Analysis of MicroRNA Expression Profiling Involved in MC-LR-Induced Cytotoxicity by High-Throughput Sequencing
Abstract
:1. Introduction
2. Result
2.1. Sequencing Summary of Small RNAs
2.2. miRNA Expression Profile in MC-LR Treated HepG2 Cells
2.3. Differentially Expressed miRNAs Validated by qPCR
2.4. Putative Target Genes of the Differentially Expressed miRNAs and the Results of GO and KEGG Pathway Analysis
2.5. Novel miRNAs Predicted
3. Discussion
4. Materials and Methods
4.1. MC-LR
4.2. Cell Culture and MC-LR Exposure
4.3. RNA Extraction
4.4. High-Throughput Sequencing
4.5. Bioinformatic Analysis of Small RNA Sequences
4.6. Analysis of Differentially Expressed miRNAs
4.7. Target Prediction of Differentially Expressed miRNAs and GO and KEGG Pathway Analysis
4.8. qPCR Verification of Differentially Expressed miRNAs
4.9. qPCR Verification the Targets miRNAs
4.10. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sivonen, K.; Jones, G. Cyanobacterial toxins. In Toxic Cyanobacteria in Water: A Guide to Their Public Health Consequences, Monitoring, and Management; Chorus, I., Bartram, J., Eds.; E&FN Spon: London, UK, 1999; pp. 41–111. [Google Scholar]
- Ibelings, B.W.; Bruning, K.; De Jonge, J.; Wolfstein, K.; Pires, L.D.; Postma, J.; Burger, T. Distribution of microcystins in a lake foodweb: No evidence for biomagnification. Microb. Ecol. 2005, 49, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Bláhová, L.; Babica, P.; Adamovský, O.; Kohoutek, J.; Maršálek, B.; Bláha, L. Analyses of cyanobacterial toxins (microcystins, cylindrospermopsin) in the reservoirs of the Czech Republic and evaluation of health risks. Environ. Chem. Lett. 2008, 6, 223–227. [Google Scholar] [CrossRef]
- Codd, G.A.; Pliński, M.; Surosz, W.; Hutson, J.; Fallowfield, H.J. Publication in 1672 of animal deaths at the Tuchomskie Lake, northern Poland and a likely role of cyanobacterial blooms. Toxicon 2015, 108, 285–286. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, J.; Zhang, X.; Xie, P. A review of reproductive toxicity of microcystins. J. Hazard. Mater. 2016, 301, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, S.M.F.O.; Carmichael, W.W.; Jochimsen, E.M.; Rinehart, K.L.; Lau, S.; Shaw, G.R.; Eaglessham, G.K. Human intoxication by microcystins during renal dialysis treatment in Caruaru-Brazil. Toxicology 2002, 181, 441–446. [Google Scholar] [CrossRef]
- Ueno, Y.; Nagata, S.; Tsuysumi, T.; Hasegawa, A.; Watanabe, M.F.; Park, H.D.; Chen, G.C.; Chen, G.; Yu, S.Z. Detection of microcystins, a blue-green algal hepatotoxin, in drinking water sampled in Haimen and Fusui, endemic areas of primary liver cancer in China, by highly sensitive immunoassay. Carcinogenesis 1996, 17, 1317–1321. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.G.; Kensler, T.W. Changing rates for liver and lung cancers in Qidong, China. Chem. Res. Toxicol. 2013, 27, 3–6. [Google Scholar] [CrossRef] [PubMed]
- MacKintosh, C.; Beattie, K.A.; Lumpp, S.; Cohen, P.; Codd, G.A. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Lett. 1990, 264, 187–192. [Google Scholar] [CrossRef]
- Falconer, I.R.; Humpage, A.R. Health risk assessment of cyanobacterial (blue-green algal) toxins in drinking water. Int. J. Environ. Res. Public Health 2005, 2, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Y.; Song, L.; Liu, J. Responses of antioxidant systems in the hepatocytes of common carp (Cyprinus carpio L.) to the toxicity of microcystin-LR. Toxicon 2003, 42, 85–89. [Google Scholar] [CrossRef]
- Ma, J.; Feng, Y.; Liu, Y.; Li, X. PUMA and survivin are involved in the apoptosis of HepG2 cells induced by microcystin-LR via mitochondria-mediated pathway. Chemosphere 2016, 157, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Bouaı̈, N.; Maatouk, I. Microcystin-LR and nodularin induce intracellular glutathione alteration, reactive oxygen species production and lipid peroxidation in primary cultured rat hepatocytes. Toxicol. Lett. 2004, 148, 53–63. [Google Scholar]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Christodoulou, F.; Raible, F.; Tomer, R.; Simakov, O.; Trachana, K.; Klaus, S.; Snyman, H.; Hannon, G.I.; Bork, P.; Arendt, D. Ancient animal microRNAs and the evolution of tissue identity. Nature 2010, 463, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.; Sharp, P. Roles for microRNAs in conferring robustness to biological processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, E.; Erozenci, A.; Smit, J.; Danesi, R.; Peters, G.J. Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Crit. Rev. Oncol. Hematol. 2012, 81, 103–122. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, T.; Nakajima, M. Toxicological implications of modulation of gene expression by microRNAs. Toxicol. Sci. 2011, 123, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Koufaris, C.; Gooderham, N.J. Are differences in microrna regulation implicated in species-dependent response to toxicological exposures? Toxicol. Sci. 2013, 131, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Lema, C.; Cunningham, M.J. MicroRNAs and their implications in toxicological research. Toxicol. Lett. 2010, 198, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Qin, W.; Zhang, H.; Wang, Y.; Dou, H.; Yu, D.; Ding, Y.; Yang, L.; Wang, Y. Alterations in microRNA expression linked to microcystin-LR-induced tumorigenicity in human WRL-68 cells. Mutat. Res. 2012, 743, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xie, P.; Fan, H. Genomic profiling of microRNAs and proteomics reveals an early molecular alteration associated with tumorigenesis induced by MC-LR in mice. Environ. Sci. Technol. 2012, 46, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Brzuzan, P.; Wozny, M.; Wolinska, L.; Piasecka, A.; Luczynski, M.K. MicroRNA expression in liver of whitefish (Coregonus lavaretus) exposed to microcystin-LR. Environ. Biotechnol. 2010, 6, 53–60. [Google Scholar]
- Brzuzan, P.; Woźny, M.; Wolińska, L.; Piasecka, A. Expression profiling in vivo demonstrates rapid changes in liver microRNA levels of whitefish (Coregonus lavaretus) following microcystin-LR exposure. Aquat. Toxicol. 2012, 122–123, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Florczyk, M.; Brzuzan, P.; Krom, J.; Woźny, M.; Łakomiak, A. miR-122-5p as a plasma biomarker of liver injury in fish exposed to microcystin-LR. J. Fish Dis. 2016, 39, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Hoeger, S.J.; Hitzfeld, B.C.; Dietrich, D.R. Occurrence and elimination of cyanobacterial toxins in drinking water treatment plants. Toxicol. Appl. Pharmacol. 2005, 203, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Brzuzan, P.; Florczyk, M.; Łakomiak, A.; Woźny, M. Illumina sequencing reveals aberrant expression of microRNAs and their variants in whitefish (Coregonus lavaretus) liver after exposure to microcystin-LR. PLoS ONE 2016, 11, e0158899. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xiang, Z.; Li, D.; Han, X. Regulation of microcystin-LR-induced toxicity in mouse spermatogonia by miR-96. Environ. Sci. Technol. 2014, 48, 6383–6390. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, H.; Wang, C.; Qiu, X.; Benson, M.; Yin, X.; Xiang, Z.; Li, D.; Han, X. Roles of miRNAs in microcystin-LR-induced Sertoli cell toxicity. Toxicol. Appl. Pharm. 2015, 287, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, L.; Chen, X.; Xiang, Z.; Li, D.; Han, X. miR-541 contributes to Microcystin-LR-induced reproductive toxicity through regulating the expression of p15 in mice. Toxins 2016, 8, 260. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, Y.; Wang, J.; Wang, L.; Xiang, Z.; Li, D.; Han, X. Microcystin-Leucine Arginine causes cytotoxic effects in sertoli cells resulting in reproductive dysfunction in male mice. Sci. Rep. 2016, 6, 39238. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, X. MicroRNAs are involved in the toxicity of microcystins. Toxin Rev. 2016, 1–11. [Google Scholar] [CrossRef]
- Wang, Z. MicroRNA: A matter of life or death. World J. Biol. Chem. 2010, 1, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Elamin, B.K.; Callegari, E.; Gramantieri, L.; Sabbioni, S.; Negrini, M. MicroRNA response to environmental mutagens in liver. Mutat. Res. 2011, 717, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Wang, D.; Baccarelli, A. Environmental chemicals and microRNAs. Mutat. Res. 2011, 714, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, W.W.; Azevedo, S.M.; An, J.S.; Molica, R.J.; Jochimsen, E.M.; Lau, S.; Rinehart, K.L.; Shaw, G.R.; Eaglesham, G.K. Human fatalities from cyanobacteria: Chemical and biological evidence for cyanotoxins. Environ. Health Perspect. 2001, 109, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xiong, Q.; Xie, P. Analysis of microRNA expression in embryonic developmental toxicity induced by MC-RR. PLoS ONE 2011, 6, e22676. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, J.; Fang, Q.; Li, Y. Transcription alterations of microRNAs, cytochrome P4501A1 and 3A65, and AhR and PXR in the liver of zebrafish exposed to crude microcystins. Toxicon 2013, 73, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Toivola, D.M.; Eriksson, J.E. Toxins affecting cell signalling and alteration of cytoskeletal structure. Toxicol. In Vitro 1999, 13, 521–530. [Google Scholar] [CrossRef]
- Komatsu, M.; Furukawa, T.; Ikeda, R.; Takumi, S.; Nong, Q.; Aoyama, K.; Akiyama, S.; Keppler, D.; Takeuchi, T. Involvement of mitogen-activated protein kinase signaling pathways in microcystin-LR-induced apoptosis after its selective uptake mediated by OATP1B1 and OATP1B3. Toxicol. Sci. 2007, 97, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Takumi, S.; Komatsu, M.; Furukawa, T.; Ikeda, R.; Sumizawa, T.; Akenaga, H.; Maeda, Y.; Aoyama, K.; Arizono, K.; Ando, S.; et al. p53 Plays an important role in cell fate determination after exposure to microcystin-LR. Environ. Health Perspect. 2010, 118, 1292–1298. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Meng, G.M.; Guo, Z.L.; Xu, L.H. Regulation of heat shock protein 27 phosphorylation during microcystin-LR-induced cytoskeletal reorganization in a human liver cell line. Toxicol. Lett. 2011, 207, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, J.; Li, Y. Molecular cloning and expression determination of p38 MAPK from the liver and kidney of silver carp. J. Biochem. Mol. Toxicol. 2016, 30, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Song, L.R.; Chen, W.; Peng, L.; Wan, N.; Gan, N.Q.; Zhang, X.M. Distribution and bioaccumulation of microcystins in water columns: A systematic investigation into the environmental fate and the risks associated with microcystins in Meiliang Bay, Lake Taihu. Water Res. 2007, 41, 2853–2864. [Google Scholar] [CrossRef] [PubMed]
- Žegura, B.; Lah, T.T.; Filipič, M. Alteration of intracellular GSH levels and its role in microcystin-LR-induced DNA damage in human hepatoma HepG2 cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2006, 611, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Nong, Q.; Komatsu, M.; Izumo, K.; Indo, H.P.; Xu, B.; Aoyama, K.; Majima, H.J.; Horiuchi, M.; Morimoto, K.; Takeuchi, T. Involvement of reactive oxygen species in Microcystin-LR-induced cytogenotoxicity. Free Radic. Res. 2007, 41, 1326–1337. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, H.; Wang, B.; Chen, T.; Wang, X.; Huang, P.; Xu, L.; Guo, Z. Microcystin-LR promotes proliferation by activating Akt/S6K1 pathway and disordering apoptosis and cell cycle associated proteins phosphorylation in HL7702 cells. Toxicol. Lett. 2016, 240, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Hagenbuch, B.; Gui, C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica 2008, 38, 778–801. [Google Scholar] [CrossRef] [PubMed]
- Kalliokoski, A.; Niemi, M. Impact of OATP transporters on pharmacokinetics. Br. J. Pharmacol. 2009, 158, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Kullak-Ublick, G.A.; Beuers, U.; Paumgartner, G. Molecular and functional characterization of bile acid transport in human hepatoblastoma HepG2 cells. Hepatology 1996, 23, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Costantini, S.; Bernardo, G.D.; Cammarota, M.; Castello, G.; Colonna, G. Gene expression signature of human HepG2 cell line. Gene 2013, 518, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Kappus, H. Lipid peroxidation: Mechanisms, analysis, enzymology and biological relevance. In Oxidative Stress; Oxidants and Antioxidants; Sies, H., Ed.; Academic Press: London, UK, 1985; pp. 273–310. [Google Scholar]
- Ma, J.; Liu, Y.; Niu, D.; Li, X. Effects of chlorpyrifos on the transcription of CYP3A cDNA, activity of acetylcholinesterase, and oxidative stress response of goldfish (Carassius auratus). Environ. Toxicol. 2015, 30, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Mersch-Sundermann, V.; Knasmuller, S.; Wu, X.J.; Darroudi, F.; Kassie, F. Use of a human-derived liver cell line for the detection of cytoprotective, antigenotoxic and cogenotoxic agents. Toxicology 2004, 198, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Łakomiak, A.; Brzuzan, P.; Jakimiuk, E.; Florczyk, M.; Woźny, M. miR-34a and bcl-2 expression in whitefish (Coregonus lavaretus) after microcystin-LR exposure. Comp. Biochem. Phys. B 2016, 193, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russel, D.V. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory: New York, NY, USA, 2001; Volume 1, pp. 13–17. [Google Scholar]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ju, Z.; Li, Q.; Hou, Q.; Wang, C.; Li, J.; Li, R.; Wang, L.; Sun, T.; Hang, S.; et al. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle. Int. J. Biol. Sci. 2011, 7, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO: TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and 2 (−Delta Delta Ct) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
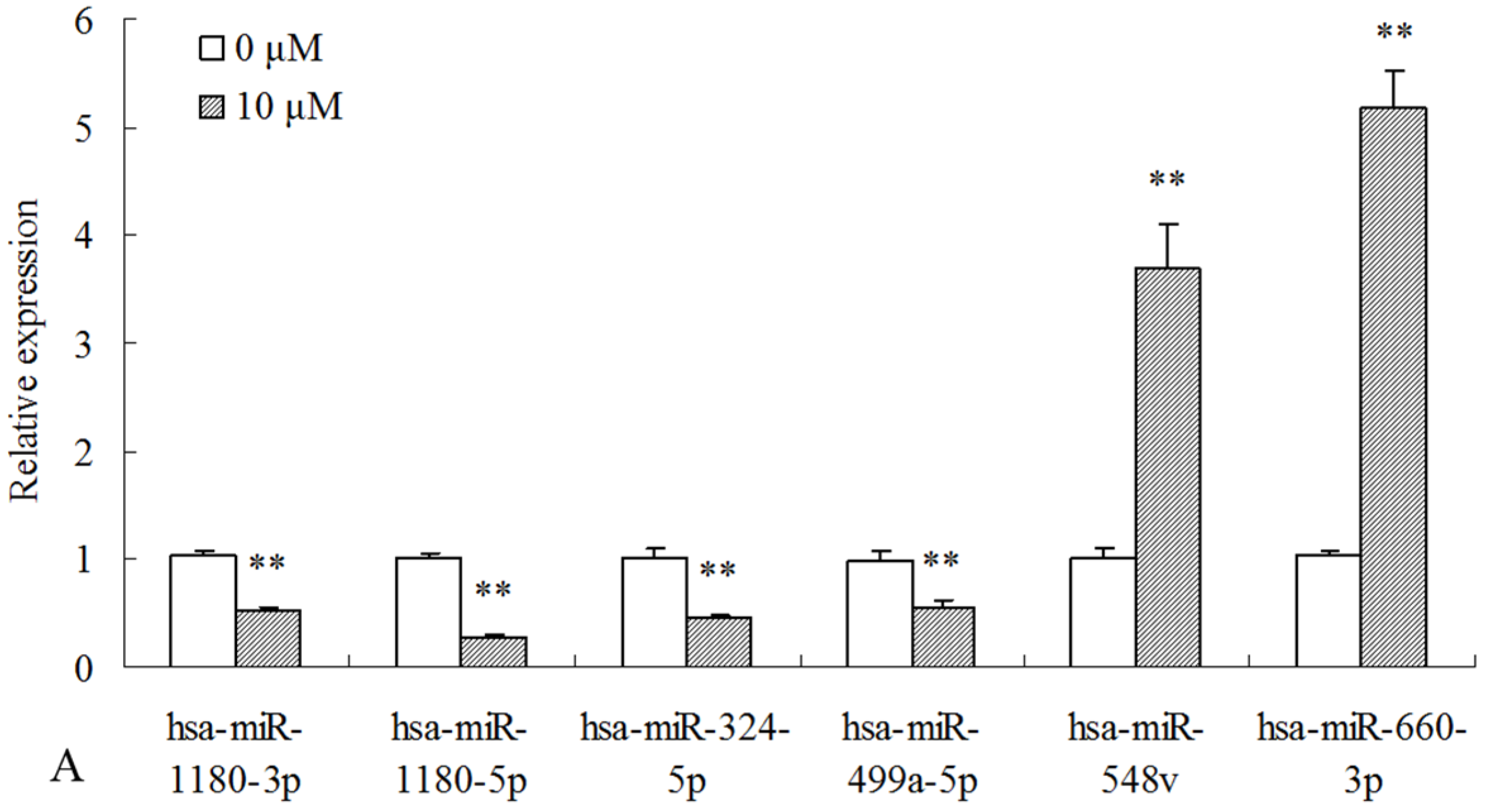
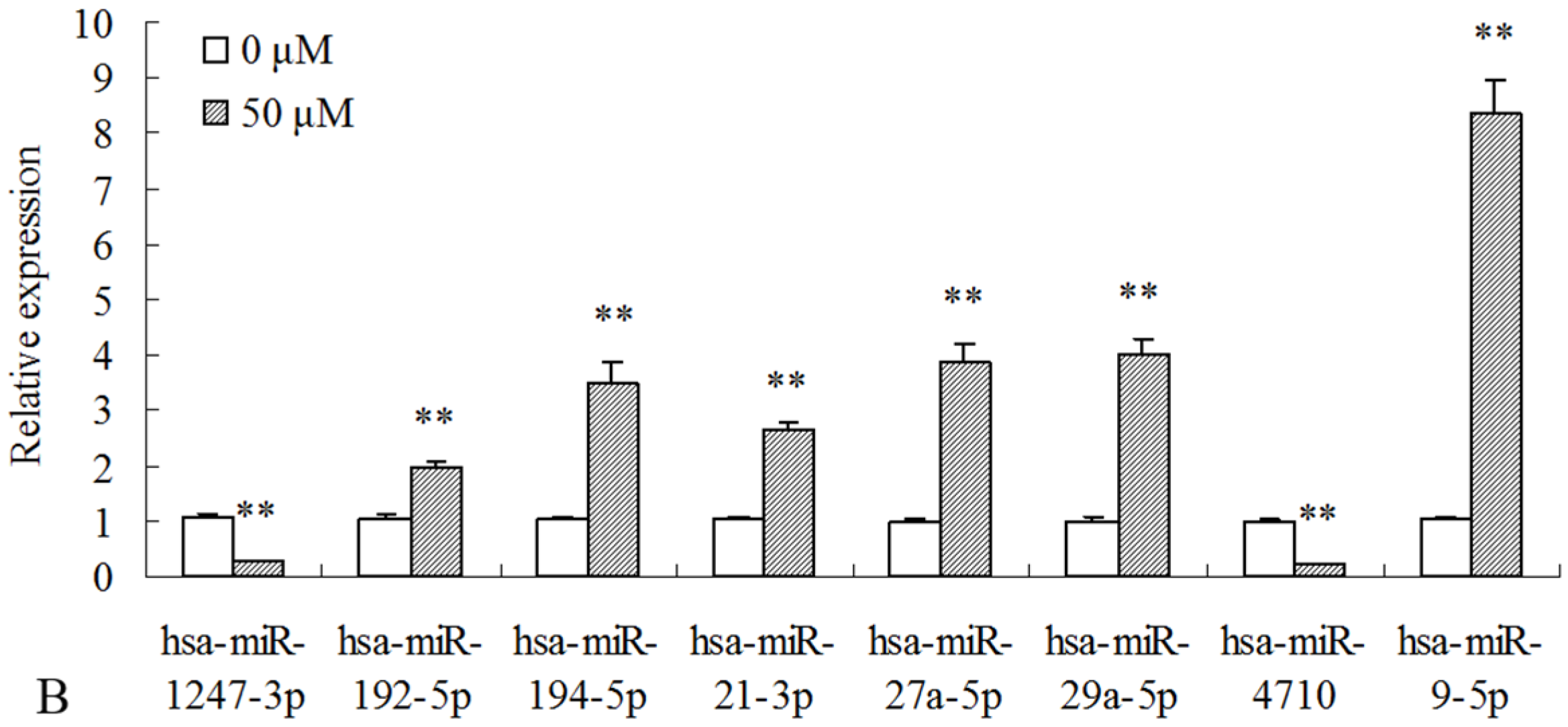
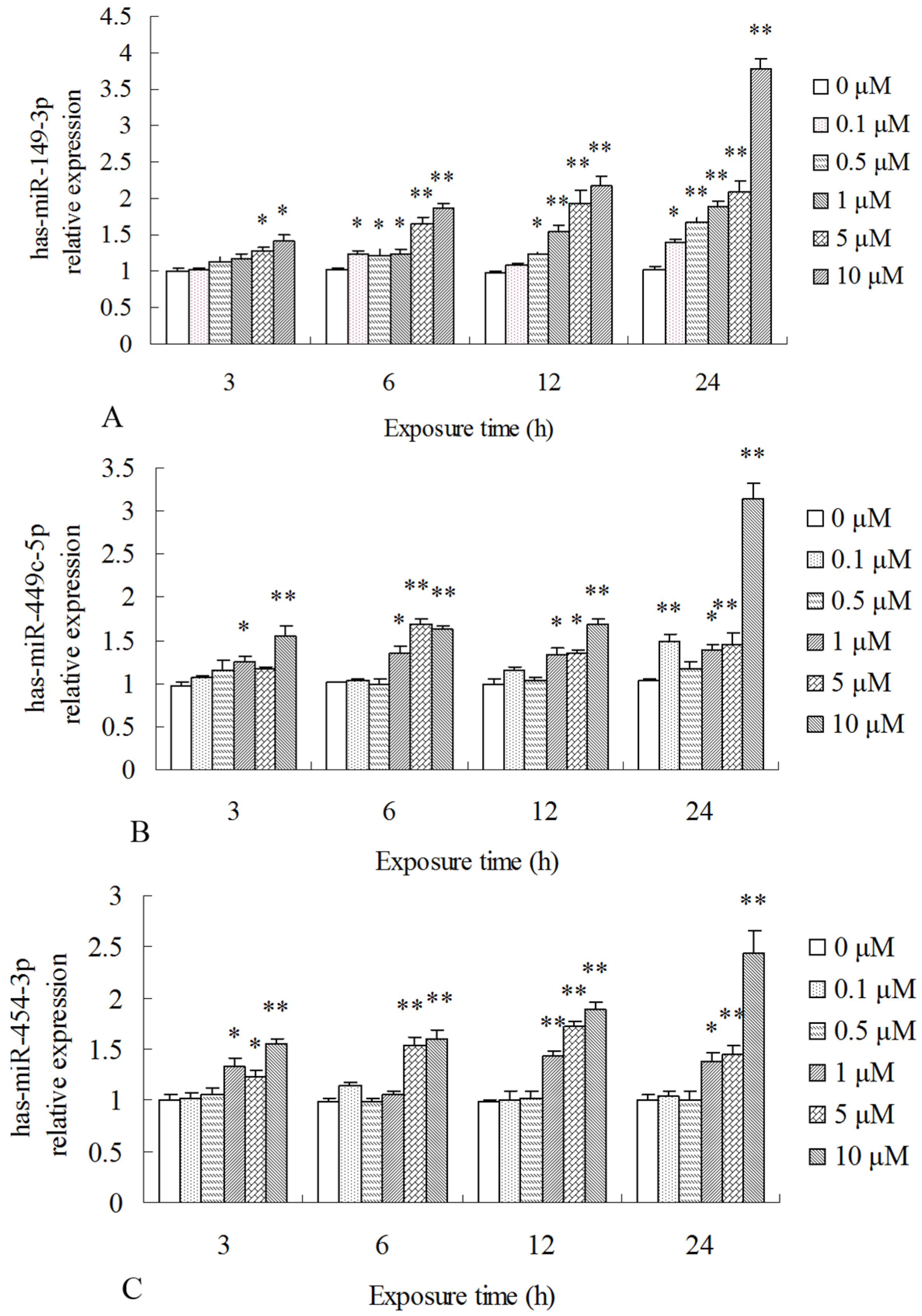
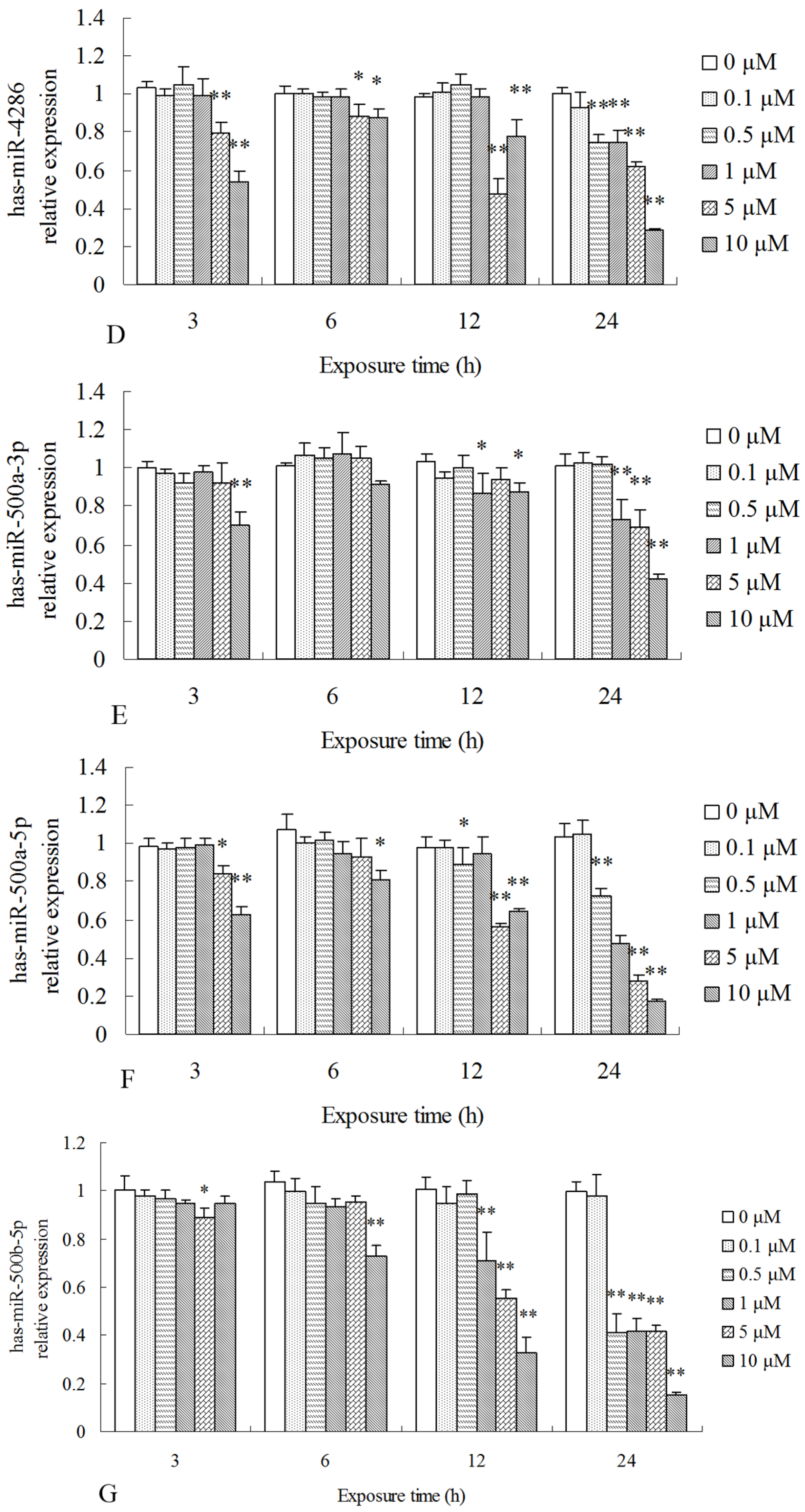
| miR_name | 0-std | 10-std | 50-std | 10 μM-vs-0 Fold-Change | 50 μM-vs-0 Fold-Change | p-Value |
|---|---|---|---|---|---|---|
| has-miR-1180-3p | 5.6930 | 2.8285 | - | −1.0092 | - | ** |
| hsa-miR-1180-5p | 3.7954 | 0.9428 | - | −2.009 | - | ** |
| hsa-miR-149-3p | 5.0605 | 10.8949 | 23.5600 | 1.1063 | 2.2189 | ** |
| hsa-miR-217 | 2.0031 | 0.8381 | - | −1.2570 | - | * |
| hsa-miR-3064-5p | 1.3705 | 0.3143 | - | −2.1245 | - | * |
| hsa-miR-324-5p | 11.3861 | 5.2379 | - | −1.1202 | - | ** |
| hsa-miR-4286 | 3.9008 | 1.4666 | 1.6205 | −1.4113 | −1.2673 | ** |
| hsa-miR-4421 | 1.7923 | 0.6285 | - | −1.5118 | - | * |
| hsa-miR-4454 | 1.3705 | 0.3143 | - | −2.1245 | - | * |
| hsa-miR-4485-3p | 1.3705 | 0.4190 | - | −1.7097 | - | * |
| hsa-miR-449c-5p | 3.3737 | 6.9140 | 7.6040 | 1.0352 | 1.1724 | ** |
| hsa-miR-454-3p | 1.0543 | 2.3047 | 2.4931 | 1.1283 | 1.2417 | * |
| hsa-miR-455-3p | 3.6899 | 1.7809 | - | −1.0509 | - | * |
| hsa-miR-499a-5p | 4.6388 | 2.3047 | - | −1.0092 | - | ** |
| hsa-miR-500a-3p | 7.2744 | 3.5618 | 3.4904 | −1.0302 | −1.0594 | ** |
| hsa-miR-500a-5p | 3.7954 | 0.6285 | 0.7479 | −2.5943 | −2.3433 | ** |
| hsa-miR-500b-5p | 3.7954 | 0.6285 | 0.7479 | −2.5943 | −2.3433 | ** |
| hsa-miR-548v | 0.5271 | 1.4666 | - | 1.4763 | - | * |
| hsa-miR-590-5p | 2.5302 | 1.2571 | - | −1.0092 | - | * |
| hsa-miR-660-3p | 0.2109 | 1.0476 | - | 2.3125 | - | * |
| hsa-miR-6858-5p | 1.0543 | 0.01 | - | −6.7201 | - | ** |
| hsa-let-7b-3p | 2.3194 | - | 0.9973 | - | −1.2177 | * |
| hsa-miR-106a-5p | 0.2109 | - | 1.4959 | - | 2.8264 | ** |
| hsa-miR-1247-3p | 2.6357 | - | 0.7479 | - | −1.8173 | ** |
| hsa-miR-1247-5p | 5.5876 | - | 2.6178 | - | −1.0939 | ** |
| hsa-miR-1256 | 1.7923 | - | 0.4986 | - | −1.8459 | * |
| hsa-miR-1276 | 0.3163 | - | 1.1219 | - | 1.8266 | * |
| hsa-miR-184 | 0.5271 | - | 4.2383 | - | 3.0073 | ** |
| hsa-miR-192-5p | 244.9062 | - | 502.3647 | - | 1.0365 | ** |
| hsa-miR-194-5p | 2.9519 | - | 9.9725 | - | 1.7563 | ** |
| hsa-miR-2116-3p | 1.3705 | - | 0.3740 | - | −1.8736 | * |
| hsa-miR-21-3p | 14.6543 | - | 30.1668 | - | 1.0416 | ** |
| hsa-miR-27a-5p | 2.5302 | - | 7.6040 | - | 1.5875 | ** |
| hsa-miR-29a-5p | 0.5271 | - | 1.4959 | - | 1.5049 | * |
| hsa-miR-3127-5p | 1.3705 | - | 4.2383 | - | 1.6288 | ** |
| hsa-miR-3162-5p | 1.2651 | - | 2.7424 | - | 1.1162 | * |
| hsa-miR-3614-5p | 0.2109 | - | 1.1219 | - | 2.4113 | * |
| hsa-miR-3615 | 0.9488 | - | 2.1192 | - | 1.1593 | * |
| hsa-miR-362-3p | 2.9519 | - | 1.3712 | - | −1.1062 | * |
| hsa-miR-3916 | 1.5814 | - | 0.3740 | - | −2.0801 | * |
| hsa-miR-4440 | 1.0543 | - | 0.01 | - | −6.7201 | ** |
| hsa-miR-4470 | 1.0543 | - | 0.2493 | - | −2.0803 | * |
| hsa-miR-4710 | 1.8977 | - | 0.4986 | - | −1.9283 | ** |
| hsa-miR-4739 | 4.1116 | - | 1.8698 | - | −1.1368 | ** |
| hsa-miR-4741 | 2.0031 | - | 4.7369 | - | 1.2417 | ** |
| hsa-miR-6515-5p | 0.3163 | - | 1.2466 | - | 1.9786 | * |
| hsa-miR-6758-5p | 1.0543 | - | 0.2493 | - | −2.0803 | * |
| hsa-miR-6805-5p | 1.8977 | - | 4.2383 | - | 1.1592 | ** |
| hsa-miR-6821-5p | 0.2109 | - | 1.1219 | - | 2.4113 | * |
| hsa-miR-766-3p | 2.5302 | - | 0.6233 | - | −2.0213 | ** |
| hsa-miR-9-5p | 0.01 | - | 1.6205 | - | 7.3403 | ** |
| miRNAs | Accession Number | Primers [5′→3′] |
|---|---|---|
| hsa-miR-1180-3p | MIMAT0005825 | GCTCGCGTGGGTGTGTAAAAA |
| hsa-miR-1180-5p | MIMAT0026735 | CTGCTGGACCCACCCGAAAA |
| hsa-miR-1247-3p | MIMAT0022721 | ACCCCGGGAACGTCGAGAAA |
| hsa-miR-149-3p | MIMAT0004609 | AGGAGGGAGGGAGGGACAAA |
| hsa-miR-192-5p | MIMAT0000222 | GGGCTCTGACCTATGAATTGAA |
| hsa-miR-194-5p | MIMAT0000460 | CGCGTAACAGCAACTCCAAAAA |
| hsa-miR-21-3p | MIMAT0004494 | GGCAACACCAGTCGATGAAAAA |
| hsa-miR-27a-5p | MIMAT0004501 | CAGGGCTTAGCTGCTTGTGAA |
| hsa-miR-29a-5p | MIMAT0004503 | GACTGATTTCTTTTGGTGTTCAG |
| hsa-miR-324-5p | MIMAT0000761 | CCCCTAGGGCATTGGTGTAAA |
| hsa-miR-4286 | MI0015894 | ACCCCACTCCTGGTACCAAAA |
| hsa-miR-449c-5p | MIMAT0010251 | CAGTGTATTGCTAGCGGCTGT |
| hsa-miR-454-3p | MIMAT0003885 | GGTGCAATATTGCTTATAGGGA |
| hsa-miR-4710 | MI0017344 | GTGGGGTGAGGGCAGGTAAA |
| hsa-miR-499a-5p | MIMAT0002870 | CGGCTGTTAAGACTTGCAGAAA |
| hsa-miR-500a-3p | MIMAT0002871 | GCAATGCACCTGGGCAAGAAA |
| hsa-miR-500a-5p | MIMAT0004773 | ATCCTTGCTACCTGGGTGAGA |
| hsa-miR-500b-5p | MIMAT0016925 | AATCCTTGCTACCTGGGTAAAA |
| hsa-miR-548v | MIMAT0015020 | CAGTTACTTTTGCACCAGCCTA |
| hsa-miR-660-3p | MIMAT0022711 | CTGTGTGCATGGATTACAGGAA |
| hsa-miR-9-5p | MIMAT0000441 | GGCTCTTTGGTTATCTAGCTGA |
| U6 | GCTTCGGCAGCACATATACTAA | |
| GCTTCACGAATTTGCGTGTCAT |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Li, Y.; Yao, L.; Li, X. Analysis of MicroRNA Expression Profiling Involved in MC-LR-Induced Cytotoxicity by High-Throughput Sequencing. Toxins 2017, 9, 23. https://doi.org/10.3390/toxins9010023
Ma J, Li Y, Yao L, Li X. Analysis of MicroRNA Expression Profiling Involved in MC-LR-Induced Cytotoxicity by High-Throughput Sequencing. Toxins. 2017; 9(1):23. https://doi.org/10.3390/toxins9010023
Chicago/Turabian StyleMa, Junguo, Yuanyuan Li, Lan Yao, and Xiaoyu Li. 2017. "Analysis of MicroRNA Expression Profiling Involved in MC-LR-Induced Cytotoxicity by High-Throughput Sequencing" Toxins 9, no. 1: 23. https://doi.org/10.3390/toxins9010023
APA StyleMa, J., Li, Y., Yao, L., & Li, X. (2017). Analysis of MicroRNA Expression Profiling Involved in MC-LR-Induced Cytotoxicity by High-Throughput Sequencing. Toxins, 9(1), 23. https://doi.org/10.3390/toxins9010023





