Droplet Microfluidics-Enabled High-Throughput Screening for Protein Engineering
Abstract
1. Introduction
2. Droplet-Based Hight-Throughput Screening
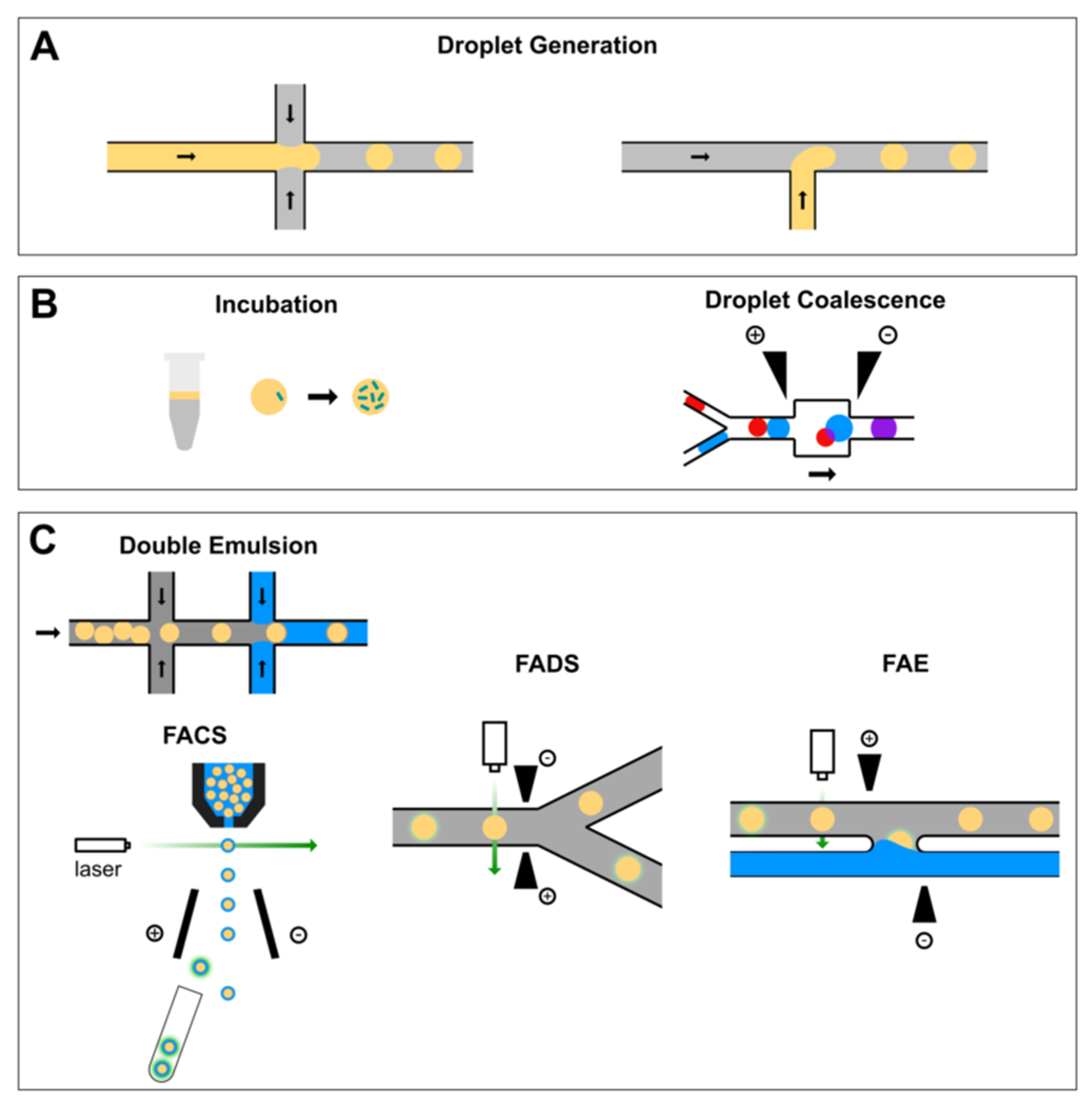
2.1. Microfluidics Droplet Generation
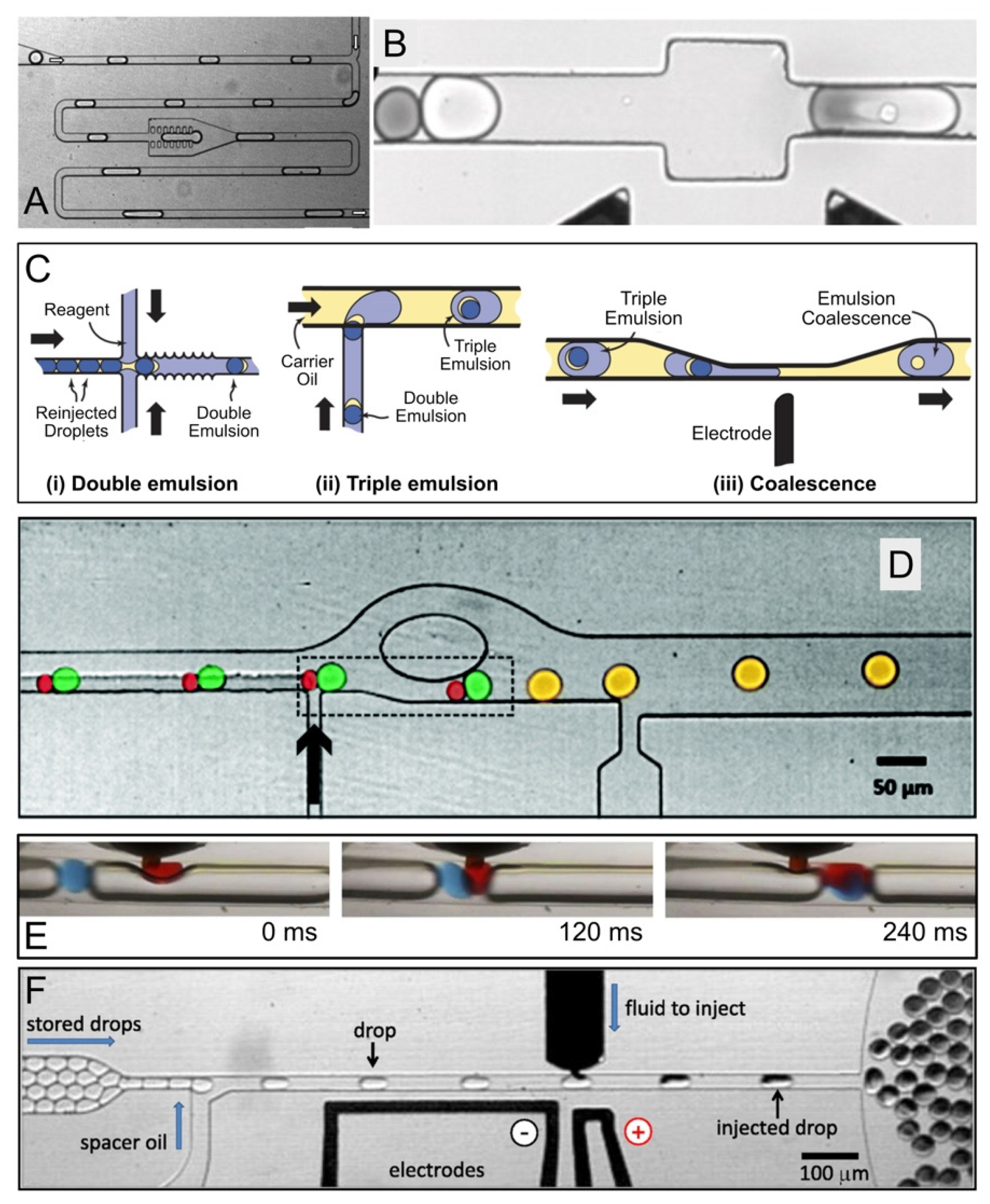
2.2. Reagent Addition in Droplet Flow
2.3. Flow Cytometric Sorting of Droplets
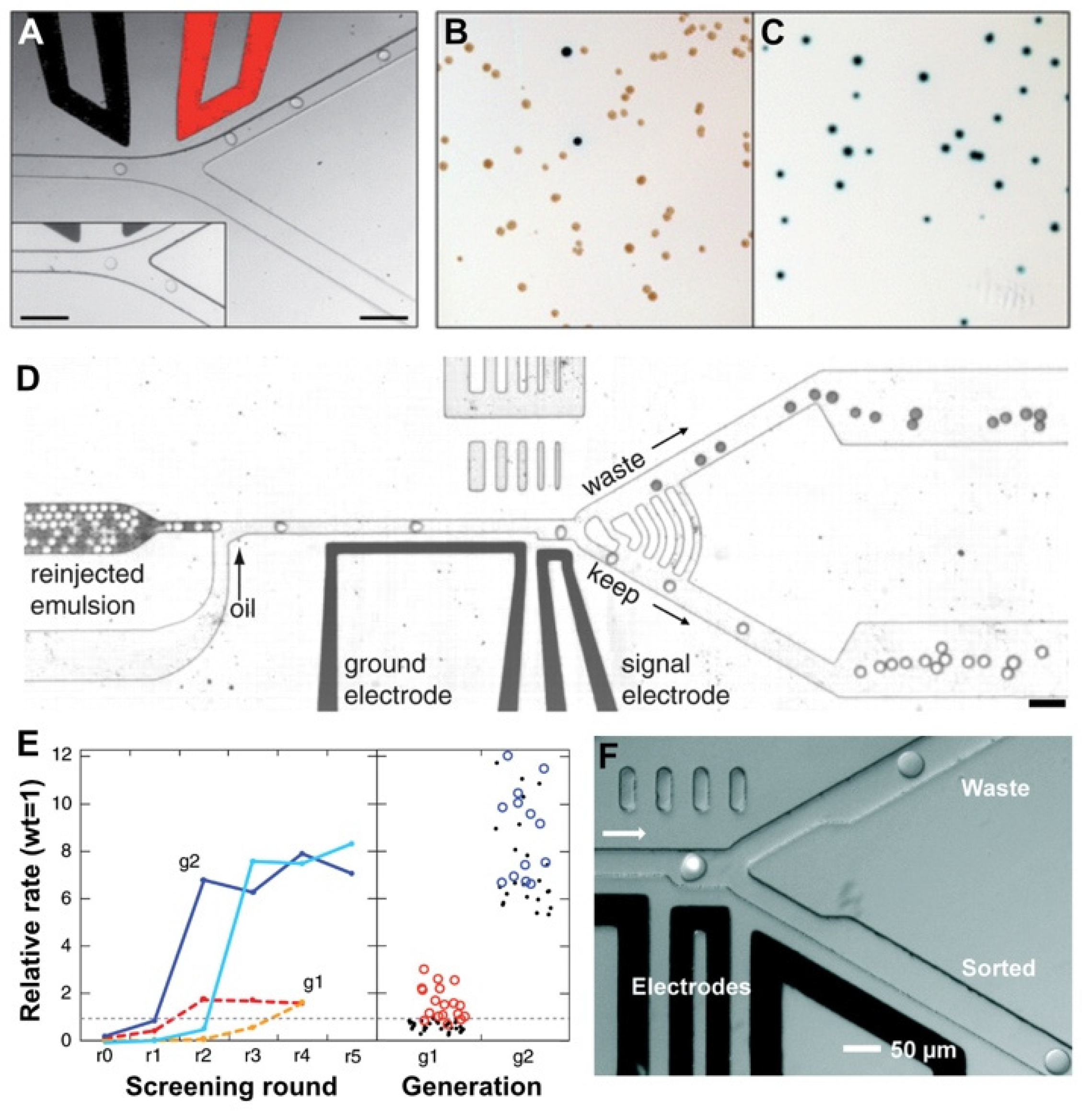
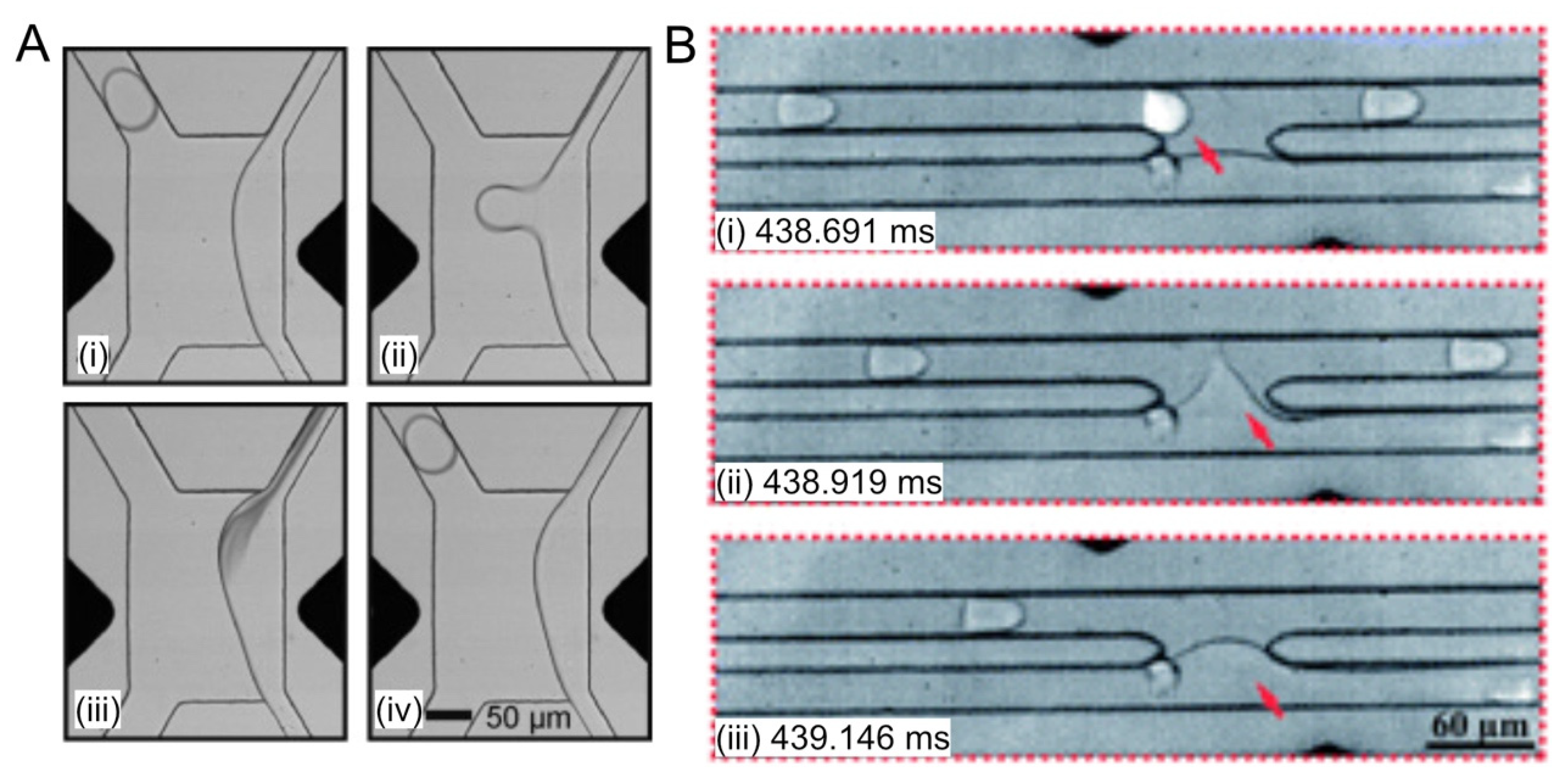
2.4. Fluorescence-Activated Droplet Sorting
2.5. Fluorescence-Activated Electrocoalescence
2.6. Mitigating Molecular Diffusion between Droplets
3. Commercialization and Commercial Activities of Droplet-based HTS in Protein Engineering
4. Challenges and Opportunities Facing Droplet Microfluidics in Protein Engineering
Funding
Conflicts of Interest
References
- Chica, R.A. Protein engineering in the 21st century. Protein Sci. 2015, 24, 431. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.K.; Wu, Z.; Arnold, F.H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 2019, 16, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Chockalingam, K.; Blenner, M.; Banta, S. Design and application of stimulus-responsive peptide systems. Protein Eng. Des. Sel. 2007, 20, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Johannes, T.W.; Zhao, H. Directed evolution of enzymes and biosynthetic pathways. Curr. Opin. Microbiol. 2006, 9, 261–267. [Google Scholar] [CrossRef]
- Schoemaker, H.E.; Mink, D.; Wubbolts, M.G. Dispelling the myths—Biocatalysis in industrial synthesis. Science 2003, 299, 1694–1697. [Google Scholar] [CrossRef]
- Nielsen, J.; Keasling, J.D. Engineering cellular metabolism. Cell 2016, 164, 1185–1197. [Google Scholar] [CrossRef]
- Wang, H.H.; Isaacs, F.J.; Carr, P.A.; Sun, Z.Z.; Xu, G.; Forest, C.R.; Church, G.M. Programming cells by multiplex genome engineering and accelerated evolution. Nature 2009, 460, 894. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA—Guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Vervoort, Y.; Linares, A.G.; Roncoroni, M.; Liu, C.; Steensels, J.; Verstrepen, K.J. High-throughput system-wide engineering and screening for microbial biotechnology. Curr. Opin. Biotechnol. 2017, 46, 120–125. [Google Scholar] [CrossRef]
- Bjork, S.M.; Joensson, H.N. Microfluidics for cell factory and bioprocess development. Curr. Opin. Biotechnol. 2019, 55, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, W.P. Rapid evolution of a protein in vitro by DNA shuffling. Nature 1994, 370, 389. [Google Scholar] [CrossRef] [PubMed]
- Crameri, A.; Raillard, S.-A.; Bermudez, E.; Stemmer, W.P. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 1998, 391, 288. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.C.; Arnold, F.H. Directed evolution of a para-nitrobenzyl esterase for aqueous-organic solvents. Nat. Biotechnol. 1996, 14, 458. [Google Scholar] [CrossRef]
- Romero, P.A.; Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009, 10, 866. [Google Scholar] [CrossRef]
- Dougherty, M.J.; Arnold, F.H. Directed evolution: New parts and optimized function. Curr. Opin. Biotechnol. 2009, 20, 486–491. [Google Scholar] [CrossRef]
- Tizei, A.G.; Csibra, E.; Torres, L.; Pinheiro, V.B. Selection platforms for directed evolution in synthetic biology. Biochem. Soc. Trans. 2016, 44, 1165. [Google Scholar] [CrossRef]
- Bassalo, M.C.; Liu, R.; Gill, R.T. Directed evolution and synthetic biology applications to microbial systems. Curr. Opin. Biotechnol. 2016, 39, 126–133. [Google Scholar] [CrossRef]
- Agresti, J.J.; Antipov, E.; Abate, A.R.; Ahn, K.; Rowat, A.C.; Baret, J.-C.; Marquez, M.; Klibanov, A.M.; Griffiths, A.D.; Weitz, D.A. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 2010, 107, 4004–4009. [Google Scholar] [CrossRef]
- Turner, N.J. Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol. 2009, 5, 567. [Google Scholar] [CrossRef]
- Zinchenko, A.; Devenish, S.R.; Kintses, B.; Colin, P.-Y.; Fischlechner, M.; Hollfelder, F. One in a million: Flow cytometric sorting of single cell-lysate assays in monodisperse picolitre double emulsion droplets for directed evolution. Anal. Chem. 2014, 86, 2526–2533. [Google Scholar] [CrossRef] [PubMed]
- Joensson, H.N.; Andersson-Svahn, H. Droplet microfluidics—A tool for protein engineering and analysis. Lab Chip 2011, 11, 4144–4147. [Google Scholar] [CrossRef] [PubMed]
- Lipovšek, D.; Antipov, E.; Armstrong, K.A.; Olsen, M.J.; Klibanov, A.M.; Tidor, B.; Wittrup, K.D. Selection of horseradish peroxidase variants with enhanced enantioselectivity by yeast surface display. Chem. Biol. 2007, 14, 1176–1185. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Thieme, K.; Chiu, C.P.; Buchini, S.; Lairson, L.L.; Chen, H.; Strynadka, N.C.; Wakarchuk, W.W.; Withers, S.G. High-throughput screening methodology for the directed evolution of glycosyltransferases. Nat. Methods 2006, 3, 609. [Google Scholar] [CrossRef] [PubMed]
- Dove, A. Screening for content—The evolution of high throughput. Nat. Biotechnol. 2003, 21, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Tawfik, D.S.; Griffiths, A.D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 1998, 16, 652. [Google Scholar] [CrossRef] [PubMed]
- Thorsen, T.; Roberts, R.W.; Arnold, F.H.; Quake, S.R. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 2001, 86, 4163. [Google Scholar] [CrossRef]
- Brouzes, E. Droplet Microfluidics for Single-Cell Analysis. In Single-Cell Analysis; Springer: Berlin/Heidelberg, Germany, 2012; pp. 105–139. [Google Scholar]
- Terekhov, S.S.; Smirnov, I.V.; Stepanova, A.V.; Bobik, T.V.; Mokrushina, Y.A.; Ponomarenko, N.A.; Belogurov, A.A.; Rubtsova, M.P.; Kartseva, O.V.; Gomzikova, M.O.; et al. Microfluidic droplet platform for ultrahigh-throughput single-cell screening of biodiversity. Proc. Natl. Acad. Sci. USA 2017, 114, 2550. [Google Scholar] [CrossRef]
- Baret, J.-C.; Miller, O.J.; Taly, V.; Ryckelynck, M.; El-Harrak, A.; Frenz, L.; Rick, C.; Samuels, M.L.; Hutchison, J.B.; Agresti, J.J.; et al. Fluorescence-activated droplet sorting (FADS): Efficient microfluidic cell sorting based on enzymatic activity. Lab Chip 2009, 9, 1850–1858. [Google Scholar] [CrossRef]
- Vallejo, D.; Nikoomanzar, A.; Paegel, B.M.; Chaput, J.C. Fluorescence-Activated Droplet Sorting for Single-Cell Directed Evolution. ACS Synth. Biol. 2019, 8, 1430–1440. [Google Scholar] [CrossRef]
- Shang, L.; Cheng, Y.; Zhao, Y. Emerging droplet microfluidics. Chem. Rev. 2017, 117, 7964–8040. [Google Scholar] [CrossRef] [PubMed]
- Joanicot, M.; Ajdari, A. Droplet control for microfluidics. Science 2005, 309, 887–888. [Google Scholar] [CrossRef] [PubMed]
- Christopher, G.F.; Anna, S.L. Microfluidic methods for generating continuous droplet streams. J. Phys. D 2007, 40, R319. [Google Scholar] [CrossRef]
- Gruner, P.; Riechers, B.; Chacòn Orellana, L.A.; Brosseau, Q.; Maes, F.; Beneyton, T.; Pekin, D.; Baret, J.-C. Stabilisers for water-in-fluorinated-oil dispersions: Key properties for microfluidic applications. Curr. Opin. Colloid Interface Sci. 2015, 20, 183–191. [Google Scholar] [CrossRef]
- Mary, P.; Dauphinot, L.; Bois, N.; Potier, M.-C.; Studer, V.; Tabeling, P. Analysis of gene expression at the single-cell level using microdroplet-based microfluidic technology. Biomicrofluidics 2011, 5, 024109. [Google Scholar] [CrossRef] [PubMed]
- Paegel, B.M.; Joyce, G.F. Microfluidic Compartmentalized Directed Evolution. Chem. Biol. 2010, 17, 717–724. [Google Scholar] [CrossRef]
- Baret, J.-C. Surfactants in droplet-based microfluidics. Lab Chip 2012, 12, 422–433. [Google Scholar] [CrossRef]
- Skhiri, Y.; Gruner, P.; Semin, B.; Brosseau, Q.; Pekin, D.; Mazutis, L.; Goust, V.; Kleinschmidt, F.; El Harrak, A.; Hutchison, J.B.; et al. Dynamics of molecular transport by surfactants in emulsions. Soft Matter 2012, 8, 10618–10627. [Google Scholar] [CrossRef]
- Bibette, J.; Morse, D.C.; Witten, T.A.; Weitz, D.A. Stability criteria for emulsions. Phys. Rev. Lett. 1992, 69, 2439–2442. [Google Scholar] [CrossRef]
- Abate, A.R.; Hung, T.; Mary, P.; Agresti, J.J.; Weitz, D.A. High-throughput injection with microfluidics using picoinjectors. Proc. Natl. Acad. Sci. USA 2010, 107, 19163. [Google Scholar] [CrossRef]
- Ahn, K.; Agresti, J.; Chong, H.; Marquez, M.; Weitz, D.A. Electrocoalescence of drops synchronized by size-dependent flow in microfluidic channels. Appl. Phys. Lett. 2006, 88, 264105. [Google Scholar] [CrossRef]
- Zagnoni, M.; Le Lain, G.; Cooper, J.M. Electrocoalescence Mechanisms of Microdroplets Using Localized Electric Fields in Microfluidic Channels. Langmuir 2010, 26, 14443–14449. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Gulati, S.; Edel, J.B.; de Mello, A.J. Pillar-induced droplet merging in microfluidic circuits. Lab Chip 2008, 8, 1837–1841. [Google Scholar] [CrossRef] [PubMed]
- Akartuna, I.; Aubrecht, D.M.; Kodger, T.E.; Weitz, D.A. Chemically induced coalescence in droplet-based microfluidics. Lab Chip 2015, 15, 1140–1144. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.M.; Barbier, I.; Schaerli, Y. Bacterial Microcolonies in Gel Beads for High-Throughput Screening of Libraries in Synthetic Biology. ACS Synth. Biol. 2017, 6, 1988–1995. [Google Scholar] [CrossRef] [PubMed]
- Sjostrom, S.L.; Bai, Y.; Huang, M.; Liu, Z.; Nielsen, J.; Joensson, H.N.; Andersson Svahn, H. High-throughput screening for industrial enzyme production hosts by droplet microfluidics. Lab Chip 2014, 14, 806–813. [Google Scholar] [CrossRef]
- Ma, F.; Chung, M.T.; Yao, Y.; Nidetz, R.; Lee, L.M.; Liu, A.P.; Feng, Y.; Kurabayashi, K.; Yang, G.-Y. Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform. Nat. Commun. 2018, 9, 1030. [Google Scholar] [CrossRef]
- Fallah-Araghi, A.; Baret, J.-C.; Ryckelynck, M.; Griffiths, A.D. A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab Chip 2012, 12, 882–891. [Google Scholar] [CrossRef]
- Sciambi, A.; Abate, A.R. Adding reagent to droplets with controlled rupture of encapsulated double emulsions. Biomicrofluidics 2013, 7, 044112. [Google Scholar] [CrossRef]
- Caen, O.; Schütz, S.; Jammalamadaka, M.S.S.; Vrignon, J.; Nizard, P.; Schneider, T.M.; Baret, J.-C.; Taly, V. High-throughput multiplexed fluorescence-activated droplet sorting. Microsyst. Nanoeng. 2018, 4, 33. [Google Scholar] [CrossRef]
- Joensson, H.N.; Uhlén, M.; Svahn, H.A. Droplet size based separation by deterministic lateral displacement—Separating droplets by cell-induced shrinking. Lab Chip 2011, 11, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Frenzel, D.; Merten, C.A. Microfluidic train station: Highly robust and multiplexable sorting of droplets on electric rails. Lab Chip 2017, 17, 1024–1030. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Lyu, F.; Tang, S.K.Y. Fluorinated Pickering Emulsions with Nonadsorbing Interfaces for Droplet-based Enzymatic Assays. Anal. Chem. 2015, 87, 7938–7943. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Rosenfeld, L.; Kim, M.; Xu, M.; Lin, E.; Derda, R.; Tang, S.K.Y. Fluorinated Pickering Emulsions Impede Interfacial Transport and Form Rigid Interface for the Growth of Anchorage-Dependent Cells. ACS Appl. Mater. Interfaces 2014, 6, 21446–21453. [Google Scholar] [CrossRef]
- Hasan, S.; Geissler, D.; Wink, K.; Hagen, A.; Heiland, J.J.; Belder, D. Fluorescence lifetime-activated droplet sorting in microfluidic chip systems. Lab Chip 2019, 19, 403–409. [Google Scholar] [CrossRef]
- Eun, Y.-J.; Utada, A.S.; Copeland, M.F.; Takeuchi, S.; Weibel, D.B. Encapsulating Bacteria in Agarose Microparticles Using Microfluidics for High-Throughput Cell Analysis and Isolation. ACS Chem. Biol. 2011, 6, 260–266. [Google Scholar] [CrossRef]
- Sandoz, P.A.; Chung, A.J.; Weaver, W.M.; Di Carlo, D. Sugar Additives Improve Signal Fidelity for Implementing Two-Phase Resorufin-Based Enzyme Immunoassays. Langmuir 2014, 30, 6637–6643. [Google Scholar] [CrossRef]
- Courtois, F.; Olguin, L.F.; Whyte, G.; Theberge, A.B.; Huck, W.T.S.; Hollfelder, F.; Abell, C. Controlling the Retention of Small Molecules in Emulsion Microdroplets for Use in Cell-Based Assays. Anal. Chem. 2009, 81, 3008–3016. [Google Scholar] [CrossRef]
- Brouzes, E.; Medkova, M.; Savenelli, N.; Marran, D.; Twardowski, M.; Hutchison, J.B.; Rothberg, J.M.; Link, D.R.; Perrimon, N.; Samuels, M.L. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA 2009, 106, 14195–14200. [Google Scholar] [CrossRef]
- Nightingale, A.M.; Phillips, T.W.; Bannock, J.H.; de Mello, J.C. Controlled multistep synthesis in a three-phase droplet reactor. Nat. Commun. 2014, 5, 3777. [Google Scholar] [CrossRef]
- Eow, J.S.; Ghadiri, M. Drop-drop coalescence in an electric field: The effects of applied electric field and electrode geometry. Colloids Surf. A 2003, 219, 253–279. [Google Scholar] [CrossRef]
- Song, H.; Li, H.-W.; Munson, M.S.; Van Ha, T.G.; Ismagilov, R.F. On-Chip Titration of an Anticoagulant Argatroban and Determination of the Clotting Time within Whole Blood or Plasma Using a Plug-Based Microfluidic System. Anal. Chem. 2006, 78, 4839–4849. [Google Scholar] [CrossRef] [PubMed]
- Um, E.; Lee, D.-S.; Pyo, H.-B.; Park, J.-K. Continuous generation of hydrogel beads and encapsulation of biological materials using a microfluidic droplet-merging channel. Microfluid. Nanofluid. 2008, 5, 541–549. [Google Scholar] [CrossRef]
- Bernath, K.; Hai, M.; Mastrobattista, E.; Griffiths, A.D.; Magdassi, S.; Tawfik, D.S. In vitro compartmentalization by double emulsions: Sorting and gene enrichment by fluorescence activated cell sorting. Anal. Biochem. 2004, 325, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Mastrobattista, E.; Taly, V.; Chanudet, E.; Treacy, P.; Kelly, B.T.; Griffiths, A.D. High-Throughput Screening of Enzyme Libraries: In Vitro Evolution of a β-Galactosidase by Fluorescence-Activated Sorting of Double Emulsions. Chem. Biol. 2005, 12, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Miller, O.J.; Bernath, K.; Agresti, J.J.; Amitai, G.; Kelly, B.T.; Mastrobattista, E.; Taly, V.; Magdassi, S.; Tawfik, D.S.; Griffiths, A.D. Directed evolution by in vitro compartmentalization. Nat. Methods 2006, 3, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Amitai, G.; Bernath, K.; Magdassi, S.; Tawfik, D.S. High-Throughput Screening of Enzyme Libraries: Thiolactonases Evolved by Fluorescence-Activated Sorting of Single Cells in Emulsion Compartments. Chem. Biol. 2005, 12, 1281–1289. [Google Scholar] [CrossRef]
- Amsden, B. Solute Diffusion within Hydrogels. Mechanisms and Models. Macromolecules 1998, 31, 8382–8395. [Google Scholar] [CrossRef]
- Wechsler, M.E.; Stephenson, R.E.; Murphy, A.C.; Oldenkamp, H.F.; Singh, A.; Peppas, N.A. Engineered microscale hydrogels for drug delivery, cell therapy, and sequencing. Biomed. Microdevices 2019, 21, 31. [Google Scholar] [CrossRef]
- Cha, C.; Oh, J.; Kim, K.; Qiu, Y.; Joh, M.; Shin, S.R.; Wang, X.; Camci-Unal, G.; Wan, K.-T.; Liao, R. Microfluidics-assisted fabrication of gelatin-silica core-shell microgels for injectable tissue constructs. Biomacromolecules 2014, 15, 283–290. [Google Scholar] [CrossRef]
- Ahn, K.; Kerbage, C.; Hunt, T.P.; Westervelt, R.M.; Link, D.R.; Weitz, D.A. Dielectrophoretic manipulation of drops for high-speed microfluidic sorting devices. Appl. Phys. Lett. 2006, 88, 024104. [Google Scholar] [CrossRef]
- Fidalgo, L.M.; Whyte, G.; Bratton, D.; Kaminski, C.F.; Abell, C.; Huck, W.T. From microdroplets to microfluidics: Selective emulsion separation in microfluidic devices. Angew. Chem. Int. Ed. 2008, 47, 2042–2045. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wijaya Gani, A.; Tang, S.K. Characterization of sensitivity and specificity in leaky droplet-based assays. Lab Chip 2012, 12, 5093–5103. [Google Scholar] [PubMed]
- Woronoff, G.; El Harrak, A.; Mayot, E.; Schicke, O.; Miller, O.J.; Soumillion, P.; Griffiths, A.D.; Ryckelynck, M. New Generation of Amino Coumarin Methyl Sulfonate-Based Fluorogenic Substrates for Amidase Assays in Droplet-Based Microfluidic Applications. Anal. Chem. 2011, 83, 2852–2857. [Google Scholar] [PubMed]
- Ramsden, W. Separation of solids in the surface-layers of solutions and ‘suspensions’ (observations on surface-membranes, bubbles, emulsions, and mechanical coagulation)—Preliminary account. Proc. R. Soc. Lond. 1904, 72, 156–164. [Google Scholar]
- Pickering, S.U. Cxcvi—Emulsions. J. Chem. Soc. Trans. 1907, 91, 2001–2021. [Google Scholar]
- Dinsmore, A.; Hsu, M.F.; Nikolaides, M.; Marquez, M.; Bausch, A.; Weitz, D. Colloidosomes: Selectively permeable capsules composed of colloidal particles. Science 2002, 298, 1006–1009. [Google Scholar] [CrossRef]
- Subramaniam, A.B.; Abkarian, M.; Stone, H.A. Controlled assembly of jammed colloidal shells on fluid droplets. Nat. Mater. 2005, 4, 553. [Google Scholar]
- Crossley, S.; Faria, J.; Shen, M.; Resasco, D.E. Solid nanoparticles that catalyze biofuel upgrade reactions at the water/oil interface. Science 2010, 327, 68–72. [Google Scholar] [CrossRef]
- Vertegel, A.A.; Siegel, R.W.; Dordick, J.S. Silica Nanoparticle Size Influences the Structure and Enzymatic Activity of Adsorbed Lysozyme. Langmuir 2004, 20, 6800–6807. [Google Scholar]
- Czeslik, C.; Winter, R. Effect of temperature on the conformation of lysozyme adsorbed to silica particles. Phys. Chem. Chem. Phys. 2001, 3, 235–239. [Google Scholar] [CrossRef]
- Preari, M.; Spinde, K.; Lazic, J.; Brunner, E.; Demadis, K.D. Bioinspired Insights into Silicic Acid Stabilization Mechanisms: The Dominant Role of Polyethylene Glycol-Induced Hydrogen Bonding. J. Am. Chem. Soc. 2014, 136, 4236–4244. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Lyu, F.; Tang, S.K.Y. Methods to coalesce fluorinated Pickering emulsions. Anal. Methods 2017, 9, 4622–4629. [Google Scholar] [CrossRef]
- Siltanen, C.A.; Cole, R.H.; Poust, S.; Chao, L.; Tyerman, J.; Kaufmann-Malaga, B.; Ubersax, J.; Gartner, Z.J.; Abate, A.R. An oil-free picodrop bioassay platform for synthetic biology. Sci. Rep. 2018, 8, 7913. [Google Scholar] [CrossRef] [PubMed]
- Cole, R.H.; Tang, S.-Y.; Siltanen, C.A.; Shahi, P.; Zhang, J.Q.; Poust, S.; Gartner, Z.J.; Abate, A.R. Printed droplet microfluidics for on demand dispensing of picoliter droplets and cells. Proc. Natl. Acad. Sci. USA 2017, 114, 8728–8733. [Google Scholar] [CrossRef]
- Sinha, R.; Shukla, P. Current Trends in Protein Engineering: Updates and Progress. Curr. Protein Pept. Sci. 2019, 20, 398–407. [Google Scholar] [CrossRef]
- Available online: https://www.zionmarketresearch.com/report/protein-engineering-market (accessed on 14 February 2019).
- Diefenbach, X.W.; Farasat, I.; Guetschow, E.D.; Welch, C.J.; Kennedy, R.T.; Sun, S.; Moore, J.C. Enabling biocatalysis by high-throughput protein engineering using droplet microfluidics coupled to mass spectrometry. ACS Omega 2018, 3, 1498–1508. [Google Scholar] [CrossRef]
- El Debs, B.; Utharala, R.; Balyasnikova, I.V.; Griffiths, A.D.; Merten, C.A. Functional single-cell hybridoma screening using droplet-based microfluidics. Proc. Natl. Acad. Sci. USA 2012, 109, 11570–11575. [Google Scholar] [CrossRef]
- Liu, X.; Painter, R.; Enesa, K.; Holmes, D.; Whyte, G.; Garlisi, C.; Monsma, F.; Rehak, M.; Craig, F.; Smith, C.A. High-throughput screening of antibiotic-resistant bacteria in picodroplets. Lab Chip 2016, 16, 1636–1643. [Google Scholar] [CrossRef]
- Edd, J.F.; Di Carlo, D.; Humphry, K.J.; Köster, S.; Irimia, D.; Weitz, D.A.; Toner, M. Controlled encapsulation of single-cells into monodisperse picolitre drops. Lab Chip 2008, 8, 1262–1264. [Google Scholar] [CrossRef]
- Lagus, T.P.; Edd, J.F. High-throughput co-encapsulation of self-ordered cell trains: Cell pair interactions in microdroplets. RSC Adv. 2013, 3, 20512–20522. [Google Scholar] [CrossRef]
- Mutlu, B.R.; Edd, J.F.; Toner, M. Oscillatory inertial focusing in infinite microchannels. Proc. Natl. Acad. Sci. USA 2018, 115, 7682–7687. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.; Graells, T.; Walldén, M.; Hjort, K. Inertial focusing with sub-micron resolution for separation of bacteria. Lab Chip 2019, 19, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef]
- Liu, L.; Dalal, C.K.; Heineike, B.M.; Abate, A.R. High throughput gene expression profiling of yeast colonies with microgel-culture Drop-seq. Lab Chip 2019, 19, 1838–1849. [Google Scholar] [CrossRef]
- Riordon, J.; Sovilj, D.; Sanner, S.; Sinton, D.; Young, E.W.K. Deep Learning with Microfluidics for Biotechnology. Trends Biotechnol. 2019, 37, 310–324. [Google Scholar] [CrossRef]
- Chu, A.; Nguyen, D.; Talathi, S.S.; Wilson, A.C.; Ye, C.; Smith, W.L.; Kaplan, A.D.; Duoss, E.B.; Stolaroff, J.K.; Giera, B. Automated detection and sorting of microencapsulation via machine learning. Lab Chip 2019, 19, 1808–1817. [Google Scholar] [CrossRef]
- Svensson, C.-M.; Shvydkiv, O.; Dietrich, S.; Mahler, L.; Weber, T.; Choudhary, M.; Tovar, M.; Figge, M.T.; Roth, M. Coding of Experimental Conditions in Microfluidic Droplet Assays Using Colored Beads and Machine Learning Supported Image Analysis. Small 2019, 15, 1802384. [Google Scholar] [CrossRef]
- Nitta, N.; Sugimura, T.; Isozaki, A.; Mikami, H.; Hiraki, K.; Sakuma, S.; Iino, T.; Arai, F.; Endo, T.; Fujiwaki, Y.; et al. Intelligent Image-Activated Cell Sorting. Cell 2018, 175, 266–276. [Google Scholar] [CrossRef]

| Oil/Surfactant | Applications |
|---|---|
| HFE-7500/PFPE-PEG | Double emulsion for flow cytometric sorting [21], bacterial microcolonies in gel beads [46], FADS [30,31,47,48], electrocoalescence [49], triple emulsification for reagent addition [50], chemically-induced droplet coalescence (PFB added to induce coalescence) [45], picoinjector [41], multiplexed FADS [51], droplet size-based separation [52] |
| FC-40/PFPE-PEG | Double emulsion for flow cytometric sorting [21], FADS [19] |
| HFE-7500/QX200 (Bio-Rad proprietary oil) | 2-way or 4-way FADS [53] |
| HFE-7500 or FC-40/fluorinated silica nanoparticles | Pickering emulsification to mitigate molecular diffusion [54,55] |
| Perfluorodecalin and perfluorooctanol (7:3 (v/v)) | Fluorescence lifetime-activated droplet sorting [56] |
| Mineral oil/Span 80 | Encapsulation of bacteria in agarose microparticles [57], emulsification of Amplex Ultra Red and horseradish peroxidase (specific sugars added to mitigate molecular diffusion) [58] |
| Mineral oil/Abil EM 90 | Emulsification of 3-O-methylfluorescein (BSA added to mitigate molecular diffusion) [59] |
| Droplet Sorting Mechanism | Advantages | Disadvantages |
|---|---|---|
| Flow cytometric sorting | Compatible with commercial instruments | Needs additional emulsification or hydrogel gelation; Droplet size is restricted by instrument specification |
| Fluorescence-activated droplet sorting (FADS) | Single emulsification; Sorted droplets are available for more rounds of sorting; Wide range of droplet sizes | Requires careful balance of “collection” and “waste” fluid flows; Needs additional step to recover the content inside the droplets |
| Fluorescence-activated electrocoalescence | Direct recovery of droplet content; Tunable conditions of the collection aqueous stream to perform additional chemistry | Sorted droplets no longer exist; Not possible to perform second round of sorting |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weng, L.; Spoonamore, J.E. Droplet Microfluidics-Enabled High-Throughput Screening for Protein Engineering. Micromachines 2019, 10, 734. https://doi.org/10.3390/mi10110734
Weng L, Spoonamore JE. Droplet Microfluidics-Enabled High-Throughput Screening for Protein Engineering. Micromachines. 2019; 10(11):734. https://doi.org/10.3390/mi10110734
Chicago/Turabian StyleWeng, Lindong, and James E. Spoonamore. 2019. "Droplet Microfluidics-Enabled High-Throughput Screening for Protein Engineering" Micromachines 10, no. 11: 734. https://doi.org/10.3390/mi10110734
APA StyleWeng, L., & Spoonamore, J. E. (2019). Droplet Microfluidics-Enabled High-Throughput Screening for Protein Engineering. Micromachines, 10(11), 734. https://doi.org/10.3390/mi10110734