Microfluidic Cell Trapping for Single-Cell Analysis
Abstract
1. Introduction
2. Experimental Setup
2.1. Materials and Methods
2.1.1. Cell Culture and Preparation
2.1.2. Cell Seeding
2.1.3. Chip Fabrications
3. Results
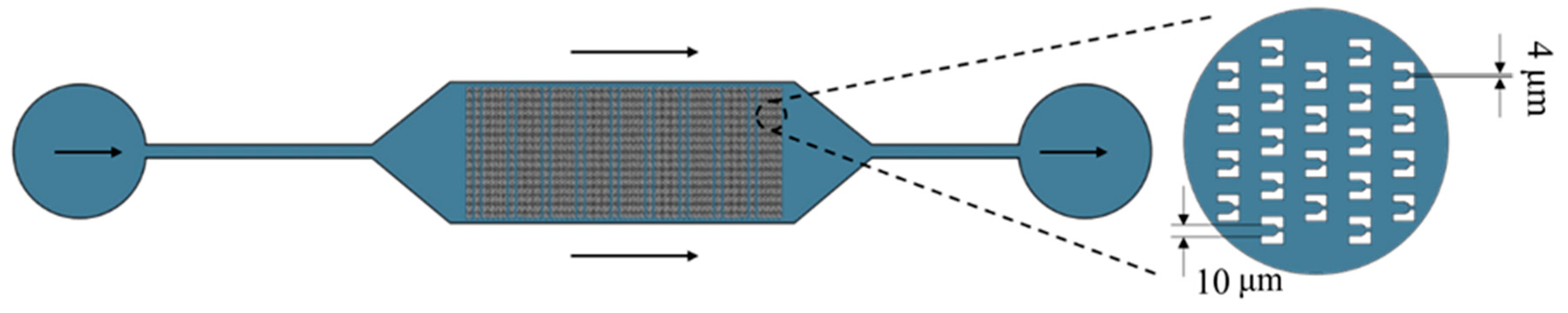
3.1. Microcavity Cell Trapping
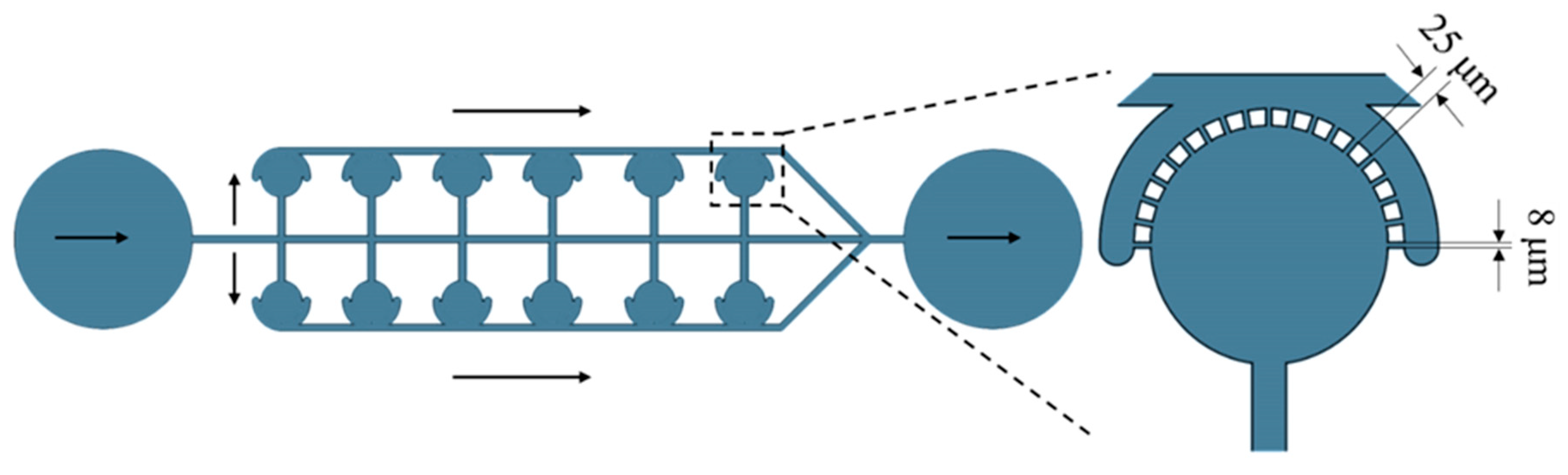
3.2. U-Shape Capture
3.3. Flow Shortcut Structure
4. Discussion and Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Iino, R.; Hayama, K.; Amezawa, H.; Sakakihara, S.; Kim, S.H.; Matsumono, Y.; Nishino, K.; Yamaguchi, A.; Noji, H. A single-cell drug efflux assay in bacteria by using a directly accessible femtoliter droplet array. Lab. Chip. 2012, 12, 3923–3929. [Google Scholar] [CrossRef] [PubMed]
- Andersson, H.; van den Berg, A. Microfluidic devices for cellomics: A review. Sens. Actuators B Chem. 2003, 92, 315–325. [Google Scholar] [CrossRef]
- Beebe, D.J.; Mensing, G.A.; Walker, G.M. Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 2002, 4, 261–286. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, P.S.; Manz, A. Lab-on-a-chip: Microfluidics in drug discovery. Nat. Rev. Drug Discov. 2006, 5, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P. Microfluidics—Downsizing large-scale biology. Nat. Biotechnol. 2001, 19, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Stone, H.A.; Kim, S. Microfluidics: Basic issues applications, and challenges. AIChE J. 2001, 47, 1250–1254. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Noshari, J.; Anderson, T.J.; Becker, F.F. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis 2009, 30, 1388–1398. [Google Scholar] [CrossRef]
- Wang, S.S.; Huang, X.Y.; Yang, C. Mixing enhancement for high viscous fluids in a microfluidic chamber. Lab Chip 2011, 11, 2081–2087. [Google Scholar] [CrossRef]
- Wunderlich, B.; Nettels, D.; Benke, S.; Clark, J.; Weidner, S.; Hofmann, H.; Pfeil, S.H.; Schuler, B. Microfluidic mixer designed for performing single-molecule kinetics with confocal detection on timescales from milliseconds to minutes. Nat. Protoc. 2013, 8, 1459–1474. [Google Scholar] [CrossRef]
- Toh, A.G.; Wang, Z.P.; Yang, C.; Nguyen, N.T. Engineering microfluidic concentration gradient generators for biological applications. Microfluid. Nanofluidics 2014, 16, 1–18. [Google Scholar] [CrossRef]
- Shields, C.W.; Reyes, C.D.; Lopez, G.P. Microfluidic cell sorting: A review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 2015, 15, 1230–1249. [Google Scholar] [CrossRef] [PubMed]
- Wagner, I.; Materne, E.M.; Brincker, S.; Süssbier, U.; Frädrich, C.; Busek, M.; Sonntag, F.; Sakharov, D.A.; Trushkin, E.V.; Tonevitsky, A.G.; et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip 2013, 13, 3538–3547. [Google Scholar] [CrossRef] [PubMed]
- Chiriacò, M.S.; Bianco, M.; Nigro, A. Lab-on-Chip for Exosomes and Microvesicles Detection and Characterization. Sensors 2018, 18, 3175. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, S.S.; Dunlay, C.J.; Diane, M. Simeonecde and Sunitha Nagrath, Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.-V.; Jen, C.-P. Impedance detection integrated with dielectrophoresis enrichment platform for lung circulating tumor cells in a microfluidic channel. Biosens. Bioelectron. 2018, 121, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.T.; Wunsch, B.H.; Dogra, N. Integrated nanoscale deterministic lateral displacement arrays for separation of extracellular vesicles from clinically-relevant volumes of biological samples. Lab Chip 2018, 18, 3913–3925. [Google Scholar] [CrossRef]
- Li, N.; Tourovskaia, A.; Folch, A. Biology on a chip: Microfabrication for studying the behavior of cultured cells. Crit. Rev. Biomed. Eng. 2003, 31, 423–488. [Google Scholar] [CrossRef]
- Cluzel, P.; Surette, M.; Leibler, S. An ultrasensitive bacterial motor revealed by monitoring. Science 2000, 287, 1652–1655. [Google Scholar] [CrossRef]
- Nourmohammadzadeh, M.; Lo, J.F.; Bochenek, M.; Mendoza-Elias, J.E.; Wang, Q.; Li, Z.; Zeng, L.; Qi, M.; Eddington, D.T.; Oberholzer, J.; et al. Microfluidic array with integrated oxygenation control for real-time live-cell imaging: Effect of hypoxia on physiology of microencapsulated pancreatic islets. Anal. Chem. 2013, 85, 11240–11249. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Khademhosseini, A. Seeking the right context for evaluating nanomedicine: From tissue models in petri dishes to microfluidic organs-on-a-chip. Nanomedicine 2015, 10, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; Heo, J.; Hua, S.Z. Spatially resolved shear distribution in microfluidic chip for studying force transduction mechanisms in cells. Lab Chip 2010, 10, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Au, S.H.; Chamberlain, M.D.; Mahesh, S.; Sefton, M.V.; Wheeler, A.R. Hepatic organoids for microfluidic drug screening. Lab Chip 2014, 14, 3290–3299. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.A.; Parashar, A.; Gibson, R.; Robertson, A.P.; Martin, R.J.; Pandey, S. A microfluidic platform for high-sensitivity, real-time drug screening on C. elegans and parasitic nematodes. Lab Chip 2011, 11, 2385–2396. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Taylor, D.; Agrawal, N.; Wang, H.; Kim, H.; Han, A.; Rege, K.; Jayaraman, A. A programmable microfluidic cell array for combinatorial drug screening. Lab Chip 2012, 12, 1813–1822. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.H.; Huang, S.B.; Lee, G.B. Microfluidic cell culture systems for drug research. Lab Chip 2010, 10, 939–956. [Google Scholar] [CrossRef] [PubMed]
- Caplin, J.D.; Granados, N.G.; James, M.R.; Montazami, R.; Hashemi, N. Microfluidic Organ-on-a-Chip Technology for Advancement of Drug Development and Toxicology. Adv. Healthc. Mater. 2015, 4, 1426–1450. [Google Scholar] [CrossRef]
- Rimann, M.; Graf-Hausner, U. Synthetic 3D multicellular systems for drug development. Curr. Opin. Biotechnol. 2012, 23, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Tay, S.; Hughey, J.J.; Lee, T.K.; Lipniacki, T.; Quake, S.R.; Covert, M.W. Single-cell NF-kappa B dynamics reveal digital activation and analogue information processing. Nature 2010, 466, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Junkin, M.; Tay, S. Microfluidic single-cell analysis for systems immunology. Lab Chip 2014, 14, 1246–1260. [Google Scholar] [CrossRef]
- Kellogg, R.A.; Tay, S. Noise facilitates transcriptional control under dynamic inputs. Cell 2015, 160, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.A.; Jiang, N.; White, R.A., 3rd; Fisher, D.S.; Quake, S.R. High-throughput sequencing of the zebrafish antibody repertoire. Science 2009, 324, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.C.; Wang, J.; Potanina, A.; Quake, S.R. Whole-genome molecular haplotyping of single cells. Nat. Biotechnol. 2011, 29, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Lecault, V.; Vaninsberghe, M.; Sekulovic, S.; Knapp, D.J.; Wohrer, S.; Bowden, W.; Viel, F.; McLaughlin, T.; Jarandehei, A.; Miller, M.; et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat. Methods 2011, 8, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Pagliara, S.; Simon, L.; Ulrich, D.; Keyser, F. Channel-Facilitated Diffusion Boosted by Particle Binding at the Channel Entrance. Phys. Rev. Lett. 2014, 113, 048102. [Google Scholar] [CrossRef] [PubMed]
- Zilionis, R.; Nainys, J.; Veres, A.; Savova, V.; Zemmour, D.; Klein, A.M.; Mazutis, L. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 2017, 2, 44–73. [Google Scholar] [CrossRef]
- Yu, F.B.; Willis, L.; Chau, R.M.W.; Zambon, A.; Horowitz, M.; Bhaya, D.; Huang, K.C.; Quake, S.R. Long-term microfluidic tracking of coccoid cyanobacterial cells reveals robust control of division timing. BMC Biol. 2017, 15, 11–28. [Google Scholar] [CrossRef]
- Robert, L.; Ollion, J.; Robert, J.; Song, X.; Matic, I.; Elez, M. Mutation dynamics and fitness effects followed in single cells. Science 2018, 359, 1283–1286. [Google Scholar] [CrossRef]
- Takayama, S.; Ostuni, E.; LeDuc, P.; Naruse, K.; Ingber, D.E.; Whitesides, G.M. Selective Chemical Treatment of Cellular Microdomains Using Multiple Laminar Streams. Chem. Biol. 2003, 10, 123–130. [Google Scholar] [CrossRef][Green Version]
- Takayama, S.; Ostuni, E.; LeDuc, P.; Naruse, K.; Ingber, D.E.; Whitesides, G.M. Laminar Flows—Subcellular Positioning of Small Molecules. Nature 2001, 411, 1016. [Google Scholar] [CrossRef]
- Tonooka, T.; Teshima, T.; Takeuchi, S. Clustering triple microbeads in adynamic microarray for timing-controllable bead-based reactions. Microflu-idics Nanofluidics 2013, 14, 1039–1048. [Google Scholar] [CrossRef]
- Matsue, T.; Matsumoto, N.; Uchida, I. Rapid Micropatterning of Living Cells by Repulsive Dielectrophoretic Force. Electrochim. Acta 1997, 42, 3251–3256. [Google Scholar] [CrossRef]
- Huang, Y.; Joo, S.; Duhon, M.; Heller, M.; Wallace, B.; Xu, X. Dielectrophoretic cellseparation and gene expression profiling on microelectronic chip arrays. Anal. Chem. 2002, 74, 3362–3371. [Google Scholar] [CrossRef] [PubMed]
- Voldman, J. Electrical forces for microscale cell manipulation. Annu. Rev. Biomed. Eng. 2006, 8, 425–454. [Google Scholar] [CrossRef] [PubMed]
- Hejazian, M.; Li, W.; Nguyen, N.-T. Lab on a chip for continuous-flow magnetic cell separation. Lab Chip 2015, 15, 959. [Google Scholar] [CrossRef] [PubMed]
- Molloy, J.E.; Padgett, M.J. Lights, action: Optical tweezers. Contemp. Phys. 2002, 43, 241–258. [Google Scholar] [CrossRef]
- Petersson, F.; Nilsson, A.; Holm, C.; Jonsson, H.; Laurell, T. Continuous separation of lipid particles from erythrocytes by means of laminar flow and acousticstanding wave forces. Lab Chip 2005, 5, 20–22. [Google Scholar] [CrossRef]
- Lenshof, A.; Jamal, A.; Dykes, J.; Urbansky, A.; Astrand-Grundström, I.; Laurell, T.; Scheding, S. Efficient purification of CD4+ lymphocytes from peripheral blood progenitor cell products using affinity bead acoustophoresis. Cytom. Part A J. Int. Soc. Anal. Cytol. 2014, 85, 933–941. [Google Scholar] [CrossRef]
- Kim, S.M.; Lee, S.H.; Suh, K.Y. Cell research with physically modified microfluidic channels: A review. Lab Chip 2008, 8, 1015–1023. [Google Scholar] [CrossRef]
- Johann, R.M. Cell trapping in microfluidic chips. Anal. Bioanal. Chem. 2006, 385, 408–412. [Google Scholar] [CrossRef]
- Di Carlo, D.; Aghdam, N.; Lee, L.P. Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays. Anal. Chem. 2006, 78, 4925–4930. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.T.; Lin, R.Z.; Chang, W.Y.; Chang, H.Y.; Liu, C.H. Rapid heterogeneous liver-cell on-chip patterning via the enhanced field-induced dielectrophotesis trap. Lab Chip 2006, 6, 724–734. [Google Scholar] [CrossRef]
- Jang, K.J.; Suh, K.Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010, 10, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Amponsah, P.K.; Al-Shatti, M.; Nie, Z.; Bandyopadhyay, B.C. Engineering of polarized tubular structures in a microfluidic device to study calcium phosphate stone formation. Lab Chip 2012, 12, 4037–4040. [Google Scholar] [CrossRef]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef]
- Huh, D.; Fujioka, H.; Tung, Y.C.; Futai, N.; Paine, R., 3rd; Grotberg, J.B.; Takayama, S. Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proc. Natl. Acad. Sci. USA 2007, 104, 18886–18891. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, B.; Wang, H.; Tan, Z.; Quan, Y. Microfluidic Cell Trapping for Single-Cell Analysis. Micromachines 2019, 10, 409. https://doi.org/10.3390/mi10060409
Deng B, Wang H, Tan Z, Quan Y. Microfluidic Cell Trapping for Single-Cell Analysis. Micromachines. 2019; 10(6):409. https://doi.org/10.3390/mi10060409
Chicago/Turabian StyleDeng, Bing, Heyi Wang, Zhaoyi Tan, and Yi Quan. 2019. "Microfluidic Cell Trapping for Single-Cell Analysis" Micromachines 10, no. 6: 409. https://doi.org/10.3390/mi10060409
APA StyleDeng, B., Wang, H., Tan, Z., & Quan, Y. (2019). Microfluidic Cell Trapping for Single-Cell Analysis. Micromachines, 10(6), 409. https://doi.org/10.3390/mi10060409



