Cross-Flow Filtration of Escherichia coli at a Nanofluidic Gap for Fast Immobilization and Antibiotic Susceptibility Testing
Abstract
:1. Introduction
2. Materials and Methods
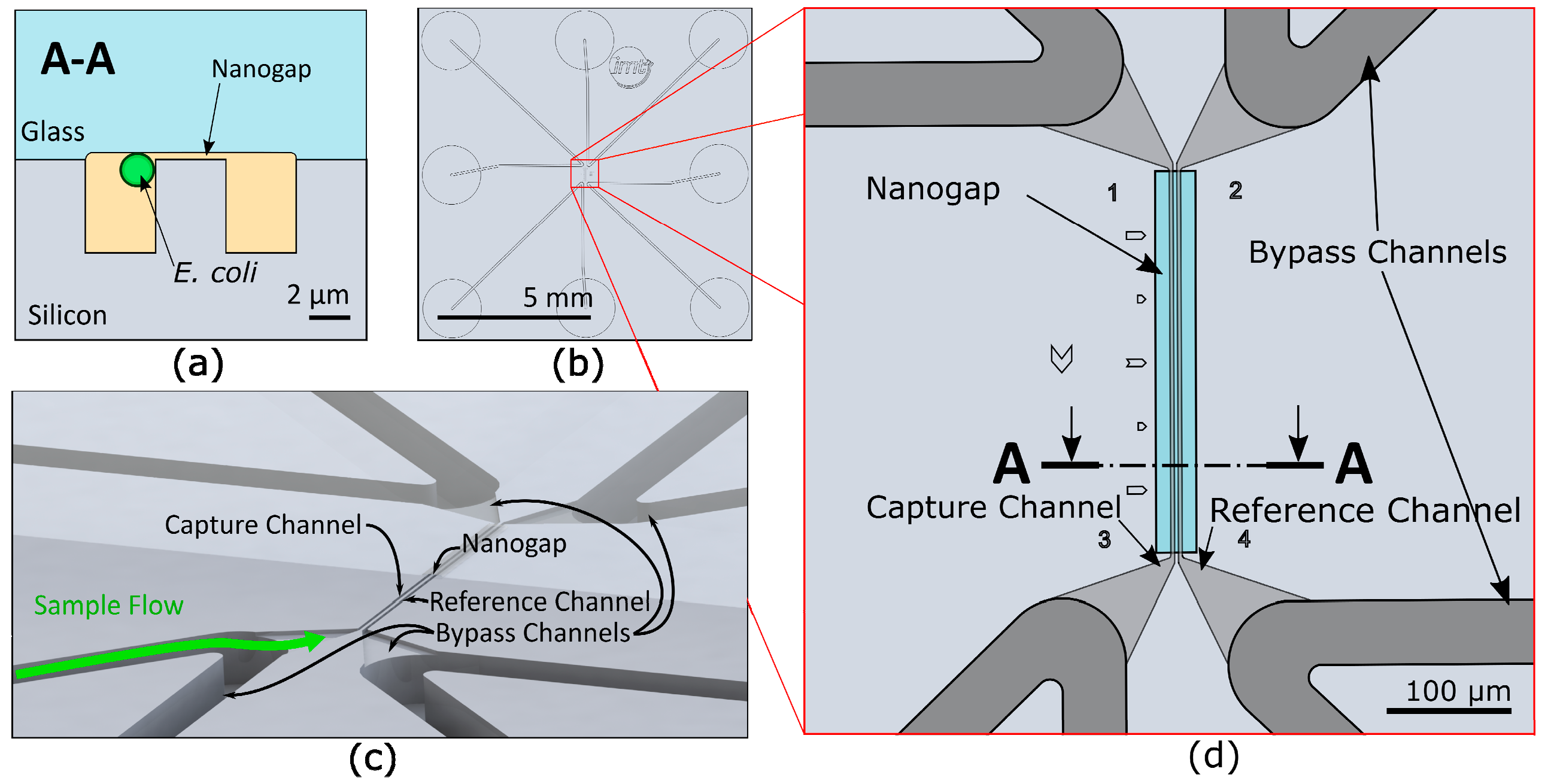
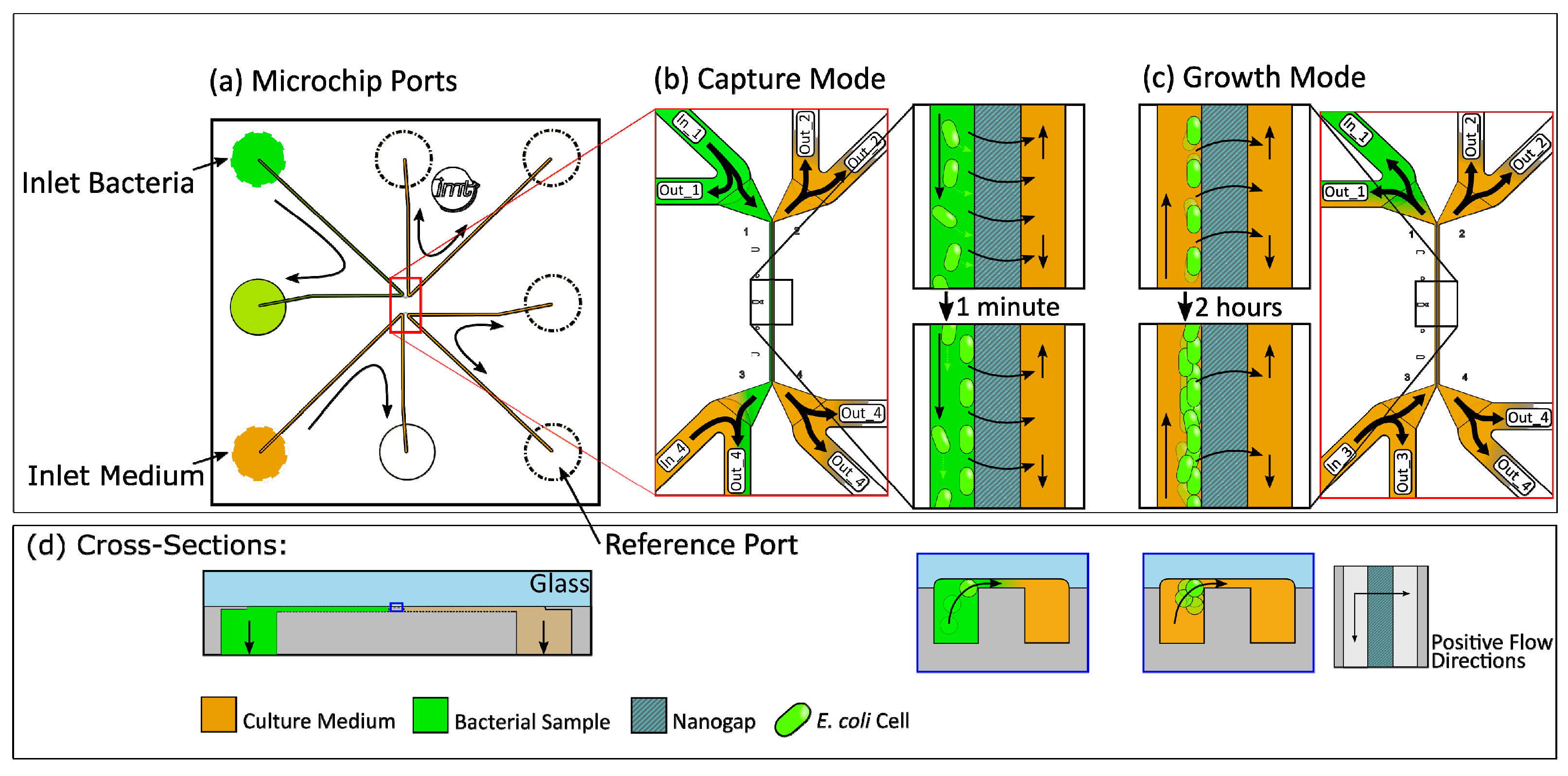
2.1. Microfluidic Chip Design
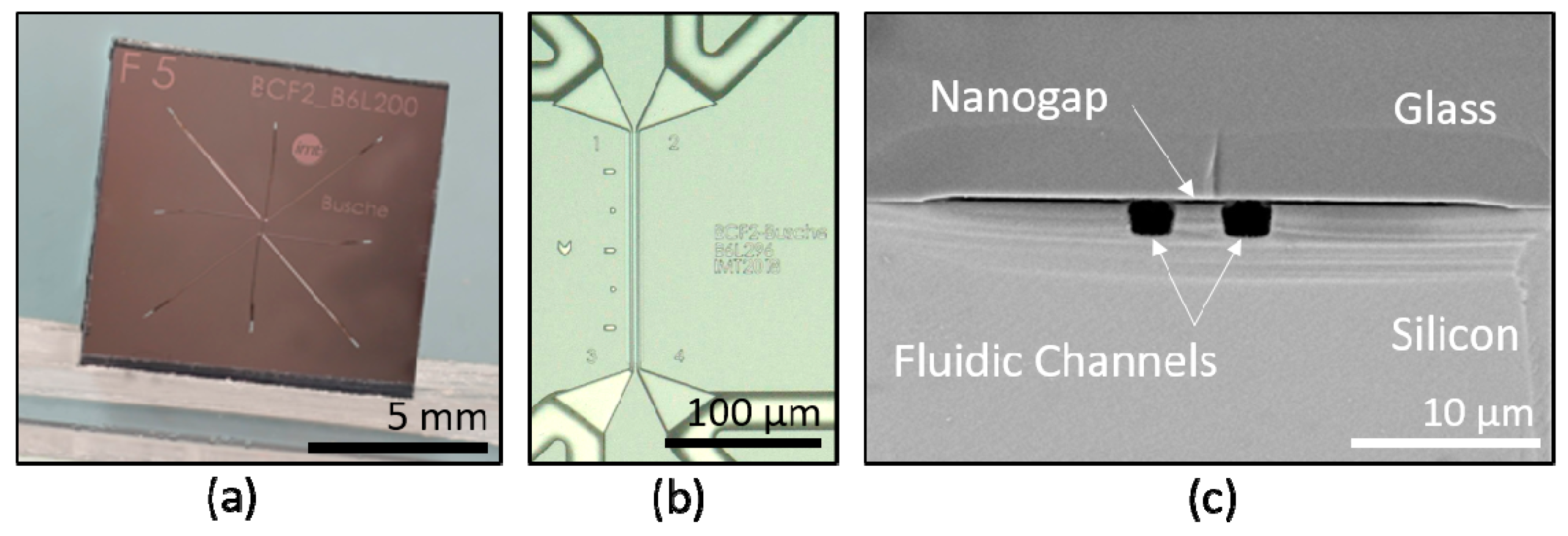
2.2. Microfabrication
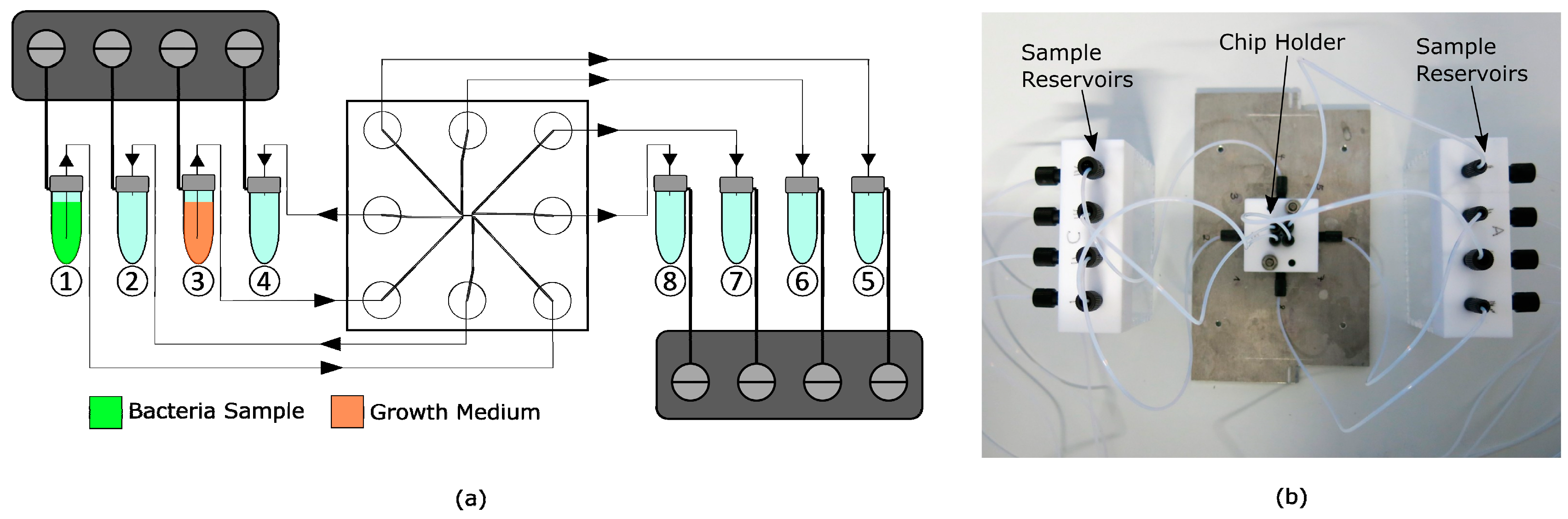
2.3. Experimental Setup
2.4. Bacterial Strain and Growth Medium
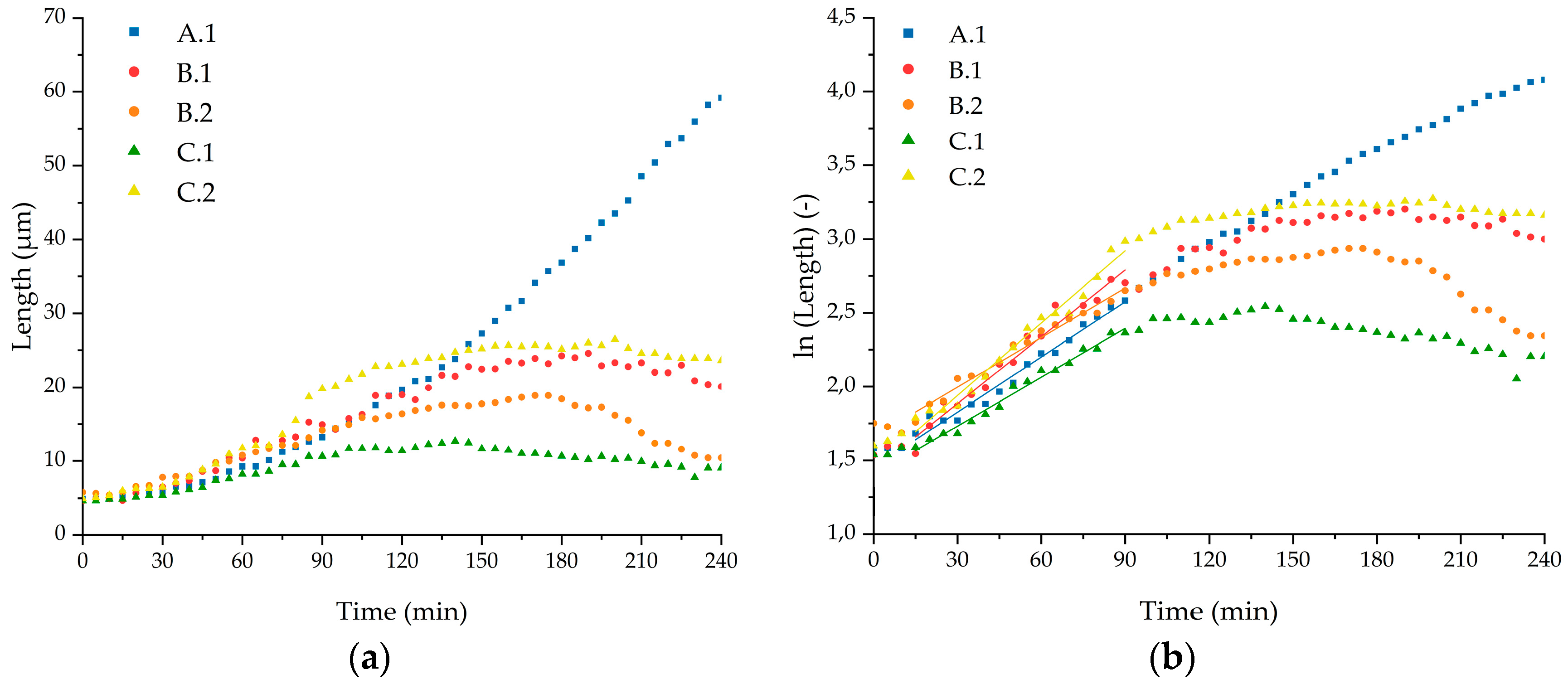
2.5. Calculation of Growth Rates
3. Results and Discussion
3.1. Fluidic Simulations of Shear Forces
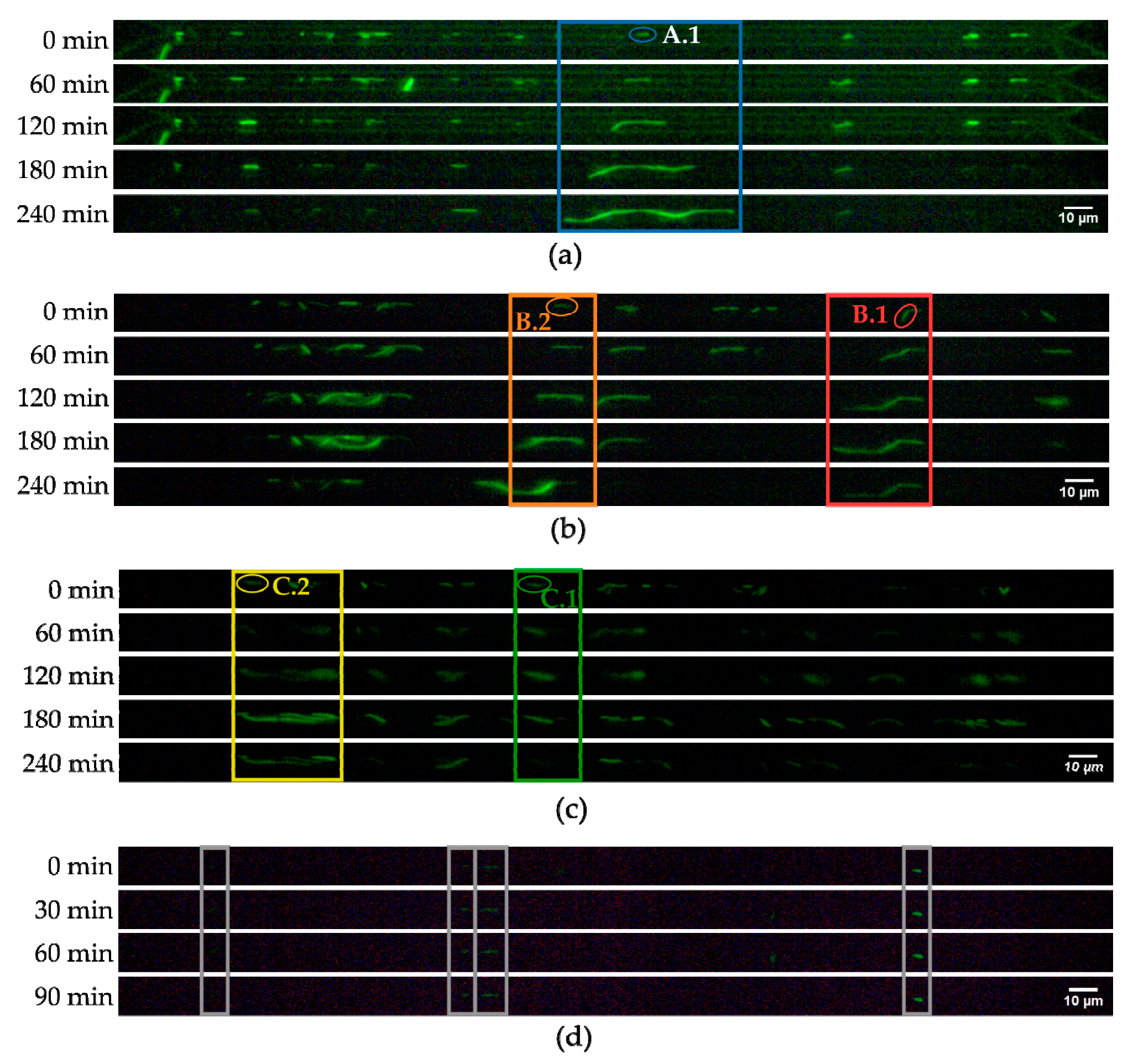
3.2. Cell Capture Experiments
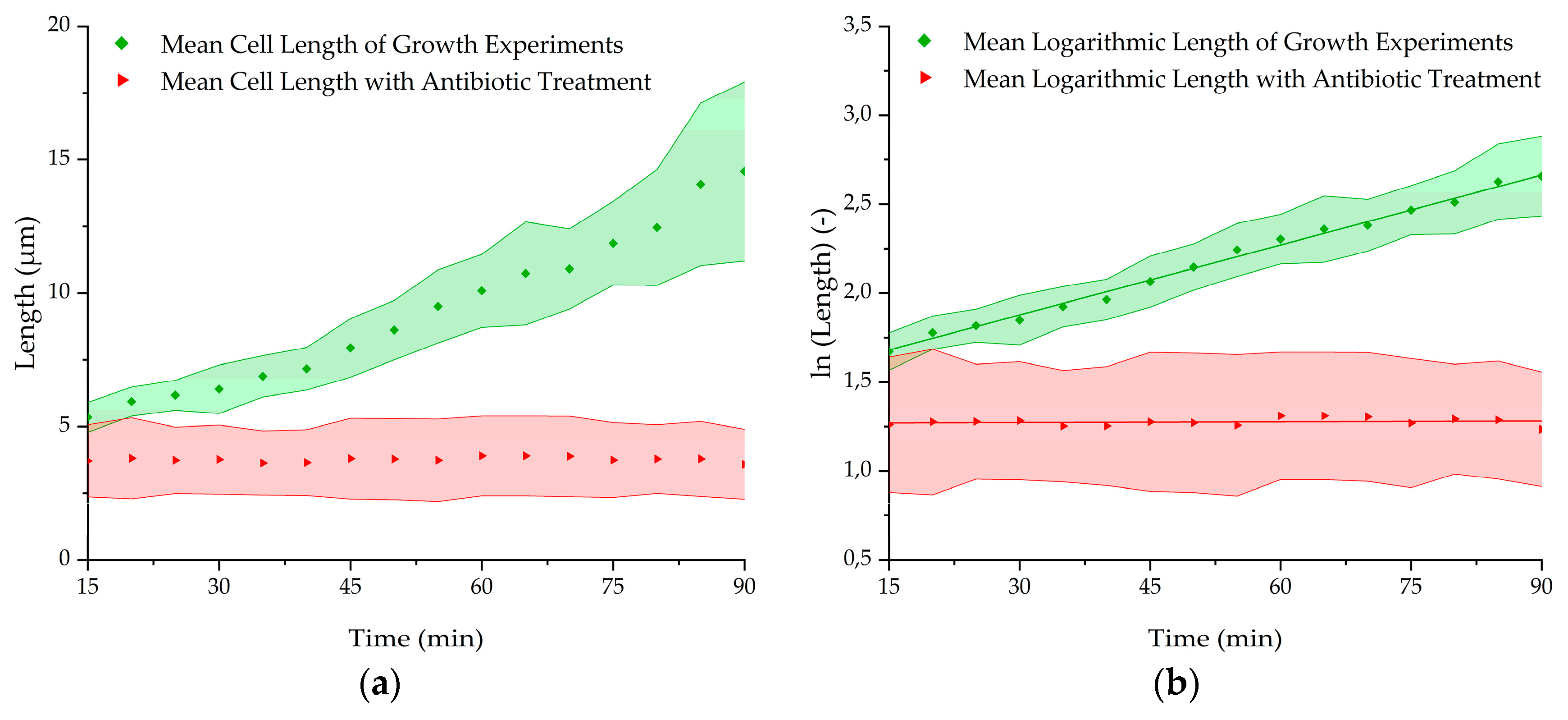
3.3. Growth Experiments
3.4. Antibiotic Treatment Experiments
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Avesar, J.; Rosenfeld, D.; Truman-Rosentsvit, M.; Ben-Arye, T.; Geffen, Y.; Bercovici, M.; Levenberg, S. Rapid phenotypic antimicrobial susceptibility testing using nanoliter arrays. Proc. Natl. Acad. Sci. USA 2017, 114, E5787–E5795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, J. Tackling drug-resistant infections globally: Final report and recommendations. Rev. Antimicrob. Res. 2017, 2017, 171–174. [Google Scholar]
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, J.H.; Ferraro, M.J. Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Stamper, P.D.; Louie, L.; Wong, H.; Simor, A.E.; Farley, J.E.; Carroll, K.C. Genotypic and phenotypic characterization of methicillin-susceptible Staphylococcus aureus isolates misidentified as methicillin-resistant Staphylococcus aureus by the BD GeneOhm MRSA assay. J. Clin. Microbiol. 2011, 49, 1240–1244. [Google Scholar] [CrossRef]
- Garcia-Prats, J.A.; Cooper, T.R.; Schneider, V.F.; Stager, C.E.; Hansen, T.N. Rapid detection of microorganisms in blood cultures of newborn infants utilizing an automated blood culture system. Pediatrics 2000, 105, 523–527. [Google Scholar] [CrossRef]
- Mishra, P.; Mishra, K.P.; Singh, D.; Ganju, L.; Kumar, B.; Singh, S.B. Advances in Rapid Detection and Antimicrobial Susceptibility Tests: A Review. Def. Life Sc. J. 2019, 4, 12–20. [Google Scholar] [CrossRef]
- Choi, J.; Jung, Y.-G.; Kim, J.; Kim, S.; Jung, Y.; Na, H.; Kwon, S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip 2013, 13, 280–287. [Google Scholar] [CrossRef]
- Hou, Z.; An, Y.; Hjort, K.; Hjort, K.; Sandegren, L.; Wu, Z. Time lapse investigation of antibiotic susceptibility using a microfluidic linear gradient 3D culture device. Lab Chip 2014, 14, 3409–3418. [Google Scholar] [CrossRef]
- Li, B.; Qiu, Y.; Glidle, A.; McIlvenna, D.; Luo, Q.; Cooper, J.; Shi, H.-C.; Yin, H. Gradient microfluidics enables rapid bacterial growth inhibition testing. Anal. Chem. 2014, 86, 3131–3137. [Google Scholar] [CrossRef]
- Wang, P.; Robert, L.; Pelletier, J.; Dang, W.L.; Taddei, F.; Wright, A.; Jun, S. Robust growth of Escherichia coli. Curr. Biol. 2010, 20, 1099–1103. [Google Scholar] [CrossRef] [PubMed]
- Baltekin, O.; Boucharin, A.; Tano, E.; Andersson, D.I.; Elf, J. Fast antibiotic susceptibility testing based on single cell growth rate measurements. BioRxiv 2017, 201, 071407. [Google Scholar]
- Baltekin, Ö.; Boucharin, A.; Tano, E.; Andersson, D.I.; Elf, J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc. Natl. Acad. Sci. USA 2017, 114, 9170–9175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Churski, K.; Kaminski, T.S.; Jakiela, S.; Kamysz, W.; Baranska-Rybak, W.; Weibel, D.B.; Garstecki, P. Rapid screening of antibiotic toxicity in an automated microdroplet system. Lab Chip 2012, 12, 1629–1637. [Google Scholar] [CrossRef]
- Baraban, L.; Bertholle, F.; Salverda, M.L.M.; Bremond, N.; Panizza, P.; Baudry, J.; de Visser, J.A.G.M.; Bibette, J. Millifluidic droplet analyser for microbiology. Lab Chip 2011, 11, 4057–4062. [Google Scholar] [CrossRef]
- Kaminski, T.S.; Scheler, O.; Garstecki, P. Droplet microfluidics for microbiology: Techniques, applications and challenges. Lab Chip 2016, 16, 2168–2187. [Google Scholar] [CrossRef]
- Shang, L.; Cheng, Y.; Zhao, Y. Emerging droplet microfluidics. Chem. Rev. 2017, 117, 7964–8040. [Google Scholar] [CrossRef]
- Liu, X.; Painter, R.E.; Enesa, K.; Holmes, D.; Whyte, G.; Garlisi, C.G.; Monsma, F.J.; Rehak, M.; Craig, F.F.; Smith, C.A. High-throughput screening of antibiotic-resistant bacteria in picodroplets. Lab Chip 2016, 16, 1636–1643. [Google Scholar] [CrossRef]
- Hesselman, M.C.; Odoni, D.I.; Ryback, B.M.; de Groot, S.; van Heck, R.G.A.; Keijsers, J.; Kolkman, P.; Nieuwenhuijse, D.; van Nuland, Y.M.; Sebus, E.; et al. A multi-platform flow device for microbial (co-) cultivation and microscopic analysis. PLoS ONE 2012, 7, e36982. [Google Scholar] [CrossRef]
- Besant, J.D.; Sargent, E.H.; Kelley, S.O. Rapid electrochemical phenotypic profiling of antibiotic-resistant bacteria. Lab Chip 2015, 15, 2799–2807. [Google Scholar] [CrossRef]
- Mohan, R.; Mukherjee, A.; Sevgen, S.E.; Sanpitakseree, C.; Lee, J.; Schroeder, C.M.; Kenis, P.J.A. A multiplexed microfluidic platform for rapid antibiotic susceptibility testing. Biosens. Bioelectron. 2013, 49, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Pitruzzello, G.; Thorpe, S.; Johnson, S.; Evans, A.; Gadêlha, H.; Krauss, T.F. Multiparameter antibiotic resistance detection based on hydrodynamic trapping of individual E. coli. Lab Chip 2019, 19, 1417–1426. [Google Scholar] [CrossRef] [PubMed]
- Kara, V.; Duan, C.; Gupta, K.; Kurosawa, S.; Stearns-Kurosawa, D.J.; Ekinci, K.L. Microfluidic detection of movements of Escherichia coli for rapid antibiotic susceptibility testing. Lab Chip 2018, 18, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Huang, X.; Chu, Q.; Ning, X.; Wang, Y.; Kong, S.-K.; Zhang, X.; Wang, G.; Ho, H.-P. A linear concentration gradient generator based on multi-layered centrifugal microfluidics and its application in antimicrobial susceptibility testing. Lab Chip 2018, 18, 1452–1460. [Google Scholar] [CrossRef]
- Kim, K.P.; Kim, Y.-G.; Choi, C.-H.; Kim, H.-E.; Lee, S.-H.; Chang, W.-S.; Lee, C.-S. In situ monitoring of antibiotic susceptibility of bacterial biofilms in a microfluidic device. Lab Chip 2010, 10, 3296–3299. [Google Scholar] [CrossRef]
- Shirure, V.S.; George, S.C. Design considerations to minimize the impact of drug absorption in polymer-based organ-on-a-chip platforms. Lab Chip 2017, 17, 681–690. [Google Scholar] [CrossRef]
- Toepke, M.W.; Beebe, D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef]
- Van Meer, B.J.; de Vries, H.; Firth, K.S.A.; van Weerd, J.; Tertoolen, L.G.J.; Karperien, H.B.J.; Jonkheijm, P.; Denning, C.; IJzerman, A.P.; Mummery, C.L. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem. Biophys. Res. Commun. 2017, 482, 323–328. [Google Scholar] [CrossRef]
- Yang, M.; Li, C.-W.; Yang, J. Cell docking and on-chip monitoring of cellular reactions with a controlled concentration gradient on a microfluidic device. Anal. Chem. 2002, 74, 3991–4001. [Google Scholar] [CrossRef]
- Bates, R.L.; Stephan Thamban, P.L.; Goeckner, M.J.; Overzet, L.J. Silicon etch using SF6/C4F8/Ar gas mixtures. J. Vac. Technol. 2014, 32, 041302. [Google Scholar] [CrossRef]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef]
- Bongaerts, G.P.; Kaptijn, G.M. Aminoglycoside phosphotransferase-II-mediated amikacin resistance in Escherichia coli. Antimicrob. Agents Chemother. 1981, 20, 344–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, H.; Taillandier, P.; Riba, J.-P. Effect of high shear stress on microbial viability. J. Chem. Technol. Biotechnol. 2001, 76, 501–505. [Google Scholar] [CrossRef]
- Dhar, N.; McKinney, J.D. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 2007, 10, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Purr, F.; Bassu, M.; Lowe, R.D.; Thürmann, B.; Dietzel, A.; Burg, T.P. Asymmetric nanofluidic grating detector for differential refractive index measurement and biosensing. Lab Chip 2017, 17, 4265–4272. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Mode | Pressure Applied (mbar) at the Inlets and Outlets | |||||||
|---|---|---|---|---|---|---|---|---|
| pIn1 | pOut1 | pIn2 | pOut2 | pIn3 | pOut3 | pIn4 | pOut4 | |
| Capture | 200 | 195 | 195 | 170 | 40 | 40 | 50 | 50 |
| Growth | 190 | 180 | 200 | 185 | 40 | 40 | 40 | 40 |
| Cell | Specific Growth Rate (min−1) | Deviation (min−1) | R2 (Fit) | Doubling Time (min) | Deviation (min) |
|---|---|---|---|---|---|
| A.1 | 0.0125 | 0.0005 | 0.9790 | 56 | 2 |
| B.1 | 0.0151 | 0.0007 | 0.9694 | 46 | 2 |
| B.2 | 0.0112 | 0.0004 | 0.9789 | 62 | 2 |
| C.1 | 0.0111 | 0.0004 | 0.9859 | 63 | 2 |
| C.2 | 0.0164 | 0.0007 | 0.9773 | 42 | 2 |
| Mean | 0.0131 | 0.0003 | 0.9928 | 53 | 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Busche, J.F.; Möller, S.; Stehr, M.; Dietzel, A. Cross-Flow Filtration of Escherichia coli at a Nanofluidic Gap for Fast Immobilization and Antibiotic Susceptibility Testing. Micromachines 2019, 10, 691. https://doi.org/10.3390/mi10100691
Busche JF, Möller S, Stehr M, Dietzel A. Cross-Flow Filtration of Escherichia coli at a Nanofluidic Gap for Fast Immobilization and Antibiotic Susceptibility Testing. Micromachines. 2019; 10(10):691. https://doi.org/10.3390/mi10100691
Chicago/Turabian StyleBusche, Jan F., Svenja Möller, Matthias Stehr, and Andreas Dietzel. 2019. "Cross-Flow Filtration of Escherichia coli at a Nanofluidic Gap for Fast Immobilization and Antibiotic Susceptibility Testing" Micromachines 10, no. 10: 691. https://doi.org/10.3390/mi10100691






