Enter the Dragon: The Dynamic and Multifunctional Evolution of Anguimorpha Lizard Venoms
Abstract
1. Introduction
2. Results
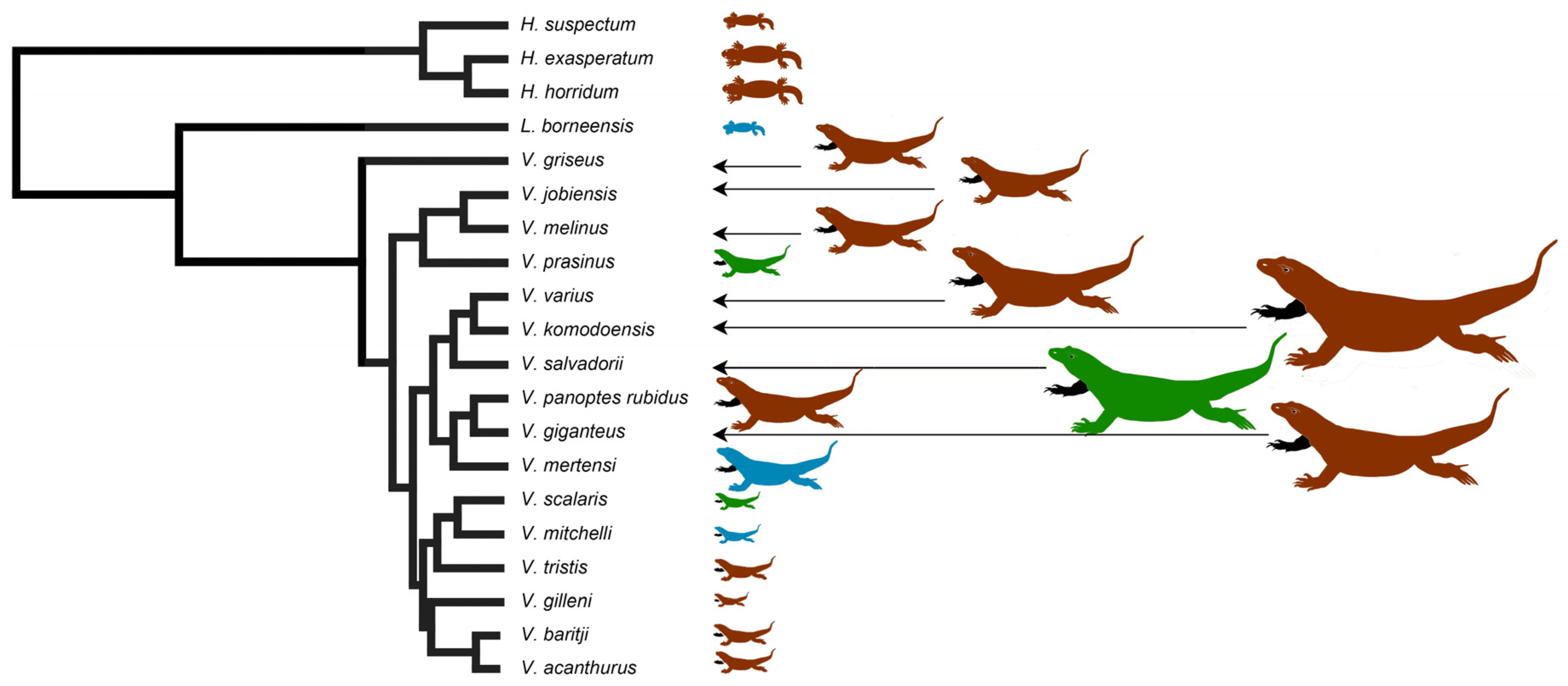
2.1. Teeth Scanning Electron Microscopy
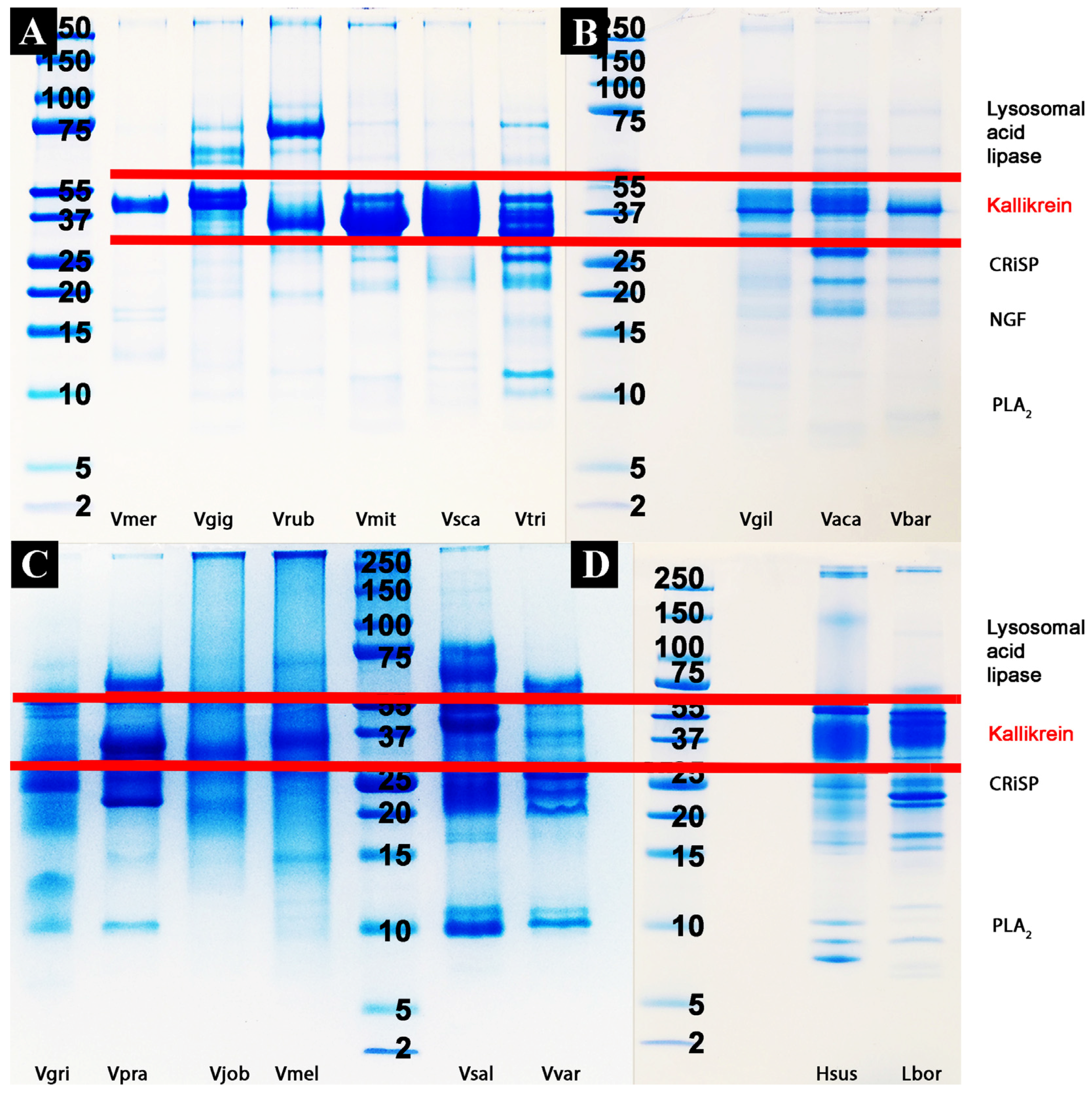
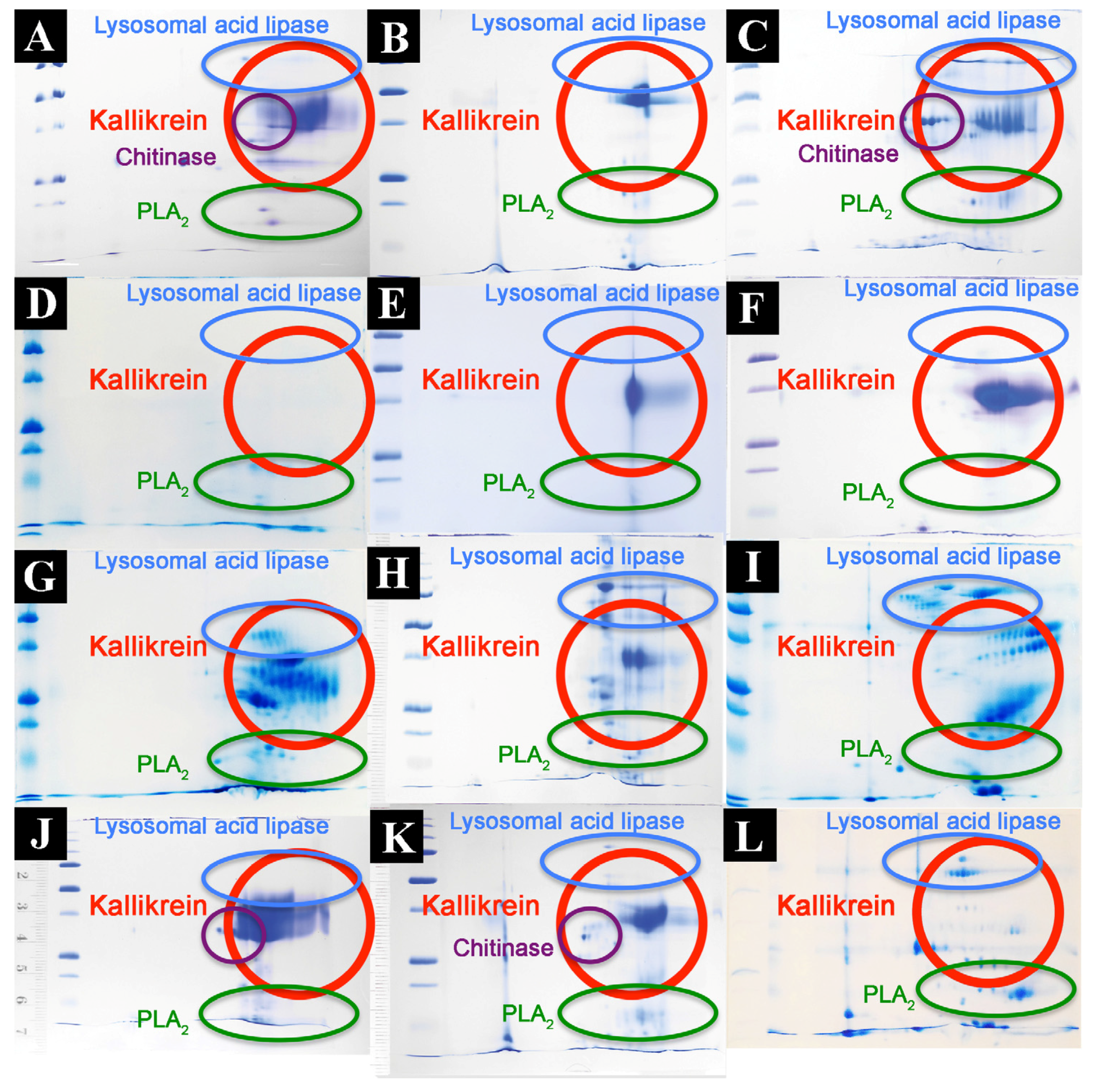
2.2. Proteomics Studies
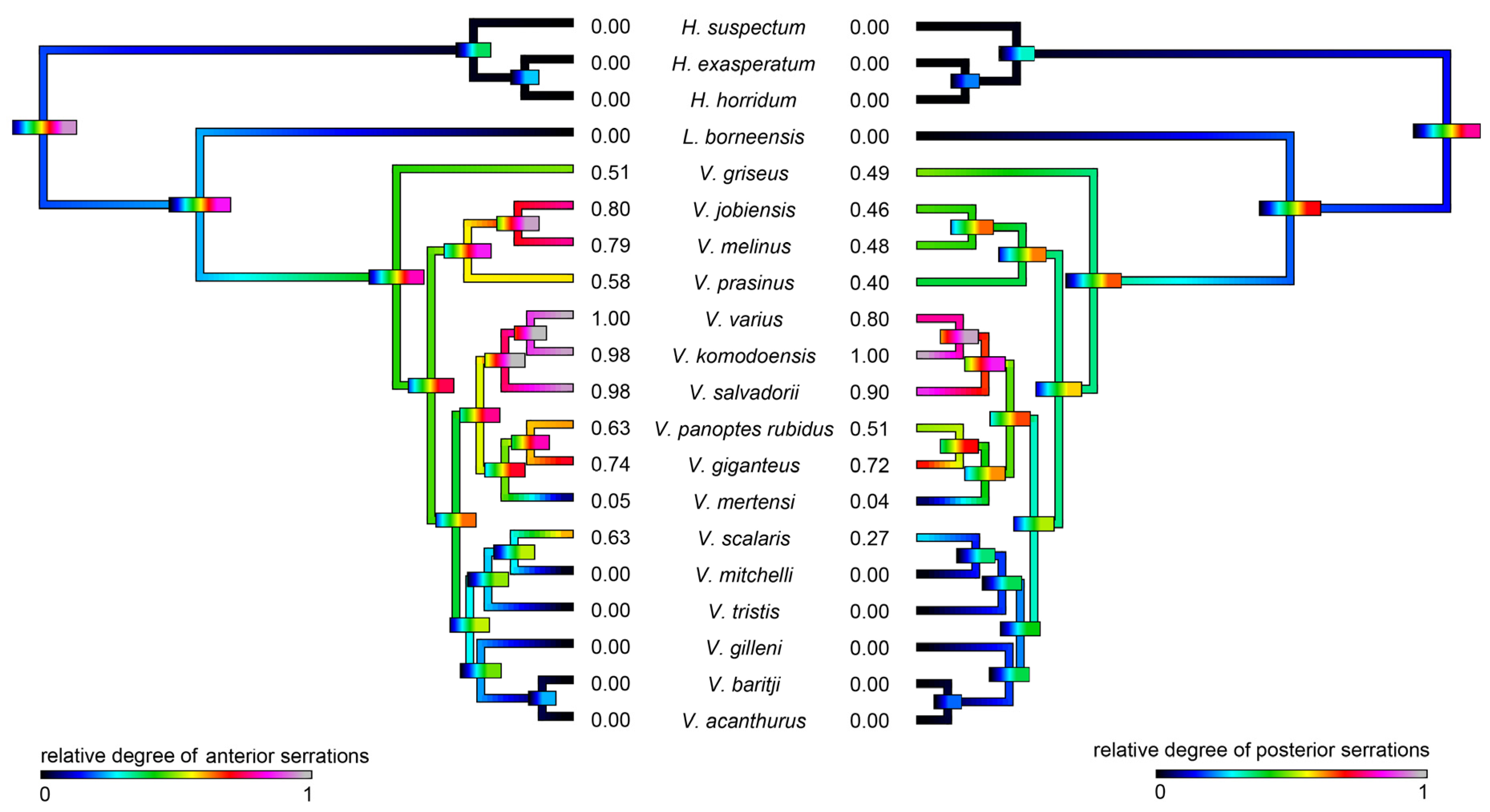
2.3. Molecular Evolution
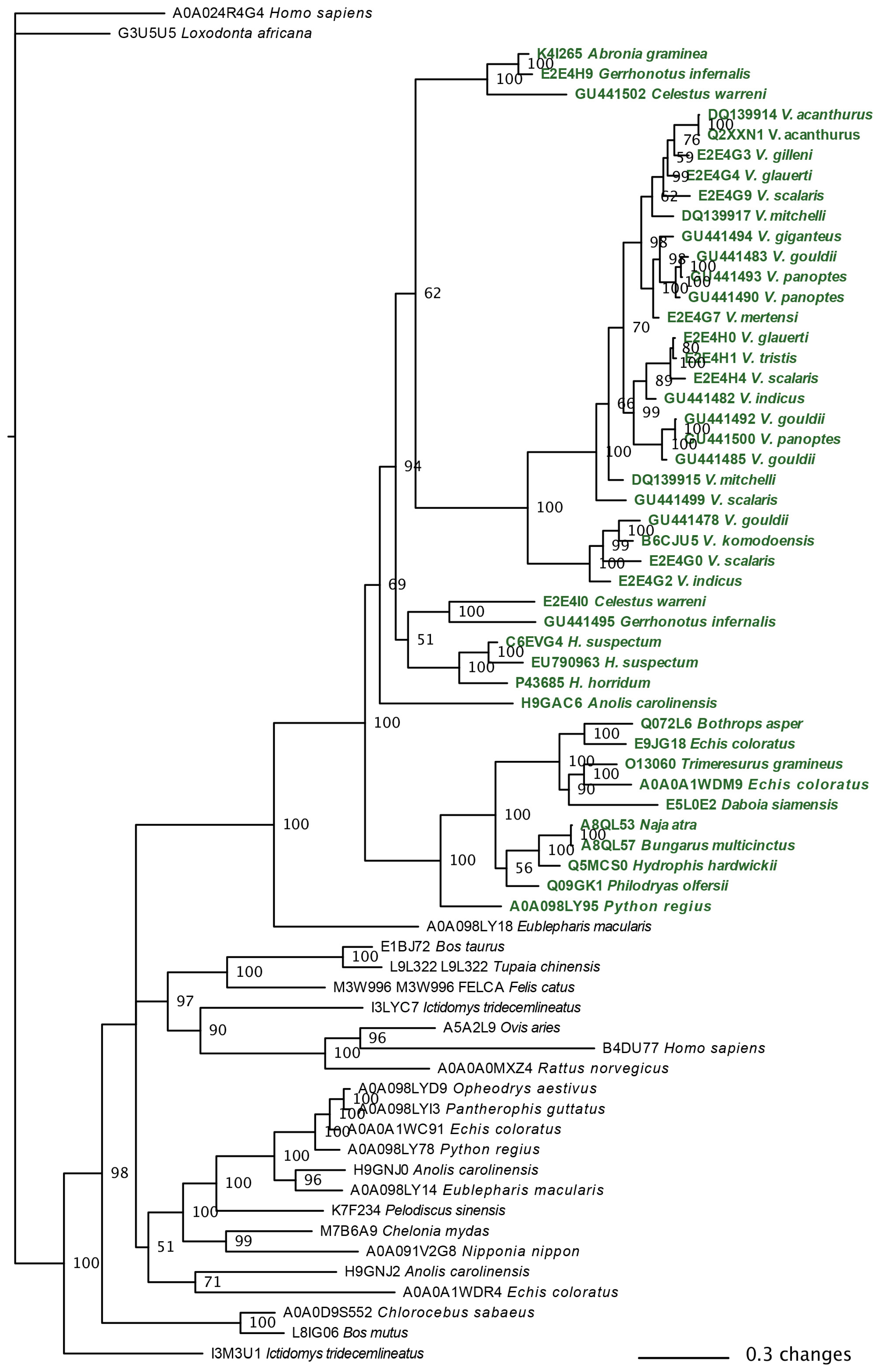
2.3.1. Kallikrein Molecular Evolution
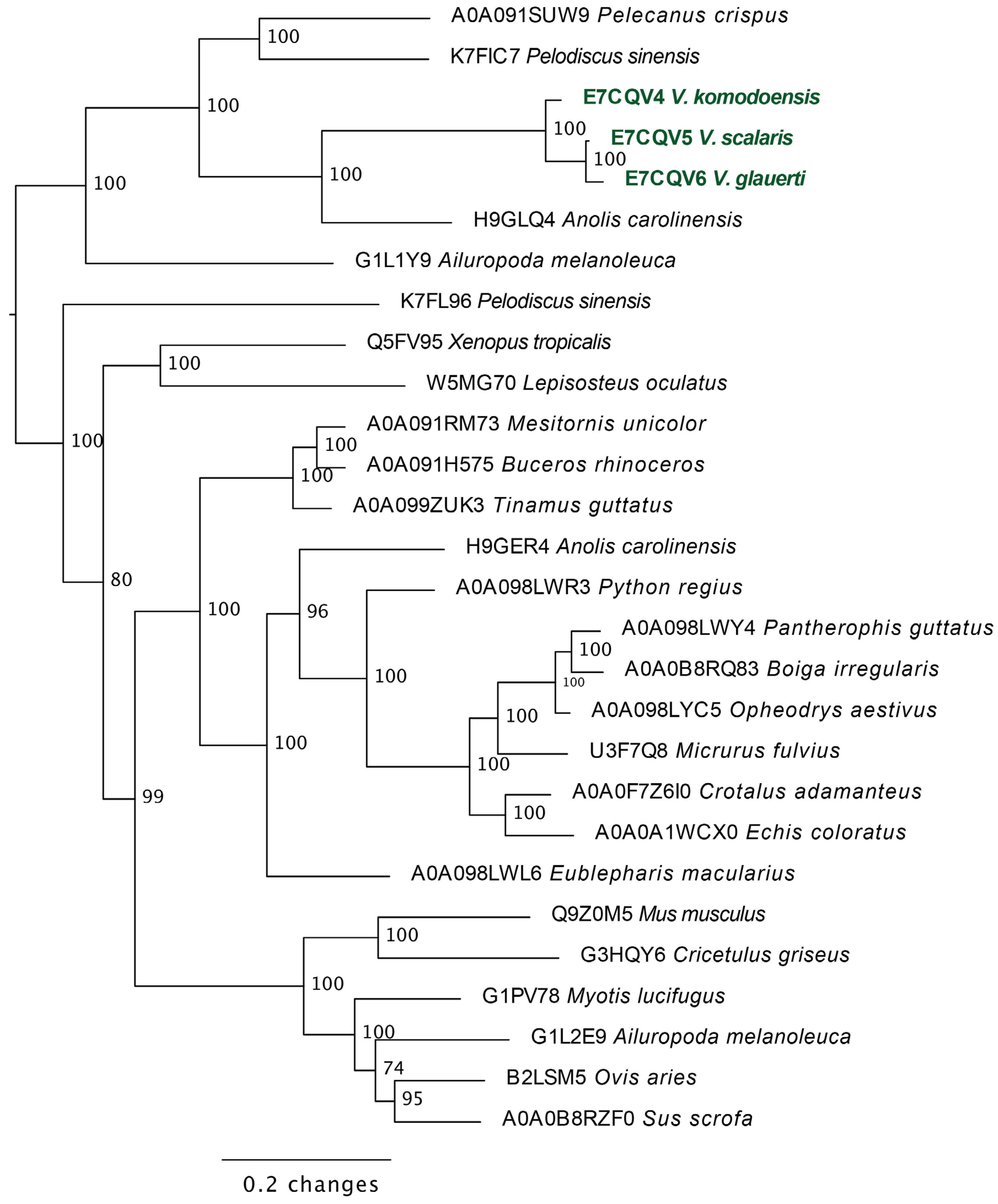
2.3.2. Lysosomal Acid Lipase Molecular Phylogeny
2.4. Bioactivity Testing
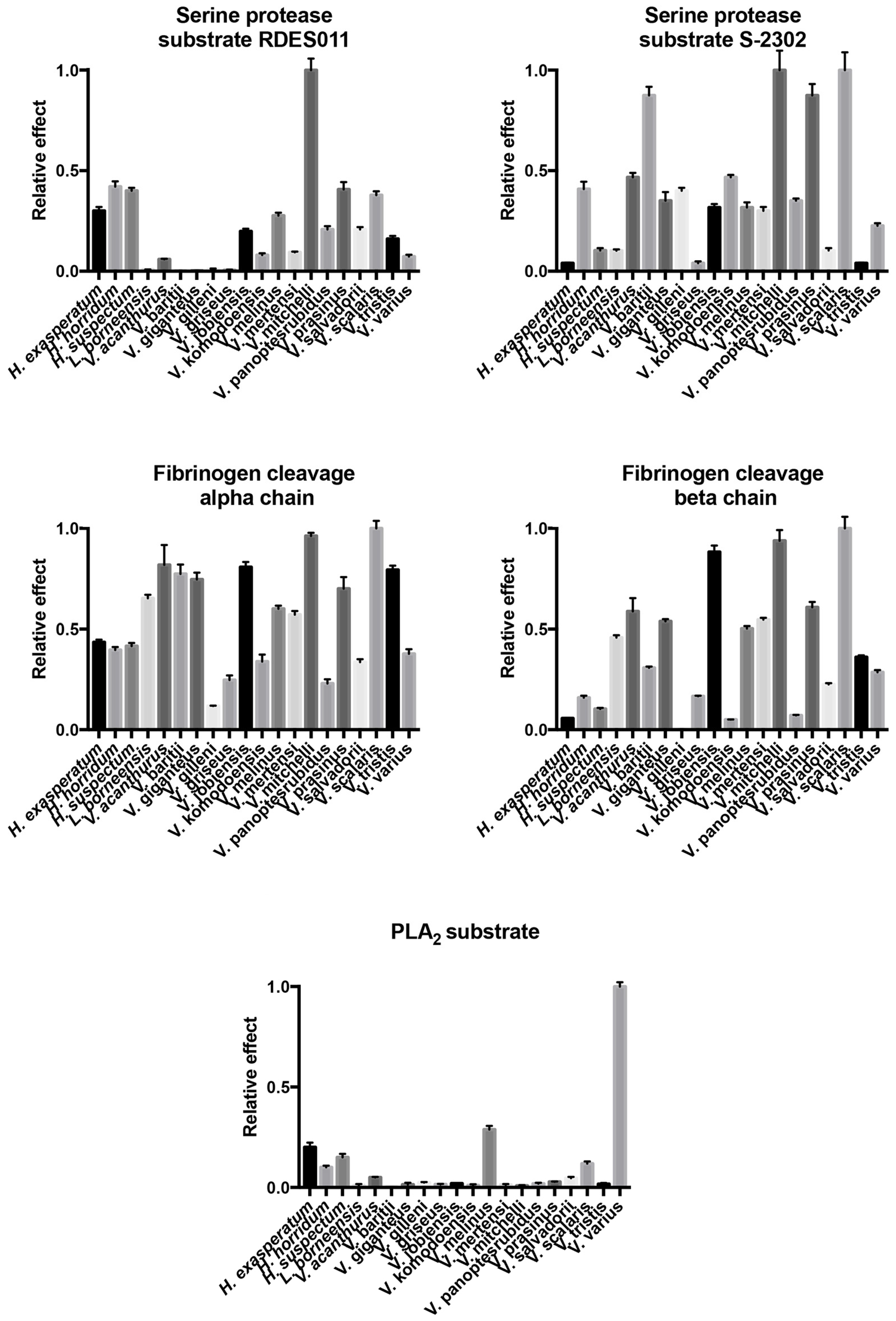
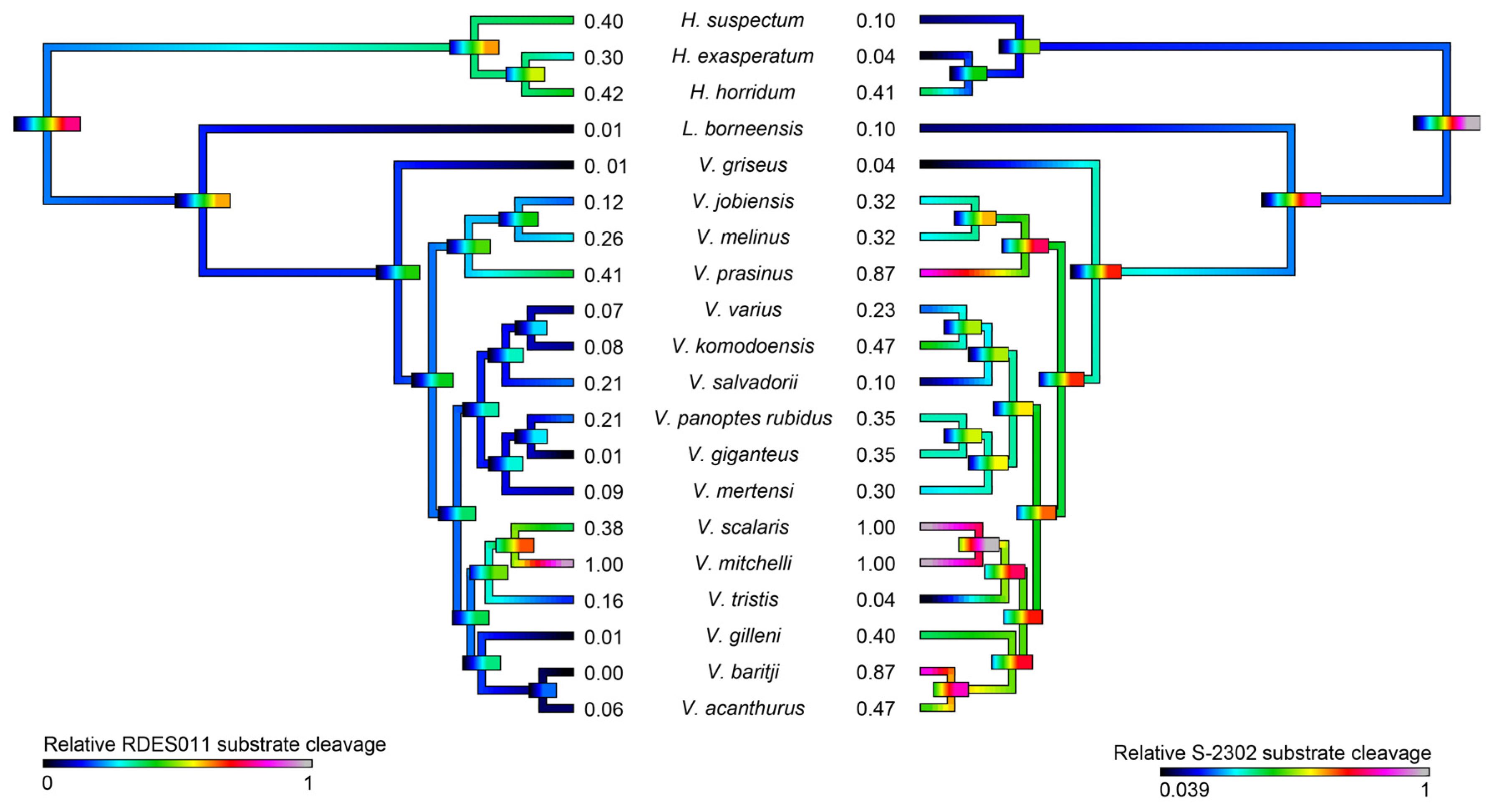
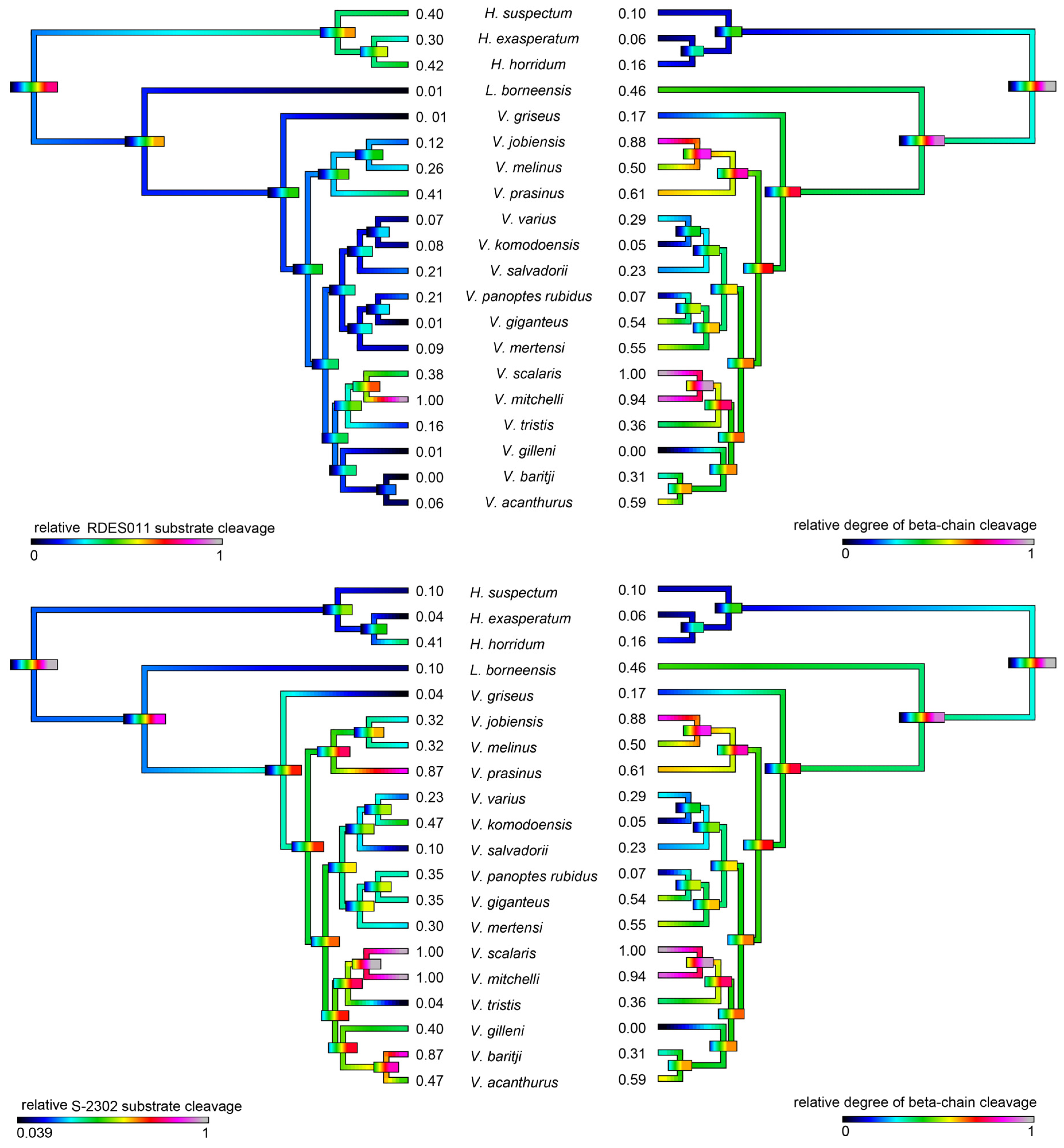
2.4.1. Kallikrein Enzymatic Activity upon Fluorescent Substrates
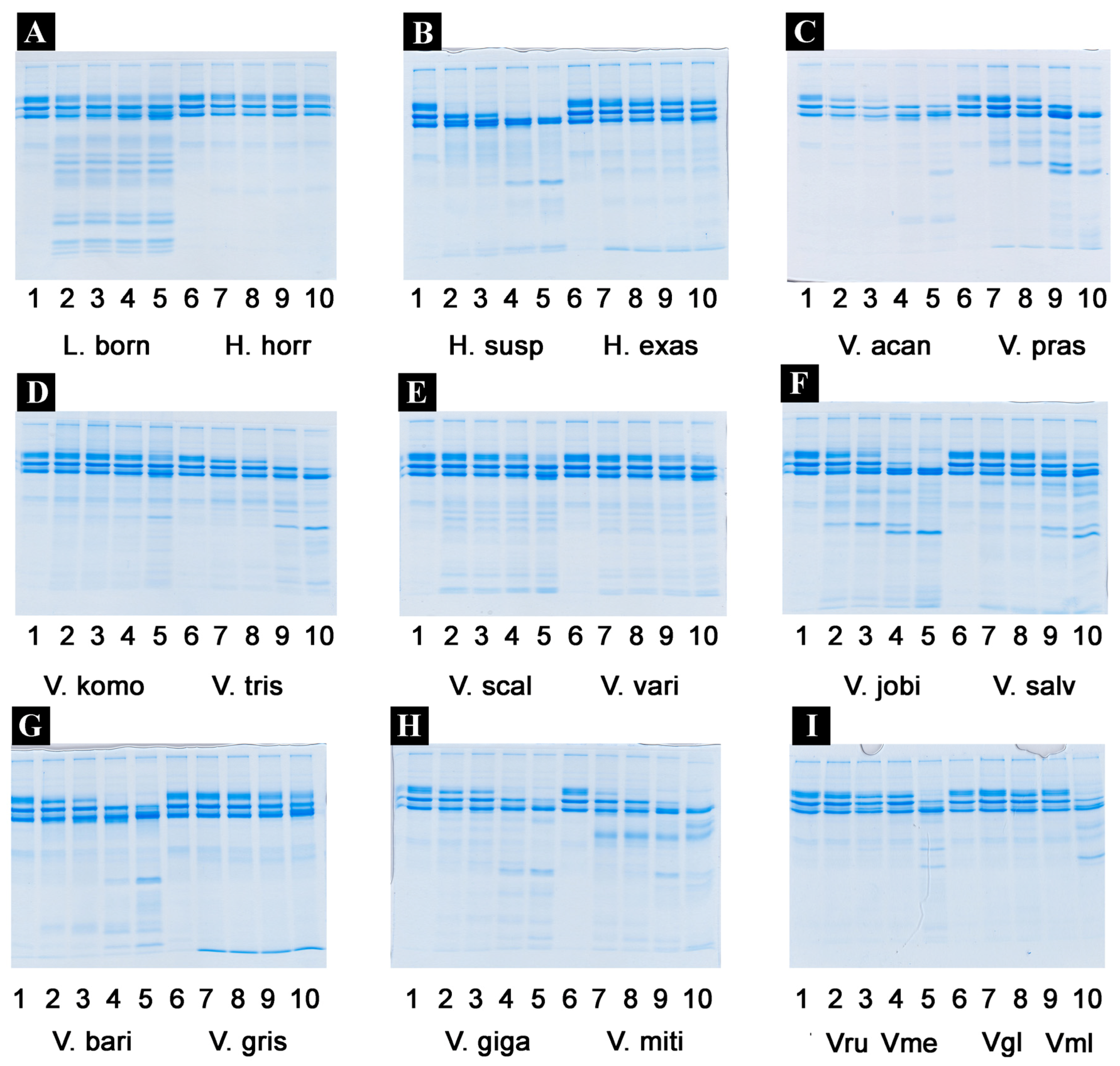
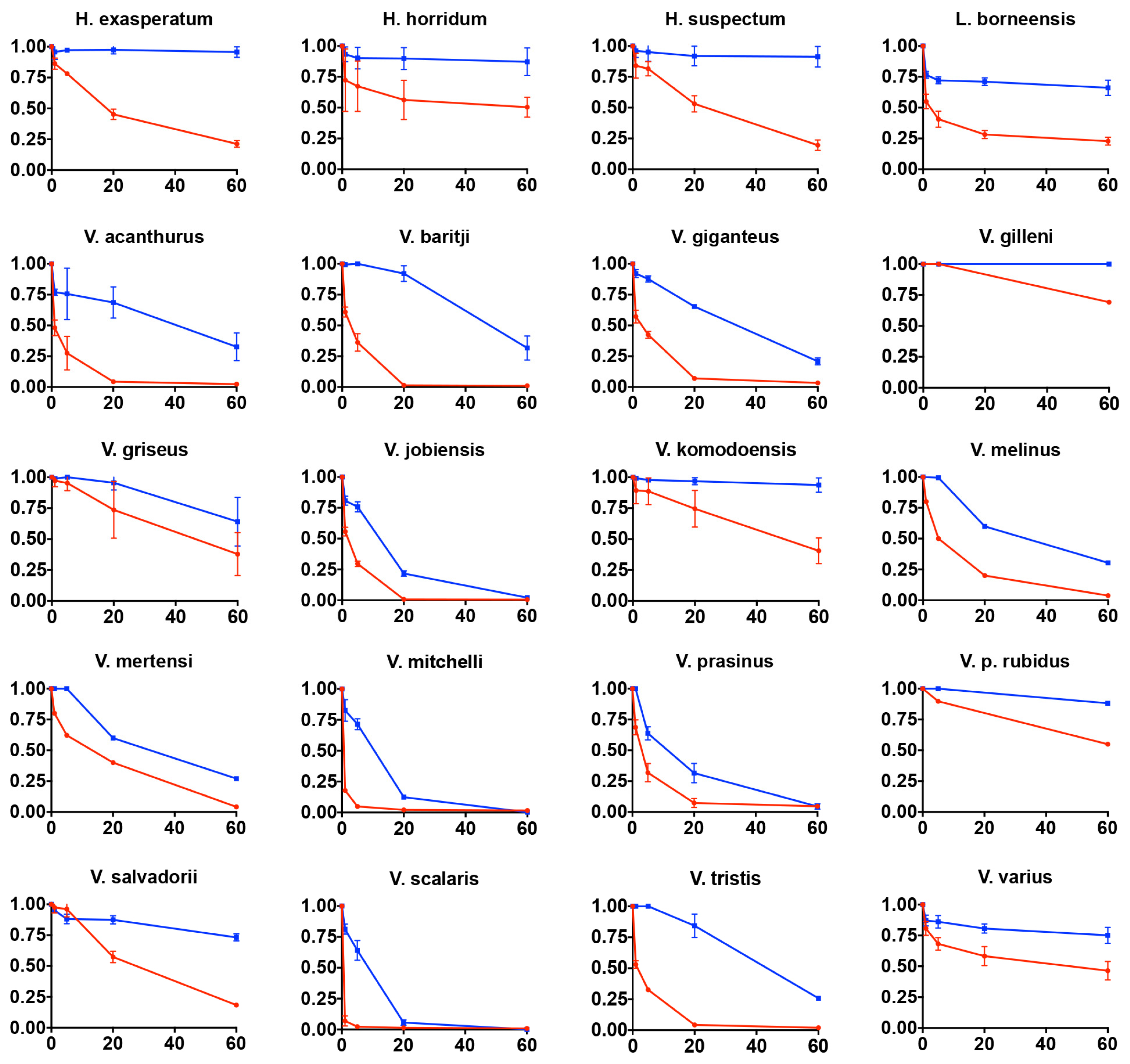
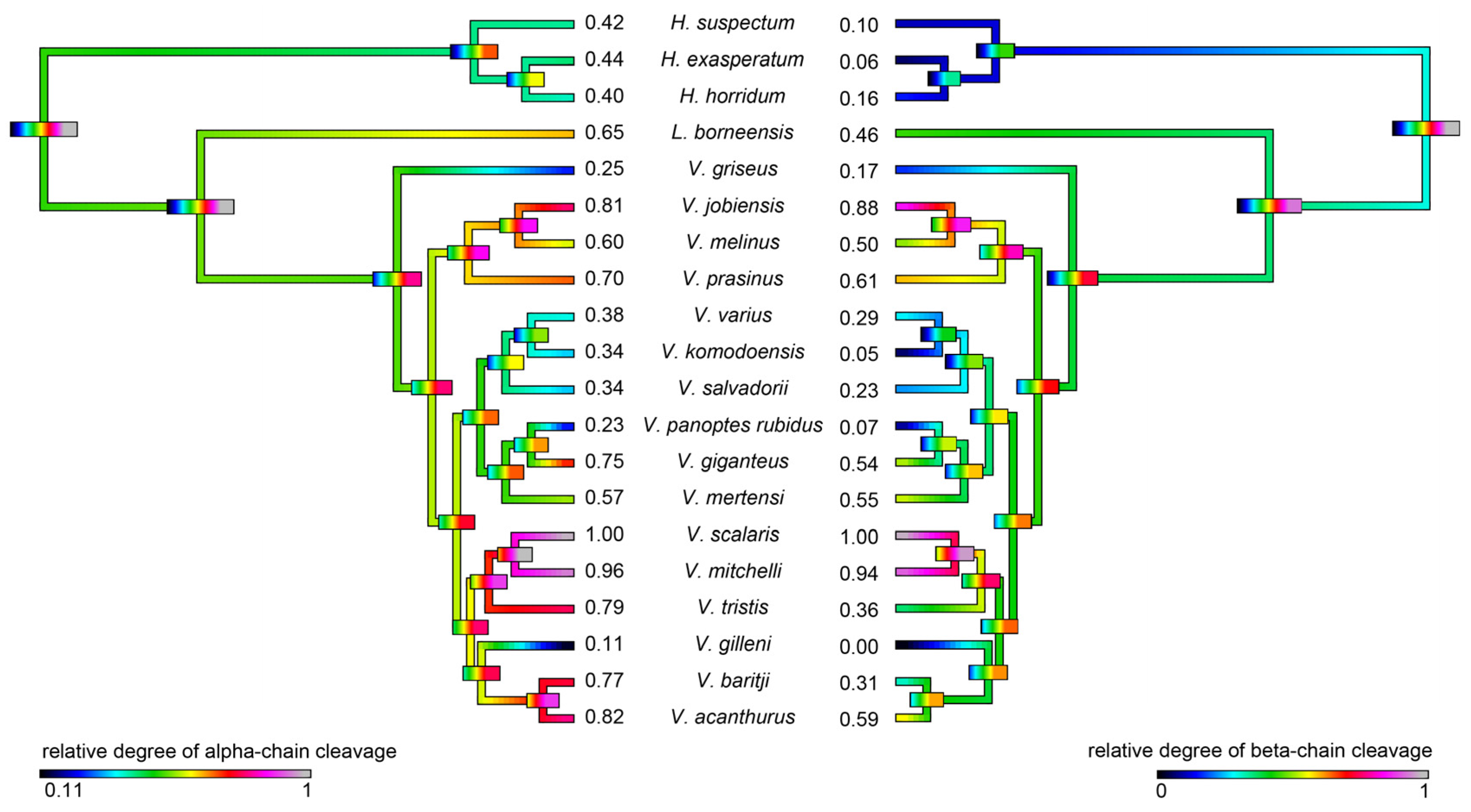
2.4.2. Fibrinogen Cleavage Gels
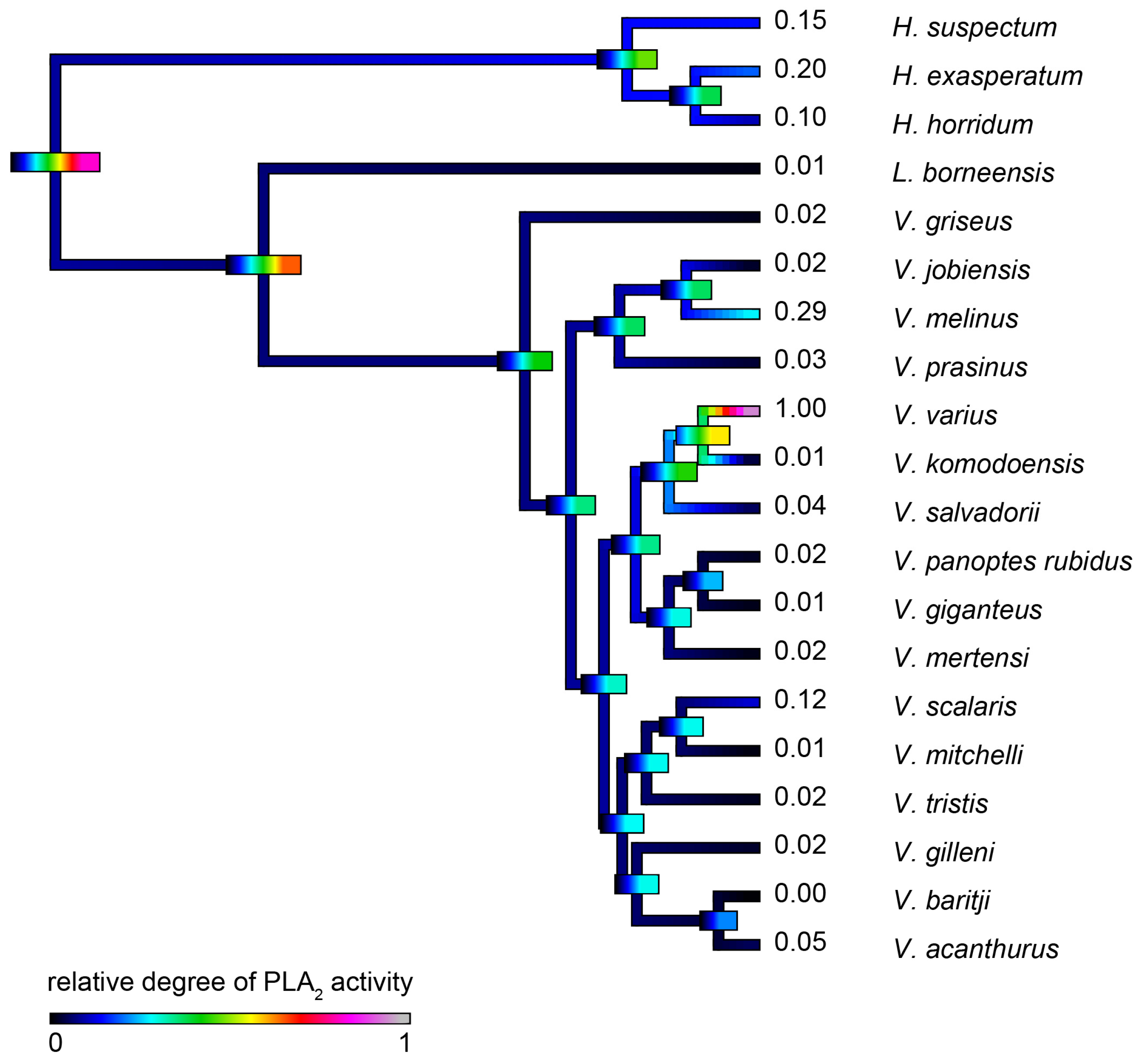
2.4.3. Phospholipase A2 Enzymatic Activity upon Fluorescent Substrate
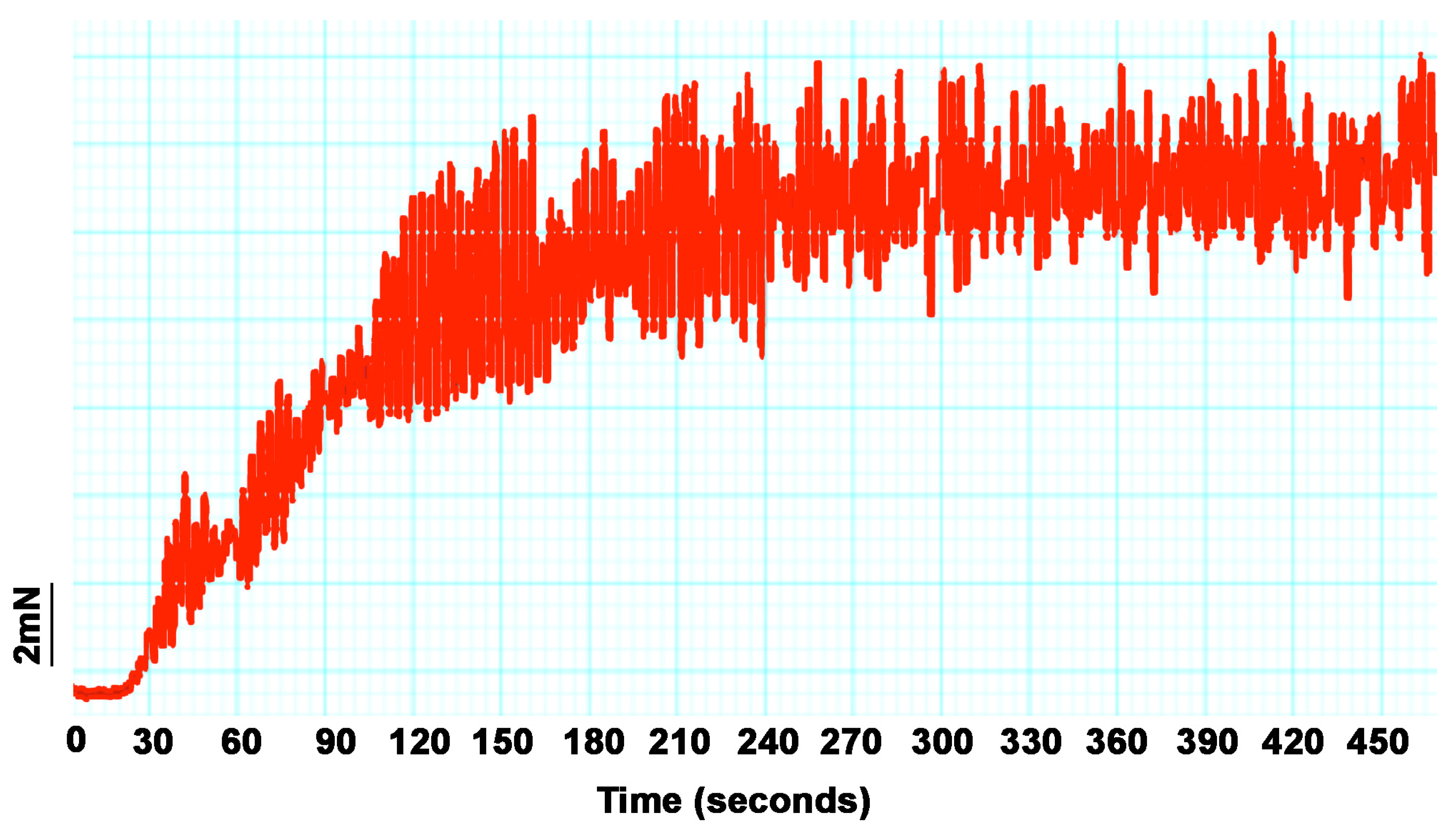
2.4.4. Rat Ileum Contraction Organ Bath Assay
3. Discussion
4. Materials and Methods
4.1. Species Studied
4.2. Scanning Electron Microscopy
4.3. Proteomics Studies
4.3.1. 1D Gel Electrophoresis
4.3.2. 2D Gel Electrophoresis
4.3.3. Shotgun Sequencing
4.3.4. LC–MS/MS
4.4. Bioactivity Studies
4.4.1. Kallikrein Activity
RDES0011 Substrate
S-2302 Substrate
4.4.2. Phsopholipase A2 Activity
4.4.3. Rat ileum Organ Bath Testings
4.4.4. Fibrinogen Gels
4.5. Bioinformatics
4.5.1. Phylogenetic Comparative Analyses
4.5.2. Phylogenetic Reconstruction
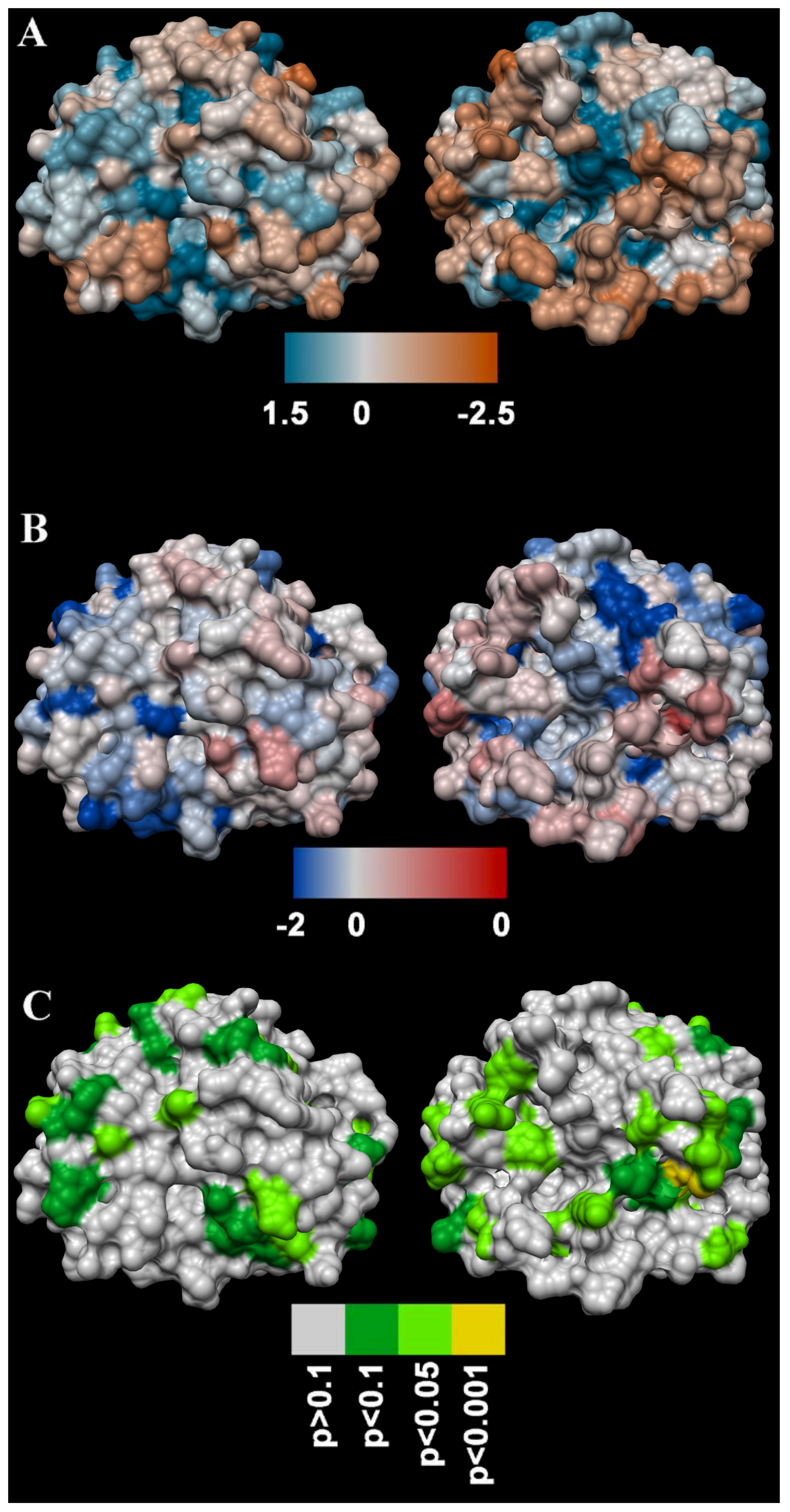
4.5.3. Molecular Modelling
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Auffenberg, W. Behavioral Ecology of the Komodo Monitor; University Presses of Florida: Gainesville, FL, USA, 1981. [Google Scholar]
- Montgomery, J.M.; Gillespie, D.; Sastrawan, P.; Fredeking, T.M.; Stewart, G.L. Aerobic salivary bacteria in wild and captive Komodo dragons. J. Wildl. Dis. 2002, 38, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, E.J.; Tyrrell, K.L.; Citron, D.M.; Cox, C.R.; Recchio, I.M.; Okimoto, B.; Bryja, J.; Fry, B.G. Anaerobic and aerobic bacteriology of the saliva and gingiva from 16 captive Komodo dragons (Varanus komodoensis): New implications for the “bacteria as venom” model. J. Zoo Wildl. Med. 2013, 44, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Hocknull, S.A.; Piper, P.J.; van den Bergh, G.D.; Due, R.A.; Morwood, M.J.; Kurniawan, I. Dragon’s paradise lost: Palaeobiogeography, evolution and extinction of the largest-ever terrestrial lizards (Varanidae). PLoS ONE 2009, 4, e7241. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Scheib, H.; Messenger, K.; Hocknull, S.; Wroe, S.; Sunagar, K.; Goldstein, E.J.C.; Tyrrell, K.L.; Citron, D.M.; Jackson, T.N.W. Poisonous snakes and bacteria as a Komodo dragon weapon: Which is a myth and which is reality? In Venomous Reptiles: Evolution, Pathophysiology and Biodiscovery; Fry, B.G., Ed.; Oxford University Press: New York, NY, USA, 2015; pp. 408–414. [Google Scholar]
- Estes, R. Charles L. Camp—An appreciation. In Phylogenetic Relationships of the Lizard Families: Essay Commemorating Charles L. Camp; Estes, R., Pregill, G., Eds.; Stanford University Press: Stanford, CA, USA, 1988; pp. 9–14. [Google Scholar]
- Losos, J.B.; Hillis, D.M.; Greene, H.W. Evolution. Who speaks with a forked tongue? Science 2012, 338, 1428–1429. [Google Scholar] [CrossRef] [PubMed]
- Sweet, S.S.; Pianka, E.R. Monitors, mammals and Wallace’s line. Mertensiella 2007, 16, 79–99. [Google Scholar]
- Vitt, L.J. Walking the Natural-History Trail. Herpetologica 2013, 69, 105–117. [Google Scholar] [CrossRef]
- Sweet, S.S. Chasing Flamingos: Toxicofera and the Misinterpretation of Venom in Varanid Lizards. In Proceedings of the 2015 Interdisciplinary World Conference on Monitor Lizards; Conta, M., Ed.; Institute for Research and Development; Suan Sunandha Rajabhat University: Bangkok, Thailand, 2016; pp. 123–149. [Google Scholar]
- Hedges, S.; Vidal, N. Lizards, snakes, and amphisbaenians (Squamata). In The Timetree of Life; Hedges, S.B., Kumar, S., Eds.; Oxford University Press: New York, NY, USA, 2009; pp. 383–389. [Google Scholar]
- Pyron, R.A.; Burbrink, F.T. Extinction, ecological opportunity, and the origins of global snake diversity. Evolution 2012, 66, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Reeder, T.W.; Townsend, T.M.; Mulcahy, D.G.; Noonan, B.P.; Wood, P.L., Jr.; Sites, J.W., Jr.; Wiens, J.J. Integrated analyses resolve conflicts over squamate reptile phylogeny and reveal unexpected placements for fossil taxa. PLoS ONE 2015, 10, e0118199. [Google Scholar] [CrossRef] [PubMed]
- Townsend, T.; Larson, A.; Louis, E.; Macey, J.R. Molecular phylogenetics of squamata: The position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst. Biol. 2004, 53, 735–757. [Google Scholar] [CrossRef] [PubMed]
- Vidal, N.; David, P. New insights into the early history of snakes inferred from two nuclear genes. Mol. Phylogenet. Evol. 2004, 31, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Vidal, N.; Hedges, S.B. The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. C. R. Biol. 2005, 328, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Vidal, N.; Hedges, S.B. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. C. R. Biol. 2009, 332, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.J.; Hutter, C.R.; Mulcahy, D.G.; Noonan, B.P.; Townsend, T.M.; Sites, J.W., Jr.; Reeder, T.W. Resolving the phylogeny of lizards and snakes (Squamata) with extensive sampling of genes and species. Biol. Lett. 2012, 8, 1043–1046. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.J.; Kuczynski, C.A.; Townsend, T.; Reeder, T.W.; Mulcahy, D.G.; Sites, J.W., Jr. Combining Phylogenomics and Fossils in Higher-Level Squamate Reptile Phylogeny: Molecular Data Change the Placement of Fossil Taxa. Syst. Biol. 2010, 59, 674–688. [Google Scholar] [CrossRef] [PubMed]
- Pyron, R.A. Novel Approaches for Phylogenetic Inference from Morphological Data and Total-Evidence Dating in Squamate Reptiles (Lizards, Snakes, and Amphisbaenians). Syst. Biol. 2017, 66, 38–56. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wiens, J.J. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 2016, 94, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Vidal, N.; Norman, J.A.; Vonk, F.J.; Scheib, H.; Ramjan, S.F.; Kuruppu, S.; Fung, K.; Hedges, S.B.; Richardson, M.K.; et al. Early evolution of the venom system in lizards and snakes. Nature 2006, 439, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G. From genome to “venome”: Molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005, 15, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Casewell, N.R.; Wuster, W.; Vidal, N.; Young, B.; Jackson, T.N. The structural and functional diversification of the Toxicofera reptile venom system. Toxicon 2012, 60, 434–448. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev.Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Roelants, K.; Winter, K.; Hodgson, W.C.; Griesman, L.; Kwok, H.F.; Scanlon, D.; Karas, J.; Shaw, C.; Wong, L.; et al. Novel venom proteins produced by differential domain-expression strategies in beaded lizards and gila monsters (genus Heloderma). Mol. Biol. Evol. 2010, 27, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Scheib, H.; van der Weerd, L.; Young, B.; McNaughtan, J.; Ramjan, S.F.; Vidal, N.; Poelmann, R.E.; Norman, J.A. Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell. Proteom. 2008, 7, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Sunagar, K.; Casewell, N.R.; Kochva, E.; Roelants, K.; Scheib, H.; Wüster, W.; Vidal, N.; Young, B.; Burbrink, F.; et al. The origin and evolution of the Toxicofera reptile venom system. In Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery; Fry, B.G., Ed.; Oxford University Press: New York, NY, USA, 2015; pp. 1–31. [Google Scholar]
- Fry, B.G.; Undheim, E.A.; Ali, S.A.; Jackson, T.N.; Debono, J.; Scheib, H.; Ruder, T.; Morgenstern, D.; Cadwallader, L.; Whitehead, D.; et al. Squeezers and leaf-cutters: Differential diversification and degeneration of the venom system in toxicoferan reptiles. Mol. Cell. Proteom. 2013, 12, 1881–1899. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Vidal, N.; van der Weerd, L.; Kochva, E.; Renjifo, C. Evolution and diversification of the Toxicofera reptile venom system. J. Proteom. 2009, 72, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Winter, K.; Norman, J.A.; Roelants, K.; Nabuurs, R.J.; van Osch, M.J.; Teeuwisse, W.M.; van der Weerd, L.; McNaughtan, J.E.; Kwok, H.F.; et al. Functional and structural diversification of the Anguimorpha lizard venom system. Mol. Cell. Proteom. 2010, 9, 2369–2390. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Wroe, S.; Teeuwisse, W.; van Osch, M.J.; Moreno, K.; Ingle, J.; McHenry, C.; Ferrara, T.; Clausen, P.; Scheib, H.; et al. A central role for venom in predation by Varanus komodoensis (Komodo Dragon) and the extinct giant Varanus (Megalania) priscus. Proc. Natl. Acad. Sci. USA 2009, 106, 8969–8974. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Wuster, W. Assembling an arsenal: Origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences. Mol. Biol. Evol. 2004, 21, 870–883. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P.; Saviola, A.J. Understanding biological roles of venoms among the caenophidia: The importance of rear-fanged snakes. Integr. Comp. Biol. 2016, 56, 1004–1021. [Google Scholar] [CrossRef] [PubMed]
- Hsiang, A.Y.; Field, D.J.; Webster, T.H.; Behlke, A.D.; Davis, M.B.; Racicot, R.A.; Gauthier, J.A. The origin of snakes: Revealing the ecology, behavior, and evolutionary history of early snakes using genomics, phenomics, and the fossil record. BMC Evol. Biol. 2015, 15, 87. [Google Scholar] [CrossRef] [PubMed]
- Sites, J.W., Jr.; Reeder, T.W.; Wiens, J.J. Phylogenetic insights on evolutionarynovelties in lizards and snakes: Sex, birth, bodies, niches, and venom. Annu. Rev. Ecol. Evolut. Syst. 2011, 42, 227–244. [Google Scholar] [CrossRef]
- Weinstein, S.A. Snake venoms: A brief treatise on etymology, origins of terminology, and definitions. Toxicon 2015, 103, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.N.W.; Young, B.; McCarthy, C.J.; Kochva, E.; Vidal, N.; Underwood, G.; Fry, B.G. Endless forms most beautiful: The evolution of ophidian oral glands, including the venom system, and the use of appropriate terminology for homologous structures. Zoomorphology 2017, 136, 107–130. [Google Scholar] [CrossRef]
- Hargreaves, A.D.; Swain, M.T.; Logan, D.W.; Mulley, J.F. Testing the Toxicofera: Comparative transcriptomics casts doubt on the single, early evolution of the reptile venom system. Toxicon 2014, 92, 140–156. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.N.; Fry, B.G. A Tricky Trait: Applying the Fruits of the “Function Debate” in the Philosophy of Biology to the “Venom Debate” in the Science of Toxinology. Toxins 2016, 8, 263. [Google Scholar] [CrossRef] [PubMed]
- Mebs, D. Venomous and Poisonous Animals: A Handbook for Biologists, Toxicologists and Toxinologists, Physicians and Pharmacists; CRC Press: Boca Raton, FL, USA; Medpharm: Stuttgart, Germany, 2002. [Google Scholar]
- Hargreaves, A.D.; Tucker, A.S.; Mulley, J.F. A Critique of the Toxicoferan Hypothesis. In Evolution of Venomous Animals and Their Toxins; Malhotra, A., Ed.; Springer Nature: Dordrech, The Netherlands, 2017; pp. 69–86. [Google Scholar]
- Fry, B.G.; Wickramaratana, J.C.; Lemme, S.; Beuve, A.; Garbers, D.; Hodgson, W.C.; Alewood, P. Novel natriuretic peptides from the venom of the inland taipan (Oxyuranus microlepidotus): Isolation, chemical and biological characterisation. Biochem. Biophys. Res. Commun. 2005, 327, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Mebs, D. Purification and properties of a kinin liberating enzyme from venom of Heloderma suspectum. Naunyn-Schmiedeberg’s Arch. Pharmakol. 1969, 264, 280–281. [Google Scholar] [CrossRef]
- Gorelov, Y.K. Concerning the Varanus griseus saliva toxicity. Izy. Akademii Turkmenistan SSR 1971, 6, 75–76. [Google Scholar]
- Sopyev, O.; Makeev, V.M.; Kudryavtsev, S.V.; Makarov, A.N. Case of intoxification from a bite of Varanus griseus. Izy. Akademii Turkmenistan SSR 1987, 87, 78. [Google Scholar]
- Ballard, V.; Antonio, F.B. Varanus griseus (Desert monitor) toxicity. Herpetol. Rev. 2001, 32, 261. [Google Scholar]
- Vikrant, S.; Verma, B.S. Monitor lizard bite-induced acute kidney injury—A case report. Ren. Fail. 2014, 36, 444–446. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Weinstein, S.A. Reply to Vikrant and Verma about “Monitor Lizard Envenoming”. Ren. Fail. 2015, 37, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Ducey, S.D.; Cooper, J.S.; Wadman, M.C. Bitten by a Dragon. Wilderness Environ. Med. 2016, 27, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Loop, M.S. The effect of relative prey size on the ingestion behavior of the Bengal monitor, Varanus bengalensis (Sauria: Varanidae). Herpetologica 1974, 30, 123–127. [Google Scholar]
- Kochva, E. Oral glands of the Reptilia. Physiology B 1978, 8, 43–162. [Google Scholar]
- Li, M.; Fry, B.G.; Kini, R.M. Eggs-Only diet: Its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii). J. Mol. Evol. 2005, 60, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Fry, B.G.; Kini, R.M. Putting the brakes on snake venom evolution: The unique molecular evolutionary patterns of Aipysuras eydouxii (Marbled sea snake) phospholipase A(2) toxins. Mol. Biol. Evol. 2005, 22, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, D.; King, G.F. The venom optimization hypothesis revisited. Toxicon 2013, 63, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Koludarov, I.; Jackson, T.N.; Sunagar, K.; Nouwens, A.; Hendrikx, I.; Fry, B.G. Fossilized venom: The unusually conserved venom profiles of Heloderma species (beaded lizards and gila monsters). Toxins 2014, 6, 3582–3595. [Google Scholar] [CrossRef] [PubMed]
- Furman, B.L. The development of Byetta (exenatide) from the venom of the Gila monster as an anti-diabetic agent. Toxicon 2012, 59, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Irwin, D.M. Origin and convergent evolution of exendin genes. Gen. Comp. Endocrinol. 2012, 175, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Kwok, H.F.; Chen, T.; O’Rourke, M.; Ivanyi, C.; Hirst, D.; Shaw, C. Helokinestatin: A new bradykinin B-2 receptor antagonist decapeptide from lizard venom. Peptides 2008, 29, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Sanggaard, K.W.; Dyrlund, T.F.; Thomsen, L.R.; Nielsen, T.A.; Brøndum, L.; Wang, T.; Thøgersen, I.B.; Enghild, J.J. Characterization of the gila monster (Heloderma suspectum suspectum) venom proteome. J. Proteom. 2015, 117, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Yang, M.; Zhou, M.; Wu, Y.; Wang, L.; Chen, T.; Ding, A.; Shaw, C. The natriuretic peptide/helokinestatin precursor from Mexican beaded lizard (Heloderma horridum) venom: Amino acid sequence deduced from cloned cDNA and identification of two novel encoded helokinestatins. Peptides 2011, 32, 1166–1171. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, H.; Wu, Y.; Zhou, M.; Lowe, G.; Wang, L.; Zhang, Y.; Chen, T.; Shaw, C. Helokinestatin-7 peptides from the venoms of Heloderma lizards. Peptides 2012, 35, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Zhou, M.; Zhou, Z.; Chen, X.; Chen, T.; Kwok, H.; Ivanyi, C.; Shaw, C. The structure of helokinestatin-5 and its biosynthetic precursor from Gila monster (Heloderma suspectum) venom: Evidence for helokinestatin antagonism of bradykinin-induced relaxation of rat tail artery smooth muscle. Peptides 2010, 31, 1555–1561. [Google Scholar] [CrossRef] [PubMed]
- Ast, J.C. Mitochondrial DNA evidence and evolution in Varanoidea (Squamata). Cladistics 2001, 17, 211–226. [Google Scholar] [CrossRef]
- Thompson, G.G.; Clemente, C.J.; Withers, P.C.; Fry, B.G.; Norman, J.A. Is body shape of varanid lizards linked with retreat choice? Aust. J. Zool. 2008, 56, 351–362. [Google Scholar] [CrossRef]
- Vidal, N.; Marin, J.; Sassi, J.; Battistuzzi, F.U.; Donnellan, S.; Fitch, A.J.; Fry, B.G.; Vonk, F.J.; Rodriguez de la Vega, R.C.; Couloux, A.; et al. Molecular evidence for an Asian origin of monitor lizards followed by Tertiary dispersals to Africa and Australasia. Biol. Lett. 2012, 8, 853–855. [Google Scholar] [CrossRef] [PubMed]
- Schweitz, H.; Pacaud, P.; Diochot, S.; Moinier, D.; Lazdunski, M. MIT1, a black mamba toxin with a new and highly potent activity on intestinal contraction. FEBS Lett. 1999, 461, 183–188. [Google Scholar] [CrossRef]
- Fry, B.G.; Sunagar, K.; Jackson, T.N.W.; Reeks, T.; Kwok, H.F. B-type natriuretic peptides. In Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery; Fry, B.G., Ed.; Oxford University Press: New York, NY, USA, 2015; pp. 312–317. [Google Scholar]
- Mochca-Morales, J.; Martin, B.M.; Possani, L.D. Isolation and characterization of helothermine, a novel toxin from Heloderma horridum horridum (Mexican beaded lizard) venom. Toxicon 1990, 28, 299–309. [Google Scholar] [CrossRef]
- Morrissette, J.; Elhayek, R.; Possani, L.; Coronado, R. Isolation and characterization of ryanodine receptor toxins from Heloderma horridum (mexican beaded lizard) venom. Biophys. J. 1994, 66, A415. [Google Scholar]
- Morrissette, J.; Krätzschmar, J.; Haendler, B.; El-Hayek, R.; Mochca-Morales, J.; Martin, B.M.; Patel, J.R.; Moss, R.L.; Schleuning, W.-D.; Coronado, R. Primary structure and properties of helothermine, a peptide toxin that blocks ryanodine receptors. Biophys. J. 1995, 68, 2280. [Google Scholar] [CrossRef]
- Nobile, M.; Magnelli, V.; Lagostena, L.; Mochca-Morales, J.; Possani, L.D.; Prestipino, G. The toxin helothermine affects potassium currents in newborn rat cerebellar granule cells. J. Membr. Biol. 1994, 139, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Nobile, M.; Noceti, F.; Prestipino, G.; Possani, L.D. Helothermine, a lizard venom toxin, inhibits calcium current in cerebellar granules. Exp. Brain Res. 1996, 110, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Grundemar, L.; Högestätt, E.D. Vascular effects of helodermin, helospectin I and helospectin II: A comparison with vasoactive intestinal peptide (VIP). Br. J. Pharmacol. 1990, 99, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Tsueshita, T.; Onyukusel, H.; Sethi, V.; Gandhi, S.; Rubinstein, I. Helospectin I and II evoke vasodilation in the intact peripheral microcirculation. Peptides 2004, 25, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Uddman, R.; Goadsby, P.J.; Jansen-Olesen, I.; Edvinsson, L. Helospectin-like peptides: Immunochemical localization and effects on isolated cerebral arteries and on local cerebral blood flow in the cat. J. Cereb. Blood Flow Metabol. 1999, 19, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Komori, Y.; Nikai, T.; Sugihara, H. Purification and characterization of a lethal toxin from the venom of Heloderma horridum horridum. Biochem. Biophys. Res. Commun. 1988, 154, 613–619. [Google Scholar] [CrossRef]
- Datta, G.; Tu, A.T. Structure and other chemical characterizations of gila toxin, a lethal toxin from lizard venom. J. Pept. Res. 1997, 50, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Mebs, D. Isolation and properties of kallikrein from venom of gila monster (Heloderma suspectum). Hoppe-Seylers Z. Physiol. Chem. 1969, 350, 821–826. [Google Scholar] [CrossRef] [PubMed]
- Nikai, T.; Imai, K.; Komori, Y.; Sugihara, H. Isolation and characterization of arginine ester hydrolase from Heloderma horridum (beaded lizard) venom. Int. J. Biochem. 1992, 24, 415–420. [Google Scholar] [PubMed]
- Nikai, T.; Imai, K.; Nagasaka, M.; Sugihara, H. Kallikrein-like enzyme from the venom of Agkistrodon p. piscivorus. Int. J. Biochem. 1988, 20, 1239–1245. [Google Scholar] [CrossRef]
- Nikai, T.; Imai, K.; Sugihara, H.; Tu, A.T. Isolation and characterization of horridum toxin with arginine ester hydrolase activity from Heloderma horridum (beaded lizard) venom. Arch. Biochem. Biophys. 1988, 264, 270–280. [Google Scholar] [CrossRef]
- Utaisincharoen, P.; Mackessy, S.P.; Miller, R.A.; Tu, A.T. Complete primary structure and biochemical properties of gilatoxin, a serine protease with kallikrein-like and angiotensin-degrading activities. J. Biol. Chem. 1993, 268, 21975–21983. [Google Scholar] [PubMed]
- Vaiyapuri, S.; Sunagar, K.; Gibbins, J.M.; Jackson, T.N.W.; Reeks, T.; Fry, B.G. Kallikrein Enzymes In Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery; Fry, B.G., Ed.; Oxford University Press: New York, NY, USA, 2015; pp. 267–280. [Google Scholar]
- Huang, T.F.; Chiang, H.S. Effect on human platelet-aggregation of phospholipase a(2) purified from Heloderma horridum (beaded lizard) venom. Biochim. Biophys. Acta Lipids Lipid Metab. 1994, 1211, 61–68. [Google Scholar] [CrossRef]
- Salemi, M.; Vandamme, A.-M. The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Aminetzach, Y.T.; Srouji, J.R.; Kong, C.Y.; Hoekstra, H.E. Convergent evolution of novel protein function in shrew and lizard venom. Curr. Biol. 2009, 19, 1925–1931. [Google Scholar] [CrossRef] [PubMed]
- Boutemy, L.S.; King, S.R.F.; Win, J.; Hughes, R.K.; Clarke, T.A.; Blumenschein, T.M.A.; Kamoun, S.; Banfield, M.J. Structures of Phytophthora RXLR Effector Proteins a conserved but adaptable fold underpins functional diversity. J. Biol. Chem. 2011, 286, 35834–35842. [Google Scholar] [CrossRef] [PubMed]
- Brodie, E.D. Convergent Evolution: Pick Your Poison Carefully. Curr. Biol. 2010, 20, R152–R154. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.T.J.; Cotton, J.A.; Kirwan, J.D.; Teeling, E.C.; Rossiter, S.J. Parallel signatures of sequence evolution among hearing genes in echolocating mammals: An emerging model of genetic convergence. Heredity 2012, 108, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Folinsbee, K.E. Evolution of venom across extant and extinct eulipotyphlans. C. R. Palevol 2013, 12, 531–542. [Google Scholar] [CrossRef]
- Garb, J.E.; Hayashi, C.Y. Molecular evolution of alpha-latrotoxin, the exceptionally potent vertebrate neurotoxin in black widow spider venom. Mol. Biol. Evol. 2013, 30, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Green, D.A.; Extavour, C.G. Convergent evolution of a reproductive trait through distinct developmental mechanisms in Drosophila. Dev. Biol. 2012, 372, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.H.; Skala, W.G.; Magdolen, V.; Briza, P.; Biniossek, M.L.; Schilling, O.; Kellermann, J.; Brandstetter, H.; Goettig, P. A Single Glycan at the 99-Loop of Human Kallikrein-related Peptidase 2 Regulates Activation and Enzymatic Activity. J. Biol. Chem. 2016, 291, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Harrington, M. Exploring the molecular underpinnings of convergent evolution. Lab. Anim. 2013, 42, 405–406. [Google Scholar] [CrossRef] [PubMed]
- Janes, D.E.; Organ, C.L.; Fujita, M.K.; Shedlock, A.M.; Edwards, S.V. Genome Evolution in Reptilia, the Sister Group of Mammals. In Annual Review of Genomics and Human Genetics; Chakravarti, A., Green, E., Eds.; 2010; Volume 11, pp. 239–264. Available online: http://www.annualreviews.org/doi/abs/10.1146/annurev-genom-082509-141646 (accessed on 6 August 2017).
- Lawrence, M.G.; Lai, J.; Clements, J.A. Kallikreins on Steroids: Structure, Function, and Hormonal Regulation of Prostate-Specific Antigen and the Extended Kallikrein Locus. Endocr. Rev. 2010, 31, 407–446. [Google Scholar] [CrossRef] [PubMed]
- Ligabue-Braun, R.; Verli, H.; Carlini, C.R. Venomous mammals: A review. Toxicon 2012, 59, 680–695. [Google Scholar] [CrossRef] [PubMed]
- Losos, J.B. Convergence, adaptation, and constraint. Evolution 2011, 65, 1827–1840. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Orgogozo, V. The loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution 2013, 67, 1235–1250. [Google Scholar] [CrossRef] [PubMed]
- Meyer, W.K.; Zhang, S.; Hayakawa, S.; Imai, H.; Przeworski, M. The convergent evolution of blue iris pigmentation in primates took distinct molecular paths. Am. J. Phys. Anthropol. 2013, 151, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Pavlopoulou, A.; Pampalakis, G.; Michalopoulos, I.; Sotiropoulou, G. Evolutionary History of Tissue Kallikreins. PLoS ONE 2010, 5, e13781. [Google Scholar] [CrossRef] [PubMed]
- Roelants, K.; Fry, B.G.; Norman, J.A.; Clynen, E.; Schoofs, L.; Bossuyt, F. Identical Skin Toxins by Convergent Molecular Adaptation in Frogs. Curr. Biol. 2010, 20, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Simmer, J.P.; Richardson, A.S.; Smith, C.E.; Hu, Y.Y.; Hu, J.C.C. Expression of kallikrein-related peptidase 4 in dental and non-dental tissues. Eur. J. Oral Sci. 2011, 119, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Song, B.X.; Wang, F.; Guo, Y.; Sang, Q.; Liu, M.; Li, D.Y.; Fang, W.; Zhang, D.L. Protein-protein interaction network-based detection of functionally similar proteins within species. Proteins-Struct. Funct. Bioinform. 2012, 80, 1736–1743. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.M.; Blanke, A.; Richter, S.; Alvarez, F.; Bleidorn, C.; Jenner, R.A. The first venomous crustacean revealed by transcriptomics and functional morphology: Remipede venom glands express a unique toxin cocktail dominated by enzymes and a neurotoxin. Mol. Biol. Evol. 2014, 31, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.S.W.; Papenfuss, A.T.; Whittington, C.M.; Warren, W.C.; Belov, K. A limited role for gene duplications in the evolution of platypus venom. Mol. Biol. Evol. 2012, 29, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Yennamalli, R.M.; Rader, A.J.; Wolt, J.D.; Sen, T.Z. Thermostability in endoglucanases is fold-specific. BMC Struct. Biol. 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.M.; Peigneur, S.; Gao, B.; Zhang, S.F.; Tytgat, J.; Zhu, S.Y. Target-driven positive selection at hot spots of scorpion toxins uncovers their potential in design of insecticides. Mol. Biol. Evol. 2016, 33, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.Y.; Gao, B.; Deng, M.C.; Yuan, Y.Z.; Luo, L.; Peigneur, S.; Xiao, Y.C.; Liang, S.P.; Tytgat, J. Drosotoxin, a selective inhibitor of tetrodotoxin-resistant sodium channels. Biochem. Pharmacol. 2010, 80, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.Y.; Peigneur, S.; Gao, B.; Lu, X.X.; Cao, C.Y.; Tytgat, J. Evolutionary diversification of mesobuthus alpha-scorpion toxins affecting sodium channels. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Schweitz, H.; Bidard, J.N.; Lazdunski, M. Purification and pharmacological characterization of peptide toxins from the black mamba (Dendroaspis polylepis) venom. Toxicon 1990, 28, 847–856. [Google Scholar] [CrossRef]
- Mertens, R. Die familie der warane (Varanidae). Abh. Senckenberg. Naturforschenden Ges. 1942, 465, 1–39. [Google Scholar]
- Rieppel, O. A functional interpretation of the varanid dentition (Reptilia, Lacertilia, Varanidae). Gegenbaurs Morphol. Jahrb. 1979, 125, 797–817. [Google Scholar] [PubMed]
- Abler, W.L. The serrated teeth of tyrannosaurid dinosaurs, and biting structures in other animals. Paleobiology 1992, 18, 161–183. [Google Scholar] [CrossRef]
- Rieppel, O.; Labhardt, L. Mandibular mechanics in Varanus niloticus (Reptilia: Lacertilia). Herpetologica 1979, 35, 158–163. [Google Scholar]
- Sweet, S.S. Comparative spatial ecology of two small arboreal monitors in northern Australia. In Advances in Monitor Research III; Horn, H.G., Boehme, W., Krebs, U., Eds.; Mertensiella 16: Rheinbach, Germany, 2007; pp. 378–402. [Google Scholar]
- Sunagar, K.; Moran, Y. The rise and fall of an evolutionary innovation: Contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 2015, 11, e1005596. [Google Scholar] [CrossRef] [PubMed]
- Pascoe, J.H.; Mulley, R.C.; Spencer, R.; Chapple, R. Diet analysis of mammals, raptors and reptiles in a complex predator assemblage in the Blue Mountains, eastern Australia. Aust. J. Zool. 2012, 59, 295–301. [Google Scholar] [CrossRef]
- Pianka, E.R.; King, D.; King, R.A. Varanoid Lizards of the World; Indiana University Press: Bloomington, IN, USA, 2004. [Google Scholar]
- Bull, J.J.; Jessop, T.S.; Whiteley, M. Deathly drool: Evolutionary and ecological basis of septic bacteria in Komodo dragon mouths. PLoS ONE 2010, 5, e11097. [Google Scholar] [CrossRef] [PubMed]
- Arbuckle, K. Ecological function of venom in Varanus, with a compilation of dietary records from the literature. Biawak 2009, 3, 46–56. [Google Scholar]
- Grant, T.R.; Temple-Smith, P.D. Field biology of the platypus (Ornithorhynchus anatinus): Historical and current perspectives. Philos. Trans. R. Soc. B Biol. Sci. 1998, 353, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Arbuckle, K. Evolutionary context of venom in animals. In Evolution of Venomous Animals and Their Toxins; Malhotra, A., Gopalakrishnakone, P., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 3–34. [Google Scholar]
- Harris, R.J.; Arbuckle, K. Tempo and mode of the evolution of venom and poison in tetrapods. Toxins 2016, 8, 193. [Google Scholar] [CrossRef] [PubMed]
- Marsh, N.; Williams, V. Practical applications of snake venom toxins in haemostasis. Toxicon 2005, 45, 1171–1181. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Baumann, K.; Jackson, T.N.; Wood, K.; Mason, S.; Undheim, E.A.; Nouwens, A.; Koludarov, I.; Hendrikx, I.; Jones, A.; et al. Proteomic comparison of Hypnale hypnale (Hump-Nosed Pit-Viper) and Calloselasma rhodostoma (Malayan Pit-Viper) venoms. J. Proteom. 2013, 91C, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Jackson, T.N.; Casewell, N.R.; Low, D.H.; Rossi, S.; Baumann, K.; Fathinia, B.; Visser, J.; Nouwens, A.; Hendrikx, I.; et al. Extreme venom variation in Middle Eastern vipers: A proteomics comparison of Eristicophis macmahonii, Pseudocerastes fieldi and Pseudocerastes persicus. J. Proteom. 2015, 116, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Low, D.H.; Sunagar, K.; Undheim, E.A.; Ali, S.A.; Alagon, A.C.; Ruder, T.; Jackson, T.N.; Pineda Gonzalez, S.; King, G.F.; Jones, A.; et al. Dracula’s children: Molecular evolution of vampire bat venom. J. Proteom. 2013, 89, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Weldon, C.L.; Mackessy, S.P. Biological and proteomic analysis of venom from the Puerto Rican Racer (Alsophis portoricensis: Dipsadidae). Toxicon 2010, 55, 558–569. [Google Scholar] [CrossRef] [PubMed]
- R-Core-Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 4 June 2017).
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Symonds, M.R.E.; Blomberg, S.P. A Primer on Phylogenetic Generalised Least Squares. In Modern Phylogenetic Comparative Methods and Their Application in Evolutionary Biology; Garamszegi, L.Z., Ed.; Springer-Verlag: Berlin, Germany, 2014; pp. 105–130. [Google Scholar]
- Orme, D.; Freckleton, R.; Thomas, G.; Petzoldt, T.; Fritz, S.; Isaac, N.; Pearse, W. The Caper Package: Comparative Analyses of Phylogenetics and Evolution in R, Version 0.5.2. 2013. Available online: https://cran.r-project.org/web/packages/caper/vignettes/caper.pdf (accessed on 6 August 2017).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2014, 42, D32–D37. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Grishin, N.V. AL2CO: Calculation of positional conservation in a protein sequence alignment. Bioinformatics 2001, 17, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky Pond, S.L.; Scheffler, K. FUBAR: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Frost, S.D.W.; Muse, S.V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef] [PubMed]


| Toxin Type | Species Recovered from | Activity | References |
|---|---|---|---|
| AVIT | V. komodoensis, V. varius | While lizard forms have yet to be tested, snake venom homologues have been shown to have potent ability to contract smooth muscle and also induce hyperalgesia | [23,33,68] |
| BNP (B-type natriuretic peptide) | C. warreni, G. infernalis, H. horridum, H. suspectum, V. glauerti, V. komodoensis, V. scalaris, V. varius | Hypotension mediated by relaxation of aortic smooth muscle | [23,27,32,33,62,69] |
| Cholecystoxin | V. varius | Binding to cholecystokinin receptor to stimulate smooth muscle contraction | [27] |
| CRiSP (cysteine rich secretory protein) | C. warreni, G. infernalis, H. horridum, H. suspectum, O. apodus, S. crocodilurus, V. acanthurus, V. albigularis, V. eremius, V. giganteus, V. gilleni, V. glauerti, V. gouldii, V. indicus, V. komodoensis, V. mertensi, V. mitchelli, V. scalaris, V. tristis, V. varius | Blockage of ryanodine receptors and potassium channels, producing lethargy, paralysis, and hypothermia | [23,32,33,69,70,71,72,73,74] |
| Celestoxin | C. warreni | Hypotension inducing | [27] |
| Exendin | H. exasperatu, H. horridum, H. suspectum | Hypotension mediated by relaxation of aortic smooth muscle | [27,75,76,77] |
| Goannatyrotoxin | V. eremius | Hypertensive/hypotensive triphasic effect | [27] |
| Helofensin | H. exasperatum, H. horridum, H. suspectum | Blockage of nerve impulse | [27,78] |
| Helokinestatin | C. warreni, G. infernalis, H. horridum, H. suspectum | Bradykinin inhibition | [23,27,32,33,60,62] |
| Kallikrein | C. warreni, G. infernalis, H. horridum, H. suspectum, O. apodus, V. gilleni, V. glauerti, V. scalaris, V. tristis, V. varius | Kinin release from kininogen, cleavage of fibrinogen | [23,32,33,45,79,80,81,82,83,84,85] |
| Type III phospholipase A2 (PLA2) | C. warren, H. horridum, H. suspectum, O. apodus, S. crododilurus, V. acanthurus, V. albigularis, V. eremius, V. giganteus, V. gilleni, V. glauerti, V. gouldii, V. indicus, V. komodoensis, V. mertensi, V. mitchelli, V. scalaris, V. tristis, V. varius | Inhibition of epinephrine-induced platelet aggregation | [23,32,86] |
| Species | AVIT | Chitinase | CRiSP | ESP | Kallikrein | Lysosomal Acid Lipase | Natriuretic Peptide | PLA2 |
|---|---|---|---|---|---|---|---|---|
| V. acanthurus | + | + | + | + | ||||
| V. baritji | + | + | + | + | ||||
| V. giganteus | + | + | + | + | + | + | ||
| V. gilleni | + | + | + | + | + | + | + | |
| V. griseus | + | + | + | + | + | |||
| V. jobiensis | + | + | + | + | + | + | ||
| V. komodoensis | + | + | + | + | + | + | ||
| V. melinus | + | + | + | + | + | |||
| V. mertensi | + | + | + | + | + | + | ||
| V. mitchelli | + | + | + | + | + | |||
| V. panoptes rubidus | + | + | + | |||||
| V. prasinus | + | + | + | + | ||||
| V. salvadorii | + | + | + | + | + | + | ||
| V. scalaris | + | + | + | + | + | |||
| V. tristis | + | + | + | + | + | + | ||
| V. varius | + | + | + | + | + | + | + |
| Species | Substrate RDSE011 | Substrate S-2302 | Fibrinogen (Alpha-Chain) | Fibrinogen (Beta-Chain) | PLA2 |
|---|---|---|---|---|---|
| H. exasperatum | 0.300 ± 0.019 | 0.04 ± 0.003 | 0.435 ± 0.013 | 0.056 ± 0.002 | 0.200 ± 0.023 |
| H. horridum | 0.420 ± 0.027 | 0.408 ± 0.037 | 0.396 ± 0.015 | 0.159 ± 0.01 | 0.100 ± 0.009 |
| H. suspectum | 0.400 ± 0.015 | 0.103 ± 0.012 | 0.416 ± 0.015 | 0.103 ± 0.006 | 0.150 ± 0.017 |
| L. borneensis | 0.008 ± 0.001 | 0.102 ± 0.007 | 0.652 ± 0.018 | 0.458 ± 0.012 | 0.010 ± 0.007 |
| V. acanthurus | 0.059 ± 0.003 | 0.467 ± 0.022 | 0.818 ± 0.099 | 0.587 ± 0.066 | 0.050 ± 0.003 |
| V. baritji | 0.001 ± 0.001 | 0.874 ± 0.043 | 0.773 ± 0.047 | 0.308 ± 0.006 | 0.001 ± 0.001 |
| V. giganteus | 0.002 ± 0.001 | 0.351 ± 0.043 | 0.746 ± 0.034 | 0.538 ± 0.011 | 0.014 ± 0.009 |
| V. gilleni | 0.010 ± 0.003 | 0.400 ± 0.015 | 0.118 ± 0.002 | 0.001 ± 0.001 | 0.024 ± 0.003 |
| V. griseus | 0.006 ± 0.002 | 0.040 ± 0.009 | 0.247 ± 0.023 | 0.166 ± 0.003 | 0.015 ± 0.003 |
| V. jobiensis | 0.198 ± 0.013 | 0.317 ± 0.017 | 0.807 ± 0.026 | 0.882 ± 0.032 | 0.019 ± 0.002 |
| V. komodoensis | 0.080 ± 0.009 | 0.467 ± 0.012 | 0.339 ± 0.035 | 0.050 ± 0.001 | 0.011 ± 0.005 |
| V. melinus | 0.276 ± 0.015 | 0.317 ± 0.025 | 0.600 ± 0.016 | 0.503 ± 0.013 | 0.288 ± 0.019 |
| V. mertensi | 0.094 ± 0.004 | 0.300 ± 0.019 | 0.571 ± 0.018 | 0.546 ± 0.009 | 0.012 ± 0.005 |
| V. mitchelli | 1.000 ± 0.057 | 1.000 ± 0.097 | 0.962 ± 0.016 | 0.938 ± 0.053 | 0.008 ± 0.003 |
| V. panoptes rubidus | 0.208 ± 0.016 | 0.351 ± 0.011 | 0.230 ± 0.021 | 0.071 ± 0.003 | 0.019 ± 0.004 |
| V. prasinus | 0.407 ± 0.036 | 0.874 ± 0.057 | 0.700 ± 0.058 | 0.607 ± 0.027 | 0.028 ± 0.002 |
| V. salvadorii | 0.208 ± 0.012 | 0.103 ± 0.012 | 0.336 ± 0.015 | 0.225 ± 0.006 | 0.046 ± 0.007 |
| V. scalaris | 0.378 ± 0.019 | 1.000 ± 0.089 | 1.000 ± 0.037 | 1.000 ± 0.057 | 0.119 ± 0.011 |
| V. tristis | 0.161 ± 0.015 | 0.040 ± 0.003 | 0.793 ± 0.021 | 0.361 ± 0.009 | 0.017 ± 0.005 |
| V. varius | 0.072 ± 0.009 | 0.226 ± 0.013 | 0.377 ± 0.023 | 0.286 ± 0.011 | 1.000 ± 0.022 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koludarov, I.; Jackson, T.N.; Brouw, B.o.d.; Dobson, J.; Dashevsky, D.; Arbuckle, K.; Clemente, C.J.; Stockdale, E.J.; Cochran, C.; Debono, J.; et al. Enter the Dragon: The Dynamic and Multifunctional Evolution of Anguimorpha Lizard Venoms. Toxins 2017, 9, 242. https://doi.org/10.3390/toxins9080242
Koludarov I, Jackson TN, Brouw Bod, Dobson J, Dashevsky D, Arbuckle K, Clemente CJ, Stockdale EJ, Cochran C, Debono J, et al. Enter the Dragon: The Dynamic and Multifunctional Evolution of Anguimorpha Lizard Venoms. Toxins. 2017; 9(8):242. https://doi.org/10.3390/toxins9080242
Chicago/Turabian StyleKoludarov, Ivan, Timothy NW Jackson, Bianca op den Brouw, James Dobson, Daniel Dashevsky, Kevin Arbuckle, Christofer J. Clemente, Edward J. Stockdale, Chip Cochran, Jordan Debono, and et al. 2017. "Enter the Dragon: The Dynamic and Multifunctional Evolution of Anguimorpha Lizard Venoms" Toxins 9, no. 8: 242. https://doi.org/10.3390/toxins9080242
APA StyleKoludarov, I., Jackson, T. N., Brouw, B. o. d., Dobson, J., Dashevsky, D., Arbuckle, K., Clemente, C. J., Stockdale, E. J., Cochran, C., Debono, J., Stephens, C., Panagides, N., Li, B., Manchadi, M.-L. R., Violette, A., Fourmy, R., Hendrikx, I., Nouwens, A., Clements, J., ... Fry, B. G. (2017). Enter the Dragon: The Dynamic and Multifunctional Evolution of Anguimorpha Lizard Venoms. Toxins, 9(8), 242. https://doi.org/10.3390/toxins9080242






_Kwok.png)


