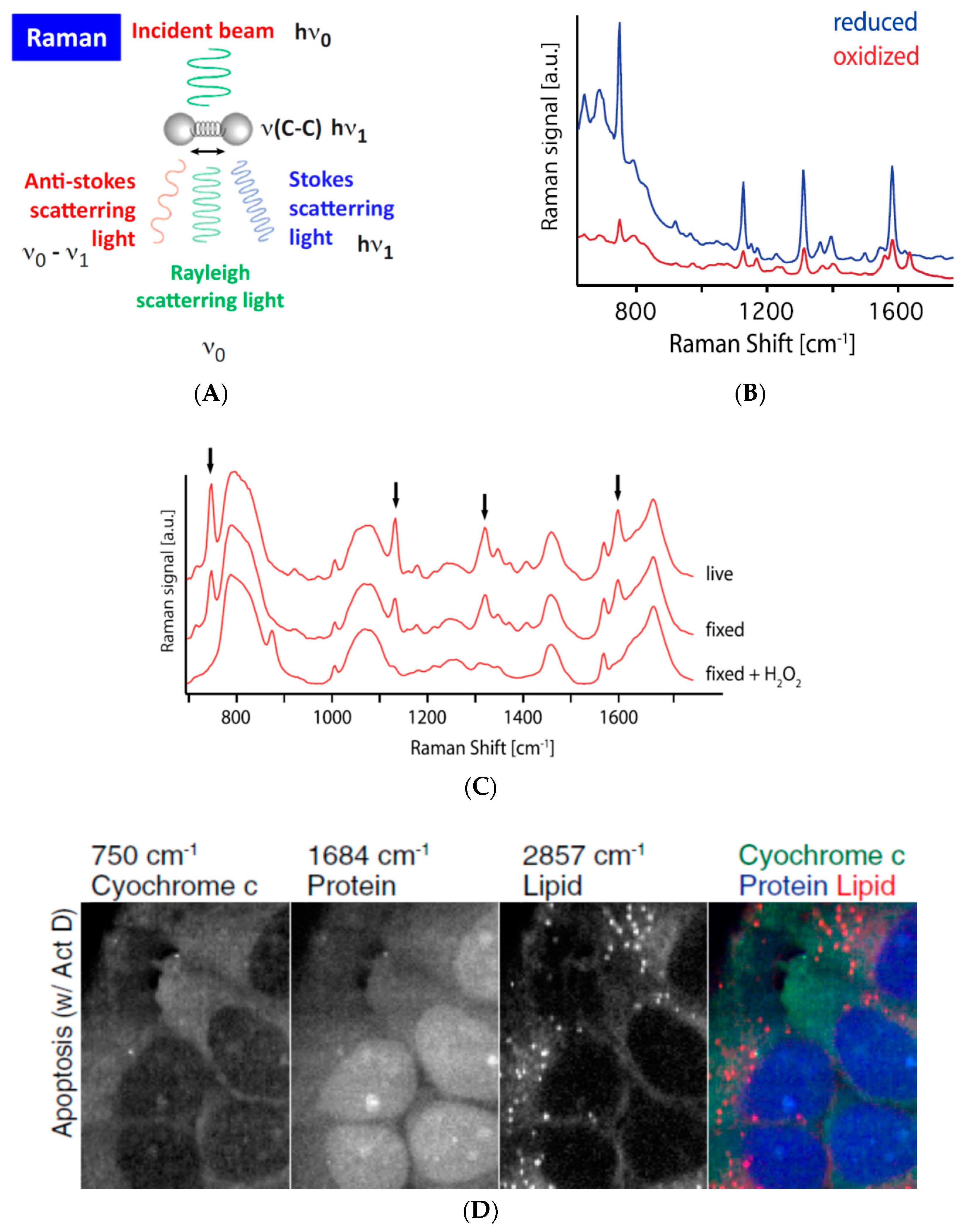
3.4. Assessing (Bio-)Chemical Properties of Intracellular Organelles by Raman Spectroscopy-Based Methods
4. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Moeendarbary, E.; Harris, A.R. Cell mechanics: Principles, practices, and prospects. Wiley Interdiscip. Rev. Syst. Biol. Med. 2014, 6, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Wall, M.; Butler, D.; Haj, A.E.; Bodle, J.C.; Loboa, E.G.; Banes, A.J. Key developments that impacted the field of mechanobiology and mechanotransduction. J. Orthop. Res. 2018, 36, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Eyckmans, J.; Chen, C.S. Designer biomaterials for mechanobiology. Nat. Mater. 2017, 16, 1164–1168. [Google Scholar] [CrossRef] [PubMed]
- Jansen, K.A.; Donato, D.M.; Balcioglu, H.E.; Schmidt, T.; Danen, E.H.J.; Koenderink, G.H. A guide to mechanobiology: Where biology and physics meet. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 3043–3052. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; van Drogen, F.; Dechant, R.; Oh, S.; Jeon, N.L.; Lee, S.S.; Peter, M. Protein kinase C and calcineurin cooperatively mediate cell survival under compressive mechanical stress. Proc. Natl. Acad. Sci. USA 2017, 114, 13471–13476. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Kornmann, B. Mechanical forces on cellular organelles. J. Cell Sci. 2018, 131, jcs218479. [Google Scholar] [CrossRef] [PubMed]
- Raab, M.; Gentili, M.; Belly, H.; Thiam, H.R.; Vargas, P.; Jimenez, A.J.; Lautenschlaeger, F.; Voituriez, R.; Lennon-Duménil, A.M.; Manel, N.; et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 2016, 352, 359–362. [Google Scholar] [CrossRef]
- Thiam, H.-R.; Vargas, P.; Carpi, N.; Crespo, C.L.; Raab, M.; Terric, E.; King, M.C.; Jecobelli, J.; Alberts, A.S.; Stradal, T.; et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun. 2016, 7, 10997. [Google Scholar] [CrossRef]
- Denais, C.M.; Gilbert, R.M.; Isermann, P.; McGregor, A.L.; Lindert, M.; Weigelin, B.; Davidson, P.M.; Friedl, P.; Wolf, K.; Lammerding, J. Nuclear envelope rupture and repair during cancer cell migration. Science 2016, 352, 353–358. [Google Scholar] [CrossRef]
- Helle, S.C.J.; Feng, Q.; Aebersold, M.J.; Hirt, L.; Gruter, R.R.; Vahid, A.; Sirianni, A.; Mostowy, S.; Snedeker, J.G.; Saric, A.; et al. Mechanical force induces mitochondrial fission. Elife 2017, 6, 1–26. [Google Scholar] [CrossRef]
- Mahecic, D.; Carlini, L.; Kleele, T.; Colom, A.; Goujon, A.; Matile, S.; Roux, A.; Manley, S. Membrane bending energy and tension govern mitochondrial division. BioRxiv 2018. [Google Scholar] [CrossRef]
- Yang, C.; Svitkina, T.M. Ultrastructure and dynamics of the actin-myosin II cytoskeleton during mitochondrial fission. Nat. Cell Biol. 2019, 21, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.A.; Nekimken, A.L.; Fechner, S.; O’Brien, L.E.; Pruitt, B.L. Microfluidics for Mechanobiology of Model Organisms; Academic Press: New York, NY, USA, 2018; Volume 146, pp. 217–259. [Google Scholar]
- Kurth, F.; Eyer, K.; Franco-Obregón, A.; Dittrich, P.S. A new mechanobiological era: Microfluidic pathways to apply and sense forces at the cellular level. Curr. Opin. Chem. Biol. 2012, 16, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, K.; Lenne, P.-F.; Graner, F. Measuring forces and stresses in situ in living tissues. Development 2016, 143, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Millet, M.; Ben Messaoud, R.; Luthold, C.; Bordeleau, F. Coupling Microfluidic Platforms, Microfabrication, and Tissue Engineered Scaffolds to Investigate Tumor Cells Mechanobiology. Micromachines 2019, 10, 418. [Google Scholar] [CrossRef]
- Song, J.W.; Gu, W.; Futai, N.; Warner, K.A.; Nor, J.E.; Takayama, S. Computer-Controlled Microcirculatory Support System for Endothelial Cell Culture and Shearing. Anal. Chem. 2005, 77, 3993–3999. [Google Scholar] [CrossRef] [PubMed]
- Tkachenko, E.; Gutierrez, E.; Ginsberg, M.H.; Groisman, A. An easy to assemble microfluidic perfusion device with a magnetic clamp. Lab. Chip 2009, 9, 1085. [Google Scholar] [CrossRef]
- Van der Meer, A.D.; Poot, A.A.; Feijen, J.; Vermes, I. Analyzing shear stress-induced alignment of actin filaments in endothelial cells with a microfluidic assay. Biomicrofluidics 2010, 4, 011103. [Google Scholar] [CrossRef]
- Cui, Y.; Hameed, F.M.; Yang, B.; Lee, K.; Pan, C.Q.; Park, S.; Sheetz, M. Cyclic stretching of soft substrates induces spreading and growth. Nat. Commun. 2015, 6, 6333. [Google Scholar] [CrossRef]
- Mann, J.M.; Lam, R.H.W.; Weng, S.; Sun, Y.; Fu, J. A silicone-based stretchable micropost array membrane for monitoring live-cell subcellular cytoskeletal response. Lab. Chip 2012, 12, 731–740. [Google Scholar] [CrossRef]
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006, 16, R551–R560. [Google Scholar] [CrossRef] [PubMed]
- Breda, C.N.S.; Davanzo, G.G.; Basso, P.J.; Saraiva Câmara, N.O.; Moraes-Vieira, P.M.M. Mitochondria as central hub of the immune system. Redox Biol. 2019, 26, 101255. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Diebold, L.P.; Kong, H.; Schieber, M.; Huang, H.; Hensley, C.T.; Mehta, M.M.; Wang, T.; Santos, J.H.; Woychik, R.; et al. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Mol. Cell 2016, 61, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Wada, J.; Nakatsuka, A. Mitochondrial Dynamics and Mitochondrial Dysfunction in Diabetes. Acta Med. Okayama 2016, 70, 151–158. [Google Scholar] [PubMed]
- Morciano, G.; Pedriali, G.; Sbano, L.; Iannitti, T.; Giorgi, C.; Pinton, P. Intersection of mitochondrial fission and fusion machinery with apoptotic pathways: Role of Mcl-1. Biol. Cell 2016, 67, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Guedes-Dias, P.; Pinho, B.R.; Soares, T.R.; Proença, J.; Duchen, M.R.; Oliveira, J.M.A. Mitochondrial dynamics and quality control in Huntington’s disease. Neurobiol. Dis. 2016, 90, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.R.; Burke, N.; Dongworth, R.K.; Hausenloy, D.J. Mitochondrial fusion and fission proteins: Novel therapeutic targets for combating cardiovascular disease. Br. J. Pharmacol. 2014, 171, 1890–1906. [Google Scholar] [CrossRef] [PubMed]
- Biala, A.K.; Dhingra, R.; Kirshenbaum, L.A. Mitochondrial dynamics: Orchestrating the journey to advanced age. J. Mol. Cell. Cardiol. 2015, 83, 37–43. [Google Scholar] [CrossRef]
- Van der Bliek, A.M.; Shen, Q.; Kawajiri, S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 2013, 5, a011072. [Google Scholar] [CrossRef]
- Otera, H.; Ishihara, N.; Mihara, K. New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 1256–1268. [Google Scholar] [CrossRef]
- Maiuri, P.; Terriac, E.; Paul-Gilloteaux, P.; Vignaud, T.; McNally, K.; Onuffer, J.; Thorn, K.; Nguyen, P.A.; Georgoulia, N.; Soong, D.; et al. The first World Cell Race. Curr. Biol. 2012, 22, R673–R675. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.P. Biophysical Tools for Cellular and Subcellular Mechanical Actuation of Cell Signaling. Biophys. J. 2016, 111, 1112–1118. [Google Scholar] [CrossRef] [PubMed]
- De Vlaminck, I.; Dekker, C. Recent advances in magnetic tweezers. Annu. Rev. Biophys. 2012, 41, 453–472. [Google Scholar] [CrossRef] [PubMed]
- Kilinc, D.; Lee, G.U. Advances in magnetic tweezers for single molecule and cell biophysics. Integr. Biol. 2014, 6, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Valberg, P.A.; Feldman, H.A. Magnetic particle motions within living cells. Measurement of cytoplasmic viscosity and motile activity. Biophys. J. 1987, 52, 551–561. [Google Scholar] [CrossRef]
- Bausch, A.R.; Möller, W.; Sackmann, E. Measurement of local viscoelasticity and forces in living cells by magnetic tweezers. Biophys. J. 1999, 76, 573–579. [Google Scholar] [CrossRef]
- Monzel, C.; Vicario, C.; Piehler, J.; Coppey, M.; Dahan, M. Magnetic control of cellular processes using biofunctional nanoparticles. Chem. Sci. 2017, 8, 7330–7338. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ho, C.; Tsatskis, Y.; Law, J.; Zhang, Z.; Zhu, M.; Dai, C.; Wang, F.; Tan, M.; Hopyan, S.; et al. Intracellular manipulation and measurement with multipole magnetic tweezers. Sci. Robot. 2019, 4, eaav6180. [Google Scholar] [CrossRef]
- Etoc, F.; Lisse, D.; Bellaiche, Y.; Piehler, J.; Coppey, M.; Dahan, M. Subcellular control of Rac-GTPase signalling by magnetogenetic manipulation inside living cells. Nat. Nanotechnol. 2013, 8, 193–198. [Google Scholar] [CrossRef]
- Norregaard, K.; Jauffred, L.; Berg-Sørensen, K.; Oddershede, L.B. Optical manipulation of single molecules in the living cell. Phys. Chem. Chem. Phys. 2014, 16, 12614–12624. [Google Scholar] [CrossRef]
- Būtaitė, U.G.; Gibson, G.M.; Ho, Y.H.D.; Taverne, M.; Taylor, J.M.; Phillips, D.B. Indirect optical trapping using light driven micro-rotors for reconfigurable hydrodynamic manipulation. Nat. Commun. 2019, 10, 1215. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Sun, Y.; Gudur, M.S.R.; Hsiao, Y.S.; Wu, Z.; Fu, J.; Deng, C.X. Two-bubble acoustic tweezing cytometry for biomechanical probing and stimulation of cells. Biophys. J. 2015, 108, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Sun, Y.; Chen, D.; Tay, D.; Chen, W.; Deng, C.X.; Fu, J. Acoustic tweezing cytometry for live-cell subcellular modulation of intracellular cytoskeleton contractility. Sci. Rep. 2013, 3, 2176. [Google Scholar] [CrossRef] [PubMed]
- Dal Molin, M.; Verolet, Q.; Colom, A.; Letrun, R.; Derivery, E.; Gonzalez-Gaitan, M.; Vauthey, E.; Roux, A.; Sakai, N.; Matile, S. Fluorescent flippers for mechanosensitive membrane probes. J. Am. Chem. Soc. 2015, 137, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Colom, A.; Derivery, E.; Soleimanpour, S.; Tomba, C.; Dal Molin, M.; Sakai, N.; González-Gaitán, M.; Matile, S.; Roux, A. A fluorescent membrane tension probe. Nat. Chem. 2018, 10, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Freikamp, A.; Cost, A.-L.; Grashoff, C. The Piconewton Force Awakens: Quantifying Mechanics in Cells. Trends Cell Biol. 2016, 26, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Sachs, F.; Meng, F. Fluorescence-based force/tension sensors: A novel tool to visualize mechanical forces in structural proteins in live cells. Antioxid. Redox Signal. 2014, 20, 986–999. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Galior, K.; Ma, V.P.-Y.; Salaita, K. Molecular Tension Probes for Imaging Forces at the Cell Surface. Acc. Chem. Res. 2017, 50, 2915–2924. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.Y.; Chin, L.K.; Ser, W.; Chen, H.F.; Hsieh, C.M.; Lee, C.H.; Sung, K.B.; Ayi, T.C.; Yap, P.H.; Liedberg, B.; et al. Cell refractive index for cell biology and disease diagnosis: Past, present and future. Lab. Chip 2016, 16, 634–644. [Google Scholar] [CrossRef]
- Park, Y.; Best, C.A.; Badizadegan, K.; Dasari, R.R.; Feld, M.S.; Kuriabova, T.; Henle, M.L.; Levine, A.J.; Popescu, G. Measurement of red blood cell mechanics during morphological changes. Proc. Natl. Acad. Sci. USA 2010, 107, 6731–6736. [Google Scholar] [CrossRef]
- Eldridge, W.J.; Steelman, Z.A.; Loomis, B.; Wax, A. Optical Phase Measurements of Disorder Strength Link Microstructure to Cell Stiffness. Biophys. J. 2017, 112, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Fang-Yen, C.; Badizadegan, K.; Oh, S.; Lue, N.; Dasari, R.R.; Feld, M.S. Tomographic phase microscopy. Nat. Methods 2007, 4, 717–719. [Google Scholar] [CrossRef] [PubMed]
- Haseda, K.; Kanematsu, K.; Noguchi, K.; Saito, H.; Umeda, N.; Ohta, Y. Significant correlation between refractive index and activity of mitochondria: Single mitochondrion study. Biomed. Opt. Express 2015, 6, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Bon, P.; Lécart, S.; Fort, E.; Lévêque-Fort, S. Fast Label-Free Cytoskeletal Network Imaging in Living Mammalian Cells. Biophys. J. 2014, 106, 1588–1595. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Park, W.S.; Na, S.; Kim, S.; Kim, T.; Heo, W.D.; Park, Y.K. Correlative three-dimensional fluorescence and refractive index tomography: Bridging the gap between molecular specificity and quantitative bioimaging. Biomed. Opt. Express 2017, 8, 5688. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Lee, S.Y.; Jung, J.H.; Shin, S.; Choi, H.G.; Cha, G.H.; Park, W.; Lee, S.; Park, Y.K. Combining Three-Dimensional Quantitative Phase Imaging and Fluorescence Microscopy for the Study of Cell Pathophysiology. Yale J. Biol. Med. 2018, 91, 267–277. [Google Scholar] [PubMed]
- Okada, M.; Smith, N.I.; Palonpon, A.F.; Endo, H.; Kawata, S.; Sodeoka, M.; Fujita, K. Label-free Raman observation of cytochrome c dynamics during apoptosis. Proc. Natl. Acad. Sci. USA 2012, 109, 28–32. [Google Scholar] [CrossRef]
- Erjavec, N.; Pinato, G.; Ramser, K. Raman spectroscopy as a tool for detecting mitochondrial fitness. J. Raman Spectrosc. 2016, 47, 933–939. [Google Scholar] [CrossRef]
- Petibois, C. 3D Quantitative Chemical Imaging of Tissues by Spectromics. Trends Biotechnol. 2017, 35, 1194–1207. [Google Scholar] [CrossRef]
- Kumamoto, Y.; Harada, Y.; Takamatsu, T.; Tanaka, H. Label-free Molecular Imaging and Analysis by Raman Spectroscopy. ACTA Histochem. Cytochem. 2018, 51, 101–110. [Google Scholar] [CrossRef]
- Scarponi, F.; Mattana, S.; Corezzi, S.; Caponi, S.; Comez, L.; Sassi, P.; Morresi, A.; Paolantoni, M.; Urbanelli, L.; Emiliani, C.; et al. High-Performance Versatile Setup for Simultaneous Brillouin-Raman Microspectroscopy. Phys. Rev. X 2017, 7, 031015. [Google Scholar] [CrossRef]
- Elsayad, K.; Polakova, S.; Gregan, J. Probing Mechanical Properties in Biology Using Brillouin Microscopy. Trends Cell Biol. 2019, 29, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Ming, W.; Zeng, H.; Zhang, Z.; Lu, H. Deep learning-based component identification for the Raman spectra of mixtures. Analyst 2019, 144, 1789–1798. [Google Scholar] [CrossRef] [PubMed]
- Gastegger, M.; Behler, J.; Marquetand, P. Machine learning molecular dynamics for the simulation of infrared spectra. Chem. Sci. 2017, 8, 6924–6935. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Q.; Lee, S.S.; Kornmann, B. A Toolbox for Organelle Mechanobiology Research—Current Needs and Challenges. Micromachines 2019, 10, 538. https://doi.org/10.3390/mi10080538
Feng Q, Lee SS, Kornmann B. A Toolbox for Organelle Mechanobiology Research—Current Needs and Challenges. Micromachines. 2019; 10(8):538. https://doi.org/10.3390/mi10080538
Chicago/Turabian StyleFeng, Qian, Sung Sik Lee, and Benoît Kornmann. 2019. "A Toolbox for Organelle Mechanobiology Research—Current Needs and Challenges" Micromachines 10, no. 8: 538. https://doi.org/10.3390/mi10080538
APA StyleFeng, Q., Lee, S. S., & Kornmann, B. (2019). A Toolbox for Organelle Mechanobiology Research—Current Needs and Challenges. Micromachines, 10(8), 538. https://doi.org/10.3390/mi10080538




