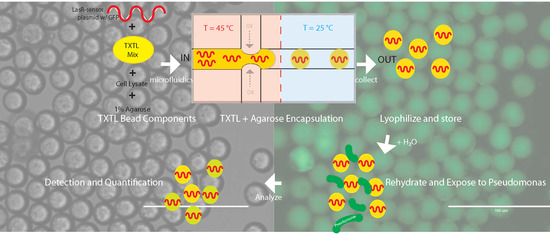
On a Robust, Sensitive Cell-Free Method for Pseudomonas Sensing and Quantification in Microfluidic Templated Hydrogels
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell-Free Lysate and TXTL Extracts
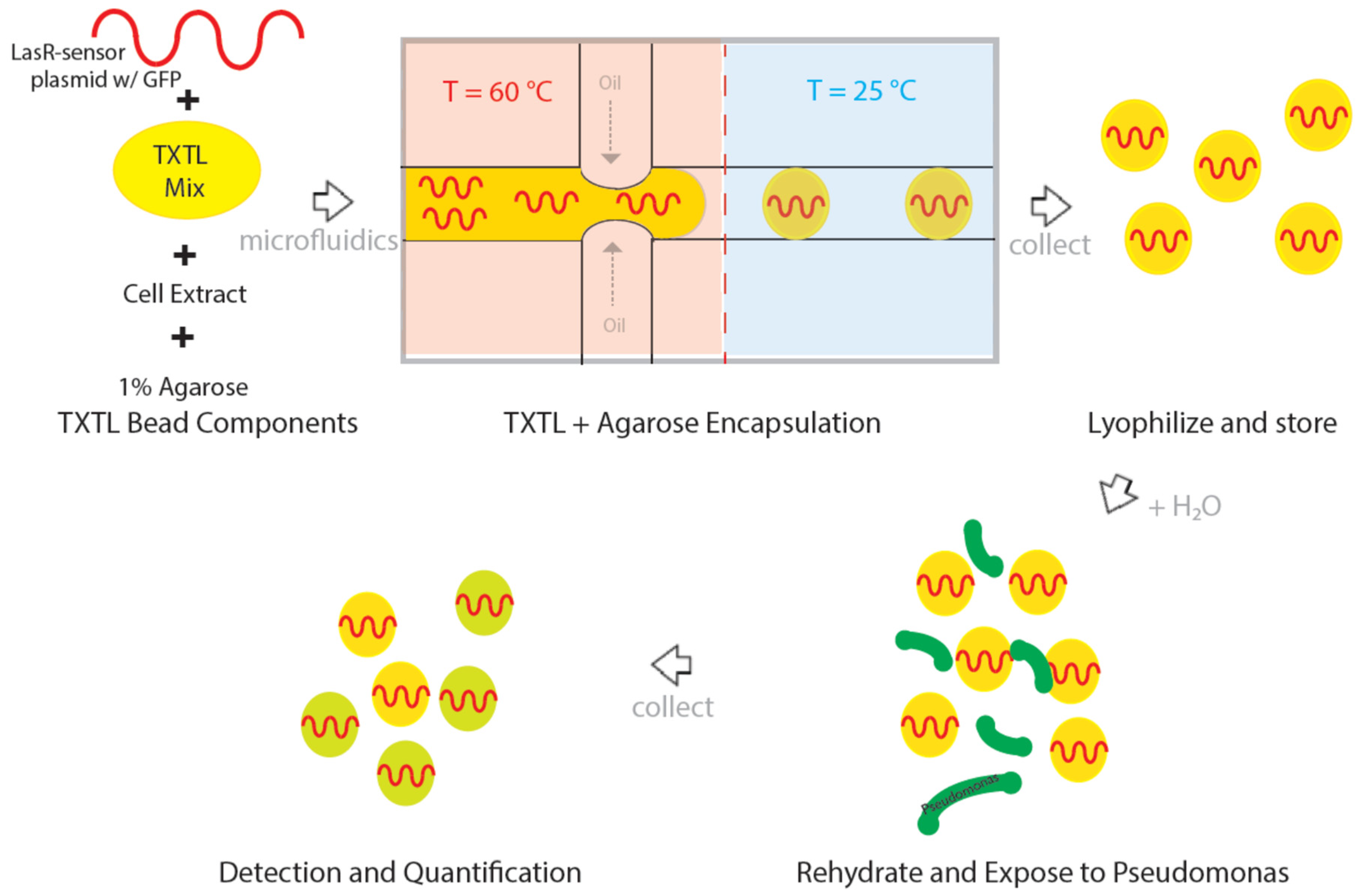
2.2. Agarose Encapsulation of TXTL
2.3. Lyophilization and Storage
2.4. Fluorescence Microscopy
2.5. RNA Analyses
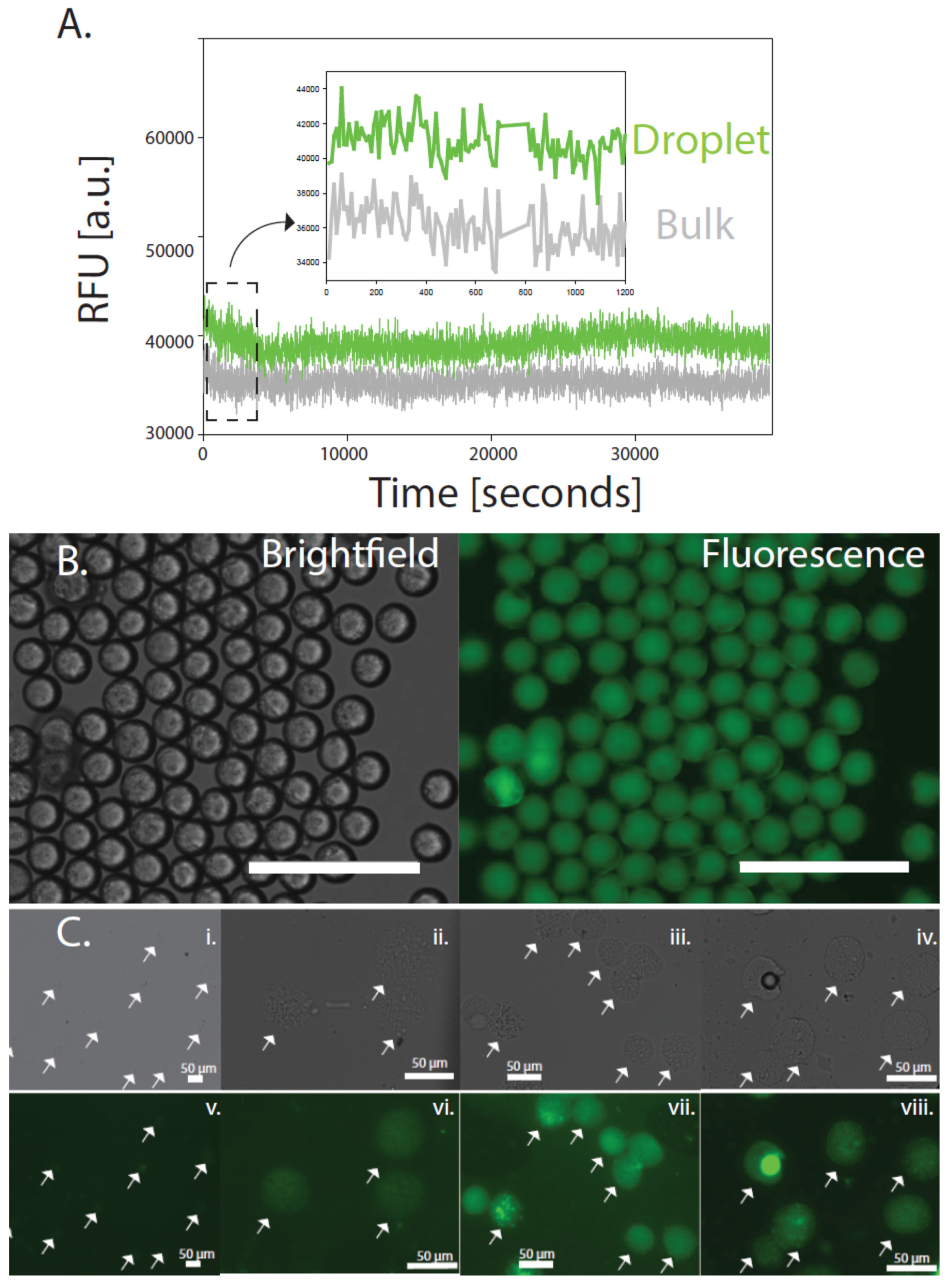
2.6. Fluorescence Plate Reader
2.7. Fluorescence Activated Cell Sorting (FACS)
2.8. 3D-Structure Illumination Microscopy (3D-SIM) and Wide-Field Deconvolution Fluorescence Microscopy
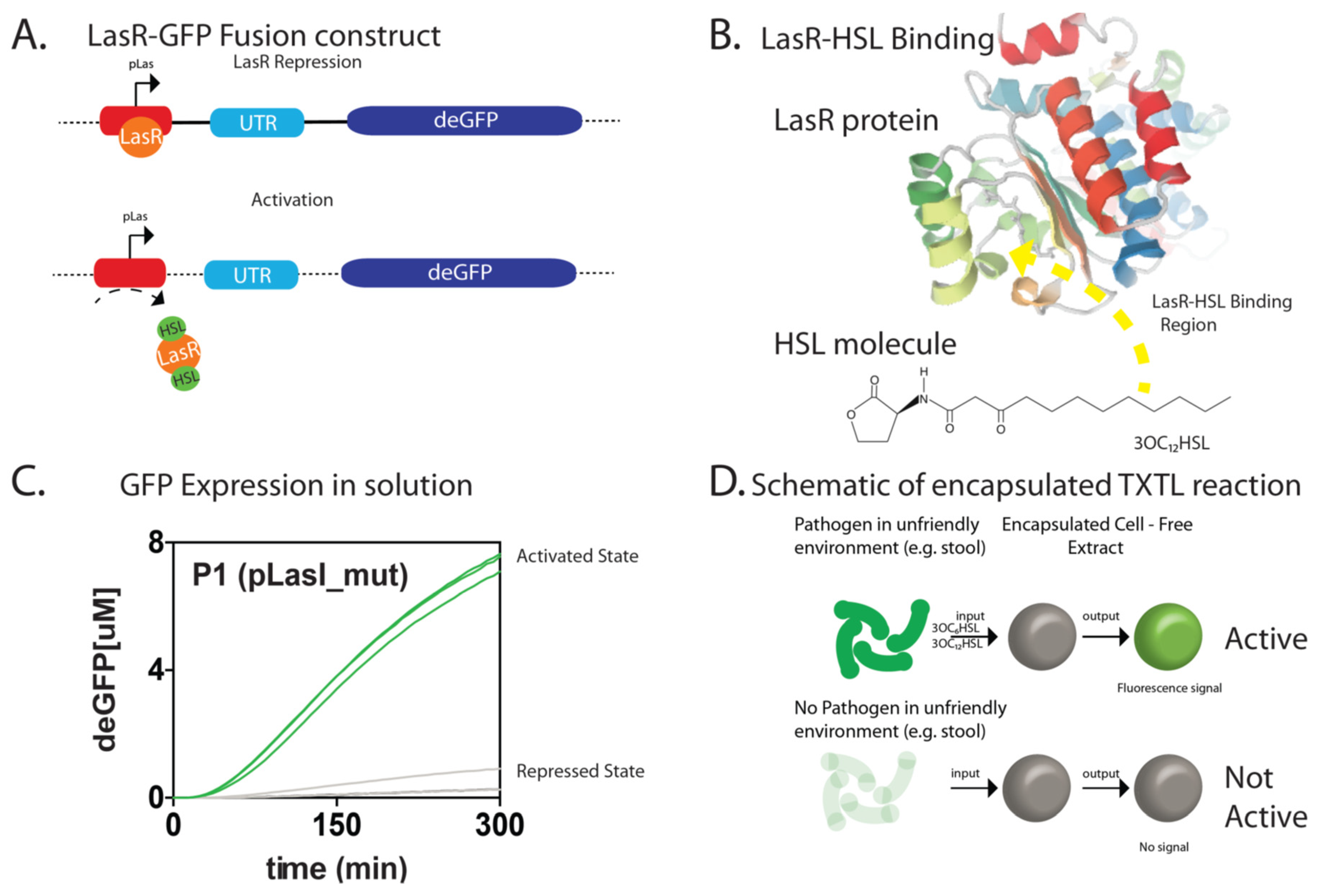
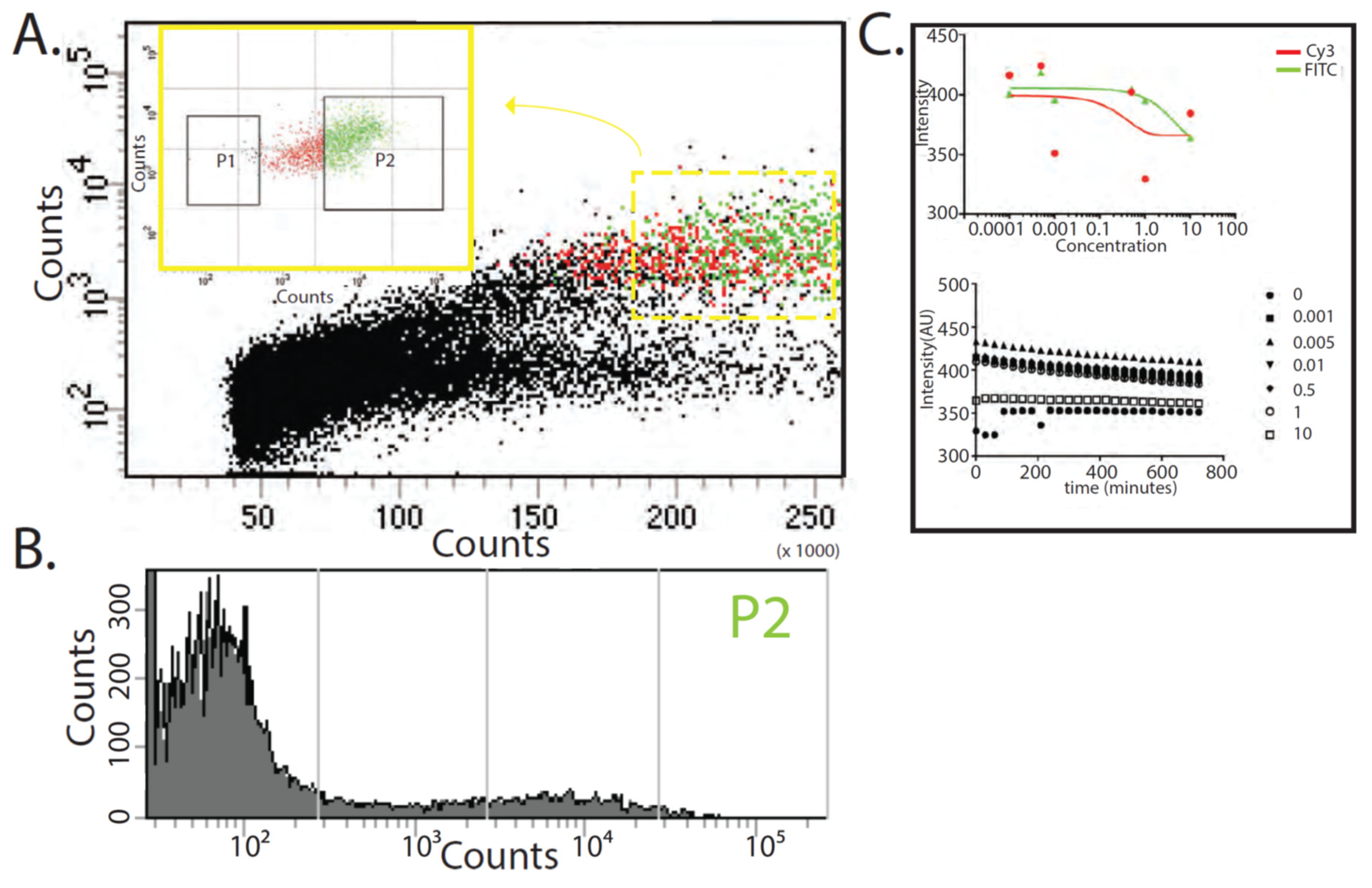
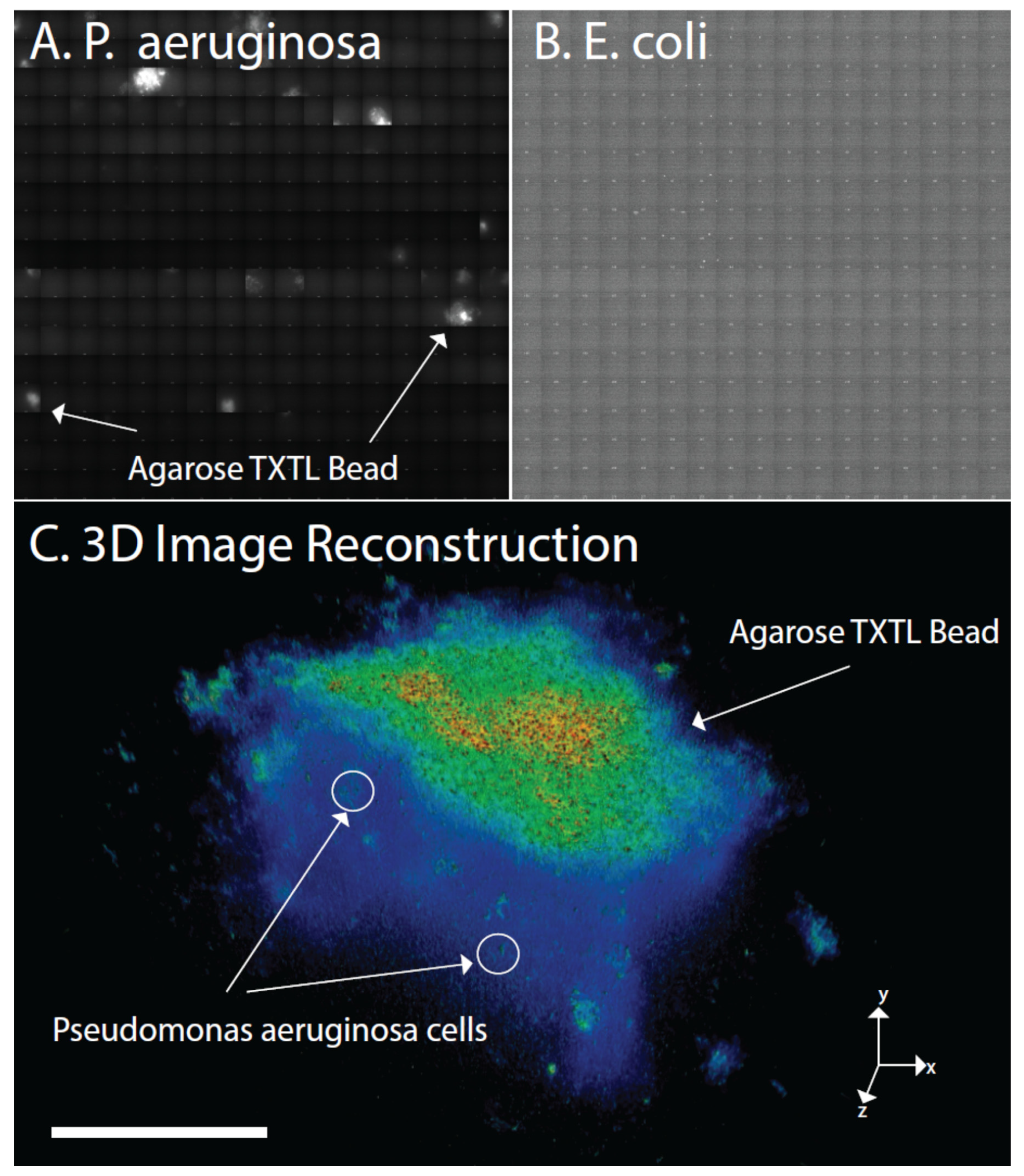
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Dicarlo, J.E.; Chavez, A.; Dietz, S.L.; Esvelt, K.M.; Church, G.M. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 2015, 33, 1250–1255. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.C.; Xie, L.; Deng, W.; Guglielmi, B.; Witkowsky, L.B.; Bosanac, L.; Zhang, E.T.; El Beheiry, M.; Masson, J.-B.; Dahan, M.; et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science 2015, 350, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Kotula, J.W.; Kerns, S.J.; Shaket, L.A.; Siraj, L.; Collins, J.J.; Way, J.C.; Silver, P.A. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. USA 2014, 111, 4838–4843. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, F.; Carr, P.A.; Wang, H.H.; Lajoie, M.J.; Sterling, B.; Kraal, L.; Tolonen, A.C.; Gianoulis, T.A.; Goodman, D.B.; Reppas, N.B.; et al. Precise Manipulation of Chromosomes in Vivo Enables Genome-Wide Code Replacement. Science 2011, 333, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Moon, T.S.; Lou, C.; Tamsir, A.; Stanton, B.C.; Voigt, C.A. Genetic programs constructed from layered logic gates in single cells. Nature 2012, 491, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Noireaux, V.; Bar-Ziv, R.; Libchaber, A. Principles of cell-free genetic circuit assembly. Proc. Natl. Acad. Sci. USA 2003, 100, 12672–12677. [Google Scholar] [CrossRef] [PubMed]
- Noireaux, V.; Libchaber, A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. USA 2004, 101, 17669–17674. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Yeung, E.; Hayes, C.A.; Noireaux, V.; Murray, R.M. Linear DNA for Rapid Prototyping of Synthetic Biological Circuits in an Escherichia coli Based TX-TL Cell-Free System. ACS Synth. Biol. 2014, 3, 387–397. [Google Scholar] [CrossRef]
- Koerber, J.; Hornsby, M.; Wells, J. An Improved Single-Chain Fab Platform for Efficient Display and Recombinant Expression. J. Mol. Biol. 2015, 427, 576–586. [Google Scholar] [CrossRef]
- Bar-Even, A.; Tawfik, D. Engineering specialized metabolic pathways—Is there a room for enzyme improvements. Curr. Opin. Biotechnol. 2013, 24, 310–319. [Google Scholar] [CrossRef]
- Jewett, M.C.; Calhoun, K.A.; Voloshin, A.; Wuu, J.J.; Swartz, J.R. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol. 2008, 4, 1–10. [Google Scholar] [CrossRef]
- Harris, D.C.; Jewett, M.C. Cell-free biology: Exploiting the interface between synthetic biology and synthetic chemistry. Curr. Opin. Biotechnol. 2012, 23, 672–678. [Google Scholar] [CrossRef]
- Albayrak, C.; Swartz, J.R. Cell-free co-production of an orthogonal transfer RNA activates efficient site-specific non-natural amino acid incorporation. Nucleic Acids Res. 2013, 41, 5949–5963. [Google Scholar] [CrossRef]
- Endo, Y.; Sawasaki, T. Cell-free expression systems for eukaryotic protein production. Curr. Opin. Biotechnol. 2006, 17, 373–380. [Google Scholar] [CrossRef]
- Goshima, N.; Kawamura, Y.; Fukumoto, A.; Miura, A.; Honma, R.; Satoh, R.; Wakamatsu, A.; Yamamoto, J.-I.; Kimura, K.; Nishikawa, T.; et al. Human protein factory for converting the transcriptome into an in vitro–expressed proteome. Nat. Methods 2008, 5, 1011–1017. [Google Scholar] [CrossRef]
- Crick, F. Central Dogma of Molecular Biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 6th ed.; Garland Science: New York, NY, USA; Abingdon, UK, 2014. [Google Scholar]
- Shrestha, P.; Holland, T.M.; Bundy, B.C. Streamlined extract preparation for Escherichia coli-based cell-free protein synthesis by sonication or bead vortex mixing. BioTechniques 2012, 53, 163–174. [Google Scholar] [CrossRef]
- Yang, W.C.; Patel, K.G.; Wong, H.E.; Swartz, J.R. Simplifying and streamlining Escherichia coli-based cell-free protein synthesis. Biotechnol. Prog. 2012, 28, 413–420. [Google Scholar] [CrossRef]
- Kwon, Y.-C.; Jewett, M.C. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep. 2015, 5, 8663. [Google Scholar] [CrossRef]
- Lentini, R.; Martini, L.; Del Bianco, C.; Spencer, A.C.; Torino, D.; Forlin, M.; Mansy, S.S. Fluorescent Proteins and in Vitro Genetic Organization for Cell-Free Synthetic Biology. ACS Synth. Biol. 2013, 2, 482–489. [Google Scholar] [CrossRef]
- Pardee, K.; Slomovic, S.; Nguyen, P.Q.; Lee, J.W.; Donghia, N.; Burrill, D.; Ferrante, T.; McSorley, F.R.; Furuta, Y.; Vernet, A.; et al. Portable, On-Demand Biomolecular Manufacturing. Cell 2016, 167, 248–259. [Google Scholar] [CrossRef]
- Pollock, N.R.; Rolland, J.P.; Kumar, S.; Beattie, P.D.; Jain, S.; Noubary, F.; Wong, V.L.; Pohlmann, R.A.; Ryan, U.S.; Whitesides, G.M. A paper-based multiplexed transaminase test for low-cost, point-of-care liver function testing. Sci. Transl. Med. 2012, 4, 152ra129. [Google Scholar] [CrossRef]
- Nath, P.; Arun, R.K.; Chanda, N. A paper based microfluidic device for the detection of arsenic using a gold nanosensor. RSC Adv. 2014, 4, 59558–59561. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Ferrante, T.; Cameron, D.E.; DaleyKeyser, A.; Yin, P.; Collins, J.J. Paper-based Synthetic Gene Networks. Cell 2014, 159, 940–954. [Google Scholar] [CrossRef]
- Streets, A.M.; Zhang, X.; Cao, C.; Pang, Y.; Wu, X.; Xiong, L.; Yang, L.; Fu, Y.; Zhao, L.; Tang, F.; et al. Microfluidic single-cell whole-transcriptome sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 7048–7053. [Google Scholar] [CrossRef]
- Niederholtmeyer, H.; Sun, Z.Z.; Hori, Y.; Yeung, E.; Verpoorte, A.; Murray, R.M.; Maerkl, S.J. Rapid cell-free forward engineering of novel genetic ring oscillators. eLife 2015, 4, 1–21. [Google Scholar] [CrossRef]
- Sun, Z.Z.; Hayes, C.A.; Shin, J.; Caschera, F.; Murray, R.M.; Noireaux, V. Protocols for Implementing an Escherichia coli Based TX-TL Cell-Free Expression System for Synthetic Biology. JoVE 2013, 79, e50762. [Google Scholar] [CrossRef]
- Abate, A.R.; Romanowsky, M.B.; Agresti, J.J.; Weitz, D.A. Valve-based flow focusing for drop formation. Appl. Phys. Lett. 2009, 94, 023503. [Google Scholar] [CrossRef]
- Holtze, C.; Rowat, A.C.; Agresti, J.J.; Hutchison, J.B.; Angilè, F.E.; Schmitz, C.H.J.; Köster, S.; Duan, H.; Humphry, K.J.; Scanga, R.A.; et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip 2008, 8, 1632. [Google Scholar] [CrossRef]
- Strauss, M.P.; Liew, A.T.F.; Turnbull, L.; Whitchurch, C.B.; Monahan, L.G.; Harry, E.J. 3D-SIM Super Resolution Microscopy Reveals a Bead-Like Arrangement for FtsZ and the Division Machinery: Implications for Triggering Cytokinesis. PLoS Biol. 2012, 10, e1001389. [Google Scholar] [CrossRef]
- Kiratisin, P.; Tucker, K.D.; Passador, L. LasR, a Transcriptional Activator of Pseudomonas aeruginosa Virulence Genes, Functions as a Multimer. J. Bacteriol. 2002, 184, 4912–4919. [Google Scholar] [CrossRef]
- Ng, W.-L.; Bassler, B.L. Bacterial Quorum-Sensing Network Architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Gambello, M.J.; Kaye, S.; Iglewski, B.H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 1993, 61, 1180–1184. [Google Scholar]
- Thaden, J.; Lory, S.; Gardner, T. Quorum-Sensing Regulation of a Copper Toxicity System in Pseudomonas aeruginosa. J. Bacteriol. 2010, 192, 2557–2568. [Google Scholar] [CrossRef]
- Didovyk, A.; Tonooka, T.; Tsimring, L.; Hasty, J. Rapid and scalable preparation of bacterial lysates for cell-free gene expression. ACS Synth. Biol. 2017, 6, 2198–2208. [Google Scholar] [CrossRef]
- Forget, A.; Pique, R.A.; Ahmadi, V.; Lüdeke, S.; Shastri, V.P. Mechanically Tailored Agarose Hydrogels through Molecular Alloying with Beta Sheet Polysaccharides. Macromol. Rapid Commun. 2014, 36, 196–203. [Google Scholar] [CrossRef]
- Jaggers, R.W.; Bon, S.A.F. Communication between hydrogel beads via chemical signalling. J. Mater. Chem. B 2017, 5, 8681–8685. [Google Scholar] [CrossRef]
- Shin, J.; Noireaux, V. Study of messenger RNA inactivation and protein degradation in an Escherichia coli cell-free expression system. J. Biol. Eng. 2010, 4, 9. [Google Scholar] [CrossRef]
- Agresti, J.; Antipov, E.; Abate, A.R.; Ahn, K.; Rowat, A.C.; Baret, J.C.; Marquez, M.; Klibanov, A.M.; Griffiths, A.D.; Weitz, D.A. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 2009, 107, 4004–4009. [Google Scholar] [CrossRef]
- Abate, A.; Hung, T.; Mary, P.; Agresti, J.J.; Weitz, D.A. High-throughput injection with microfluidics using picoinjectors. Proc. Natl. Acad. Sci. USA 2010, 107, 19163–19166. [Google Scholar] [CrossRef] [PubMed]
- Chappell, J.; Jensen, K.; Freemont, P.S. Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res. 2013, 41, 3471–3481. [Google Scholar] [CrossRef] [PubMed]
- Wen, K.Y.; Cameron, L.; Chappell, J.; Jensen, K.; Bell, D.J.; Kelwick, R.; Kopniczky, M.; Davies, J.C.; Filloux, A.; Freemont, P.S. A Cell-Free Biosensor for Detecting Quorum Sensing Molecules in P. aeruginosa-Infected Respiratory Samples. ACS Synth. Biol. 2017, 6, 2293–2301. [Google Scholar] [CrossRef] [PubMed]
- Eder, P.S.; Devine, R.J.; Dagle, J.M.; Walder, J.A. Substrate Specificity and Kinetics of Degradation of Antisense Oligonucleotides by a 3′ Exonuclease in Plasma. Antisense Res. Dev. 1991, 1, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Kerins, S.; Collins, R.; McCarthy, T. Characterization of an Endonuclease IV 3′-5′ Exonuclease Activity. J. Biol. Chem. 2003, 278, 3048–3054. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, 222–226. [Google Scholar] [CrossRef]
- Contreras-Moreira, B. 3D-footprint: A database for the structural analysis of protein-DNA complexes. Nucleic Acids Res. 2010, 38, 91–97. [Google Scholar] [CrossRef]
- Fallah-Araghi, A.; Ryckelynck, M.; Griffiths, A.D.; Baret, J.-C. A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab Chip 2012, 12, 882–891. [Google Scholar] [CrossRef]
- Hori, Y.; Kantak, C.; Murray, R.M.; Abate, A.R. Cell-free exttract based optimization of biomolecular circuits with droplet microfluidics. Lab Chip 2017, 17, 3037–3042. [Google Scholar] [CrossRef]
- Eastburn, D.J.; Sciambi, A.; Abate, A.R. Identification and genetic analysis of cancer cells with PCR-activated cell sorting. Nucleic Acids Res. 2014, 42, e128. [Google Scholar] [CrossRef]
- Gooderham, W.; Hancock, R. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMs Microbiol. Rev. 2008, 33, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Wagner, V.; Gillis, R.; Iglewski, B. Transcriptome analysis of quorum-sensing regulation and virulence factor expression in Pseudomonas aeurginosa. Vaccine 2004, 22, S15–S20. [Google Scholar] [CrossRef] [PubMed]
- LeRoux, M.; Kirkpatrick, R.L.; Montauti, E.I.; Tran, B.Q.; Peterson, S.B.; Harding, B.N.; Whitney, J.C.; Russell, A.B.; Traxler, B.; Goo, Y.A. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aerginosa. eLife 2015, 4, e05701. [Google Scholar] [CrossRef] [PubMed]
- Stanton, B.; Nielsen, A.A.; Tamsir, A.; Clancy, K.; Peterson, T.; Voigt, C.A. Genomic Mining of Prokaryotic Repressors for Orthogonal Logic Gates. Nat. Chem. Biol. 2014, 10, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Cole, R.H.; Tang, S.-Y.; Siltanen, C.A.; Shahi, P.; Zhang, J.Q.; Poust, S.; Gartner, Z.J.; Abate, A.R. Printed droplet microfluidics for on demand dispensing of picoliter droplets and cells. Proc. Natl. Acad. Sci. USA 2017, 114, 8728–8733. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.A.; Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009, 10, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Seto, J.; Romero, P. Shaping it up: Design and Engineering of biominerals and crystalline materials from the bottom up. In Biomineralization and Biomaterials: Fundamentals and Applications; Aparicio, C., Ginebra, M.P., Eds.; Woodhead Publishing: Cambridge, UK, 2016; pp. 3–50. [Google Scholar]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seto, J. On a Robust, Sensitive Cell-Free Method for Pseudomonas Sensing and Quantification in Microfluidic Templated Hydrogels. Micromachines 2019, 10, 506. https://doi.org/10.3390/mi10080506
Seto J. On a Robust, Sensitive Cell-Free Method for Pseudomonas Sensing and Quantification in Microfluidic Templated Hydrogels. Micromachines. 2019; 10(8):506. https://doi.org/10.3390/mi10080506
Chicago/Turabian StyleSeto, Jong. 2019. "On a Robust, Sensitive Cell-Free Method for Pseudomonas Sensing and Quantification in Microfluidic Templated Hydrogels" Micromachines 10, no. 8: 506. https://doi.org/10.3390/mi10080506
APA StyleSeto, J. (2019). On a Robust, Sensitive Cell-Free Method for Pseudomonas Sensing and Quantification in Microfluidic Templated Hydrogels. Micromachines, 10(8), 506. https://doi.org/10.3390/mi10080506