A Combinational Strategy upon RNA Sequencing and Peptidomics Unravels a Set of Novel Toxin Peptides in Scorpion Mesobuthus martensii
Abstract
:1. Introduction
2. Results
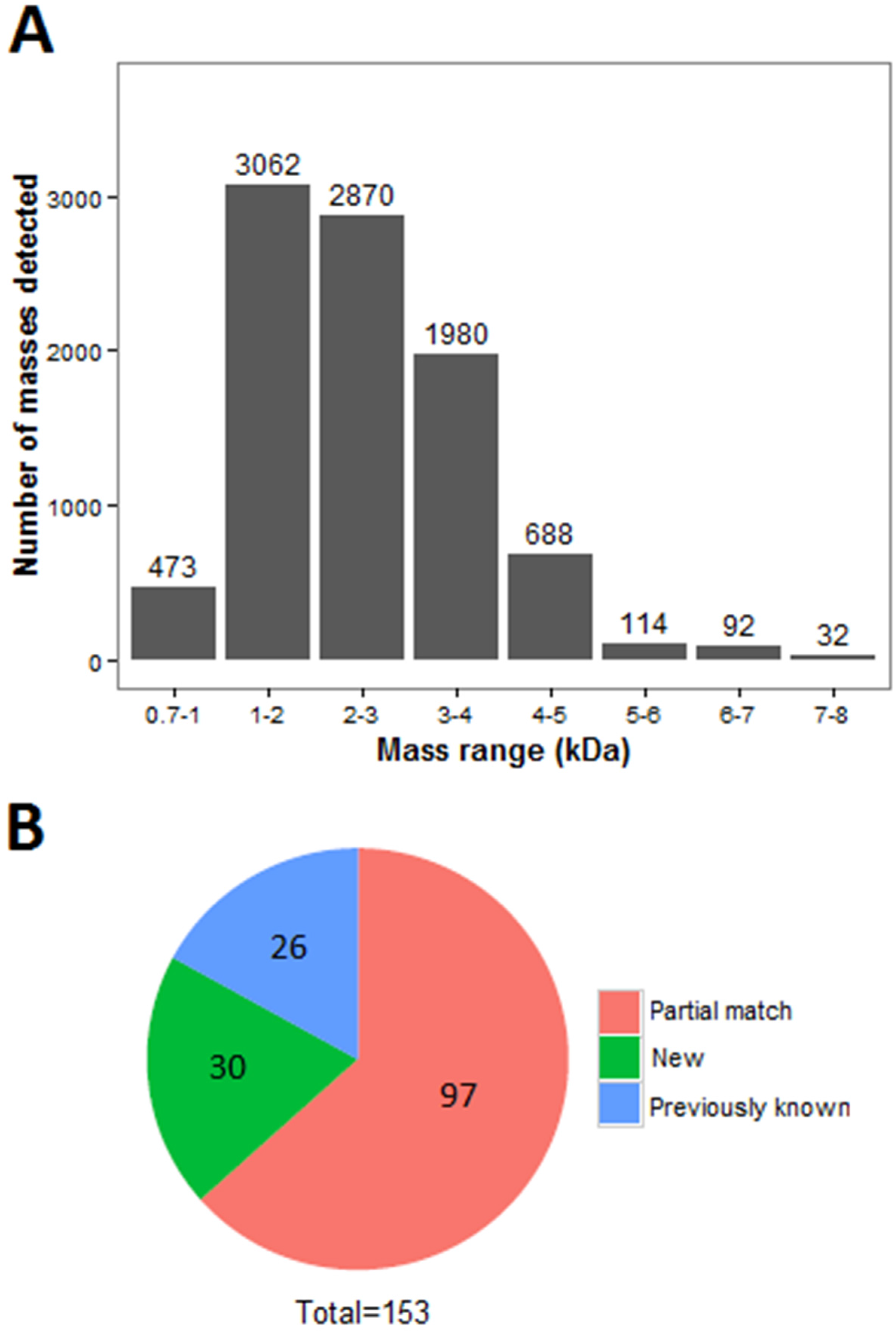
2.1. Venomics of the Scorpion Gland
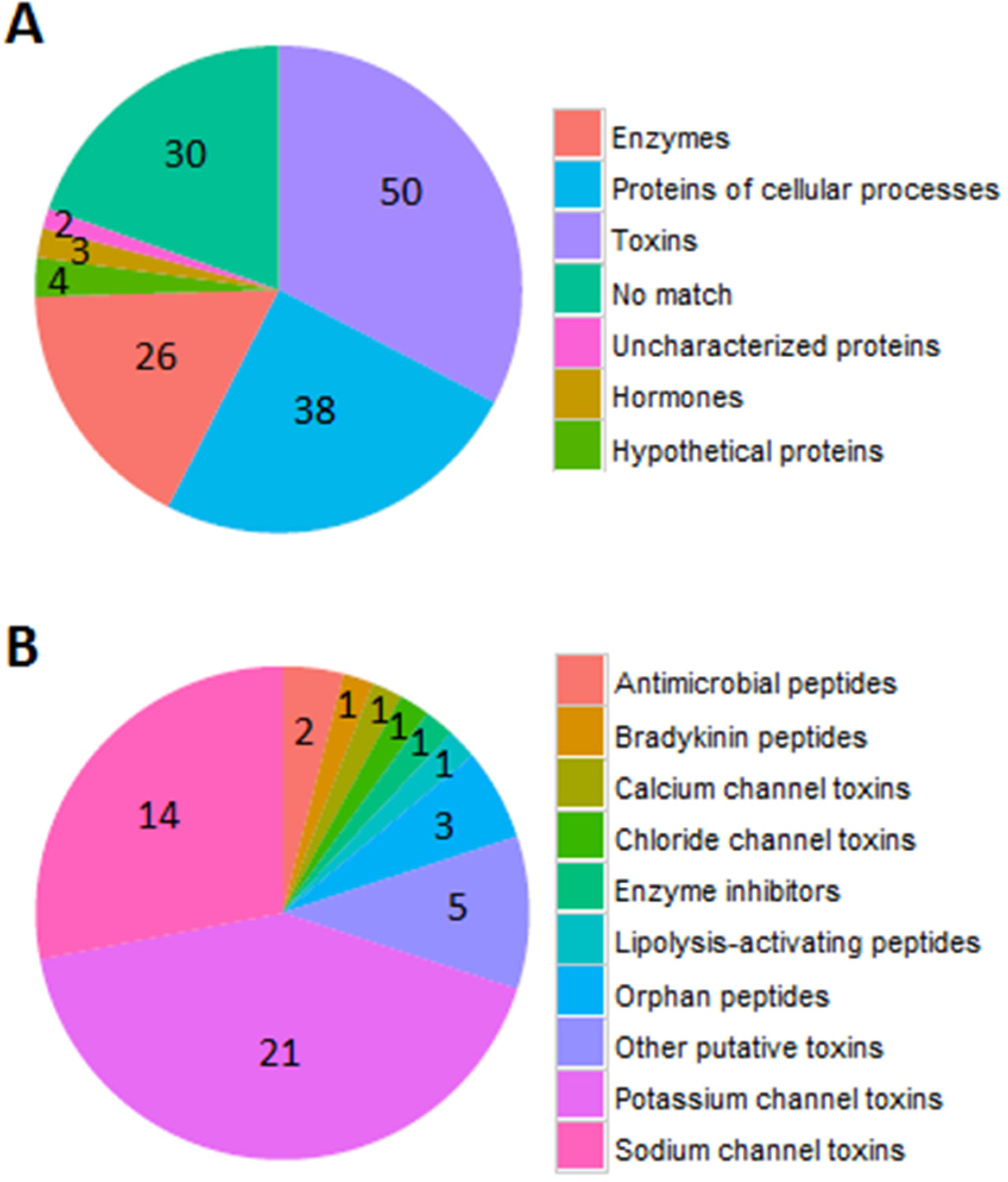
2.2. Identification of Peptides by LC-MS/MS Combined with RNA-Seq Data
2.3. Sodium Channel Toxins
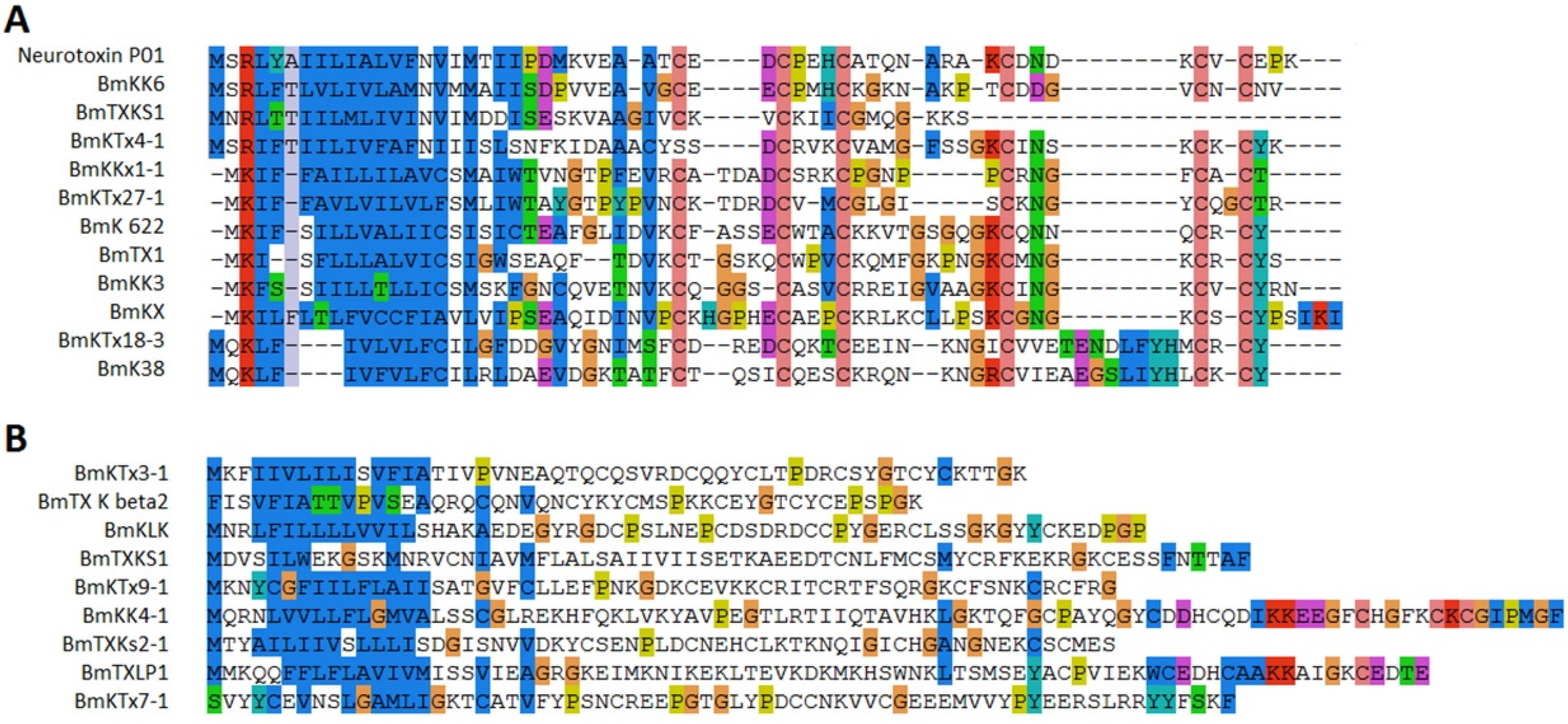
2.4. Potassium Channel Toxins
2.5. Other Functions
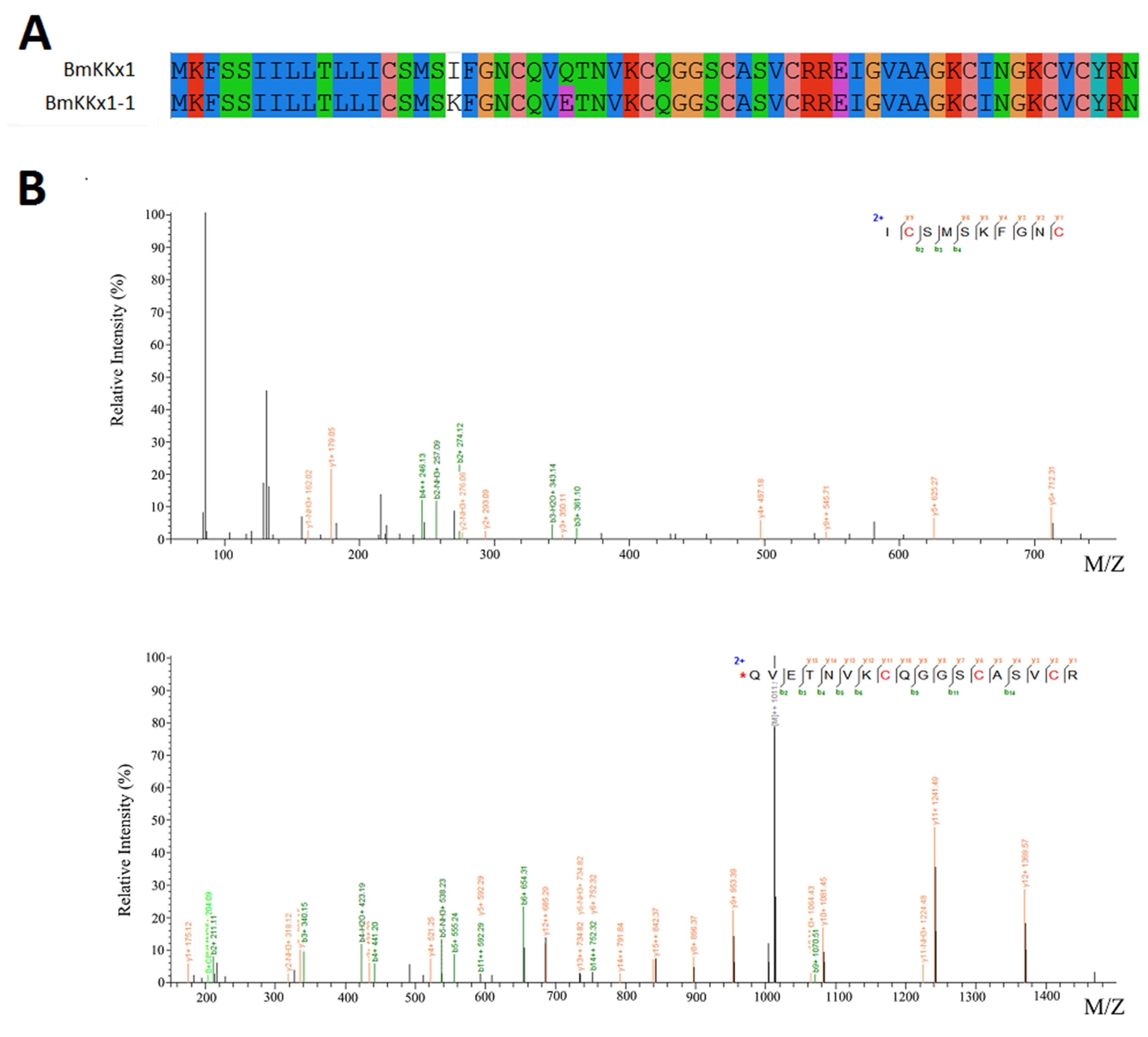
2.6. Novel Venom Peptide
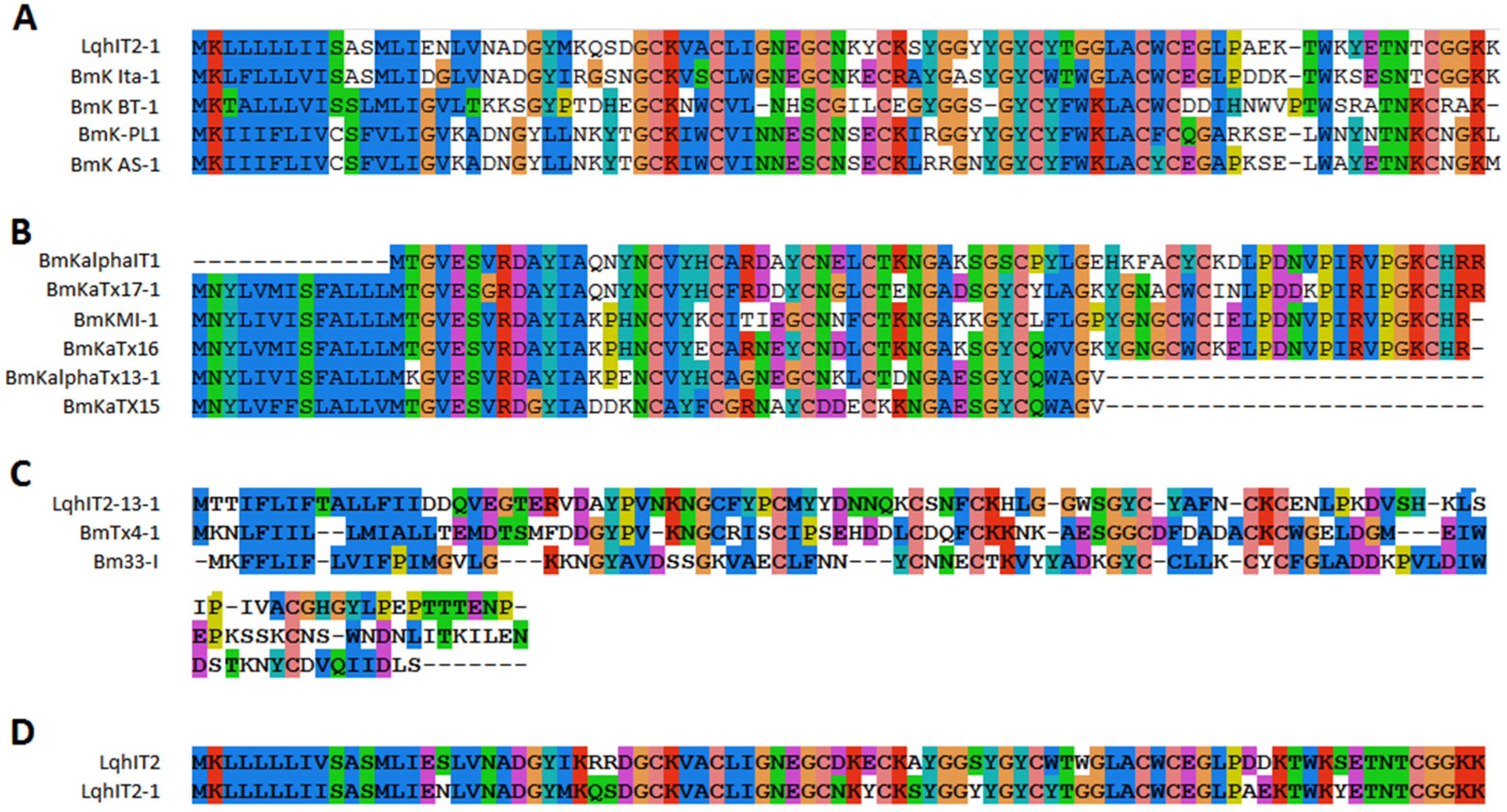
2.7. Amino Acid Variability in Peptide Toxins
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Venom Collection
5.2. Transcriptome Sequencing
5.3. Venom Sample Preparation
5.4. Solid-Phase Extraction of Venom Peptides
5.5. Nano LC-MS/MS Analysis
5.6. Peptide Identification and Bioinformatic Analysis
5.7. Ethics Statement
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| LC MS/MS | Liquid chromatography–mass spectrometry/mass spectrometry |
| 2-DE | Two-dimensional gel electrophoresis |
| SDS-PAGE | Sodium Dodecyl Sulfate Polyacrylamide gel electrophoresis |
| RP-HPLC | Reversed-Phase High-Performance Liquid Chromatography |
| ESI-Q-TOF MS | Electrospray ionization tandem quadrupole/orthogonal-acceleration time-of-flight mass spectrometer |
| MALDI-TOF MS | Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry |
References
- Utkin, Y.N. Animal venom studies: Current benefits and future developments. World J. Biol. Chem. 2015, 6, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Hernandez, V.; Jimenez-Vargas, J.M.; Gurrola, G.B.; Valdivia, H.H.; Possani, L.D. Scorpion venom components that affect ion-channels function. Toxicon 2013, 76, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Vetter, I.; Davis, J.L.; Rash, L.D.; Anangi, R.; Mobli, M.; Alewood, P.F.; Lewis, R.J.; King, G.F. Venomics: A new paradigm for natural products-based drug discovery. Amino. Acids 2011, 40, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, J.R.; Lewis, R.J.; Dutertre, S. Towards an integrated venomics approach for accelerated conopeptide discovery. Toxicon 2012, 60, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.H.; Kaas, Q.; Jones, A.; Alewood, P.F.; Lewis, R.J. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell. Proteom. 2013, 12, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Yu, Y.; Wu, Y.; Hao, P.; Di, Z.; He, Y.; Chen, Z.; Yang, W.; Shen, Z.; He, X.; et al. The genome of mesobuthus martensii reveals a unique adaptation model of arthropods. Nat. Commun. 2013, 4, 2602. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.-X.; Zhu, M.-S.; Lourenço, W.R. Redescription of mesobuthus martensii martensii (karsch, 1879)(scorpiones: Buthidae) from China. Revista Ibérica Aracnología 2004, 10, 137–144. [Google Scholar]
- Xu, X.; Duan, Z.; Di, Z.; He, Y.; Li, J.; Li, Z.; Xie, C.; Zeng, X.; Cao, Z.; Wu, Y.; et al. Proteomic analysis of the venom from the scorpion mesobuthus martensii. J. Proteom. 2014, 106, 162–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shi, W.; Zeng, X.C.; Ge, F.; Yang, M.; Nie, Y.; Bao, A.; Wu, S.; E, G. Unique diversity of the venom peptides from the scorpion androctonus bicolor revealed by transcriptomic and proteomic analysis. J. Proteom. 2015, 128, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, X.; Guo, Z.; Yan, J.; Yu, L.; Li, X.; Xue, X.; Liang, X. Short-chain peptides identification of scorpion buthus martensi karsch venom by employing high orthogonal 2D-HPLC system and tandem mass spectrometry. Proteomics 2012, 12, 3076–3084. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhao, Y.; Zhao, R.; Zhang, W.; He, Y.; Wu, Y.; Cao, Z.; Guo, L.; Li, W. Molecular diversity of toxic components from the scorpion heterometrus petersii venom revealed by proteomic and transcriptome analysis. Proteomics 2010, 10, 2471–2485. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; He, Y.; Zhao, R.; Wu, Y.; Li, W.; Cao, Z. Extreme diversity of scorpion venom peptides and proteins revealed by transcriptomic analysis: Implication for proteome evolution of scorpion venom arsenal. J. Proteom. 2012, 75, 1563–1576. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.C.; Luo, F.; Li, W.X. Molecular dissection of venom from chinese scorpion mesobuthus martensii: Identification and characterization of four novel disulfide-bridged venom peptides. Peptides 2006, 27, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.; Karbat, I.; Ilan, N.; Cohen, L.; Kahn, R.; Gilles, N.; Dong, K.; Stuhmer, W.; Tytgat, J.; Gurevitz, M. The differential preference of scorpion alpha-toxins for insect or mammalian sodium channels: Implications for improved insect control. Toxicon 2007, 49, 452–472. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.Y.; Mao, X.; Xiao, H.; Zhao, Z.Q.; Ji, Y.H. Buthus martensi karsch agonist of skeletal-muscle RYR-1, a scorpion active polypeptide: Antinociceptive effect on rat peripheral nervous system and spinal cord, and inhibition of voltage-gated Na(+) currents in dorsal root ganglion neurons. Neurosci. Lett. 2001, 297, 65–68. [Google Scholar] [CrossRef]
- Zlotkin, E.; Gurevitz, M.; Fowler, E.; Adams, M.E. Depressant insect selective neurotoxins from scorpion venom: Chemistry, action, and gene cloning. Arch. Insect Biochem. Physiol. 1993, 22, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Gurevitz, M.; Karbat, I.; Cohen, L.; Ilan, N.; Kahn, R.; Turkov, M.; Stankiewicz, M.; Stuhmer, W.; Dong, K.; Gordon, D. The insecticidal potential of scorpion β-toxins. Toxicon 2007, 49, 473–489. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.Y.; Li, W.X.; Zeng, X.C.; Liu, H.; Jiang, D.H.; Mao, X. Nine novel precursors of buthus martensii scorpion α-toxin homologues. Toxicon 2000, 38, 1653–1661. [Google Scholar] [CrossRef]
- Ji, Y.H.; Mansuelle, P.; Terakawa, S.; Kopeyan, C.; Yanaihara, N.; Hsu, K.; Rochat, H. Two neurotoxins (bmk i and bmk ii) from the venom of the scorpion buthus martensi karsch: Purification, amino acid sequences and assessment of specific activity. Toxicon 1996, 34, 987–1001. [Google Scholar] [PubMed]
- Xiong, Y.M.; Lan, Z.D.; Wang, M.; Liu, B.; Liu, X.Q.; Fei, H.; Xu, L.G.; Xia, Q.C.; Wang, C.G.; Wang, D.C.; et al. Molecular characterization of a new excitatory insect neurotoxin with an analgesic effect on mice from the scorpion buthus martensi karsch. Toxicon 1999, 37, 1165–1180. [Google Scholar] [CrossRef]
- Kozminsky-Atias, A.; Bar-Shalom, A.; Mishmar, D.; Zilberberg, N. Assembling an arsenal, the scorpion way. BMC Evol. Biol. 2008, 8, 333. [Google Scholar] [CrossRef] [PubMed]
- Zhijian, C.; Yun, X.; Chao, D.; Shunyi, Z.; Shijin, Y.; Yingliang, W.; Wenxin, L. Cloning and characterization of a novel calcium channel toxin-like gene bmca1 from chinese scorpion mesobuthus martensii karsch. Peptides 2006, 27, 1235–1240. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Zeng, X.C.; Luo, X.; Wu, S.; Zhang, L.; Cao, H.; Zhou, J.; Zhou, L. Tremendous intron length differences of the bmkbt and a novel bmkbt-like peptide genes provide a mechanical basis for the rapid or constitutive expression of the peptides. Peptides 2012, 37, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Billen, B.; Bosmans, F.; Tytgat, J. Animal peptides targeting voltage-activated sodium channels. Curr. Pharm. Des. 2008, 14, 2492–2502. [Google Scholar] [CrossRef] [PubMed]
- Huys, I.; Xu, C.Q.; Wang, C.Z.; Vacher, H.; Martin-Eauclaire, M.F.; Chi, C.W.; Tytgat, J. BmTx3, a scorpion toxin with two putative functional faces separately active on A-type K+ and HERG currents. Biochem. J. 2004, 378, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Chen, X.; Li, M.; Cao, C.; Wang, Y.; Wu, G.; Hu, G.; Wu, H. Solution structure of BmKK4, the first member of subfamily α-KTx 17 of scorpion toxins. Biochemistry 2004, 43, 12469–12476. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenkov, A.I.; Vassilevski, A.A.; Kudryashova, K.S.; Nekrasova, O.V.; Peigneur, S.; Tytgat, J.; Feofanov, A.V.; Kirpichnikov, M.P.; Grishin, E.V. Variability of potassium channel blockers in mesobuthus eupeus scorpion venom with focus on Kv1.1: An integrated transcriptomic and proteomic study. J. Biol. Chem. 2015, 290, 12195–12209. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Moran, Y. The rise and fall of an evolutionary innovation: Contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 2015, 11, e1005596. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, Z.; Xiao, Y.; Li, Y.; Rong, M.; Liang, S.; Zhang, Z.; Yu, H.; King, G.F.; Lai, R. Chemical punch packed in venoms makes centipedes excellent predators. Mol. Cell. Proteom. 2012, 11, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Cao, R.; Jiang, L.; Liang, S. A combined de novo protein sequencing and cDNA library approach to the venomic analysis of chinese spider araneus ventricosus. J. Proteom. 2013, 78, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Tashima, A.K.; Zelanis, A.; Kitano, E.S.; Ianzer, D.; Melo, R.L.; Rioli, V.; Sant’anna, S.S.; Schenberg, A.C.; Camargo, A.C.; Serrano, S.M. Peptidomics of three bothrops snake venoms: Insights into the molecular diversification of proteomes and peptidomes. Mol. Cell. Proteom. 2012, 11, 1245–1262. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Xu, S.; Zhou, R.; Zhang, B.; Wang, X.; Liu, X.; Xu, X.; Liu, S. PGA: An R/Bioconductor package for identification of novel peptides using a customized database derived from RNA-Seq. BMC Bioinform. 2016, 17, 244. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Zhou, R.; Feng, Q.; Wang, Q.; Wang, J.; Liu, S. Iquant: An automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 2014, 14, 2280–2285. [Google Scholar] [CrossRef] [PubMed]
- Brosch, M.; Yu, L.; Hubbard, T.; Choudhary, J. Accurate and sensitive peptide identification with Mascot Percolator. J. Proteom. Res. 2009, 8, 3176–3181. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. Mega: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gao, B.; Zhu, S. Target-driven evolution of scorpion toxins. Sci. Rep. 2015, 5, 14973. [Google Scholar] [CrossRef] [PubMed]
- Armon, A.; Graur, D.; Ben-Tal, N. Consurf: An algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J. Mol. Biol. 2001, 307, 447–463. [Google Scholar] [CrossRef] [PubMed]

| Name of Toxin | Identity | Evalue | Function | Sequnce |
|---|---|---|---|---|
| BmCa-1 | 100 | 2.76E-41 | Calcium channel toxin | MNTFVVVFLLLTAILCHAEHALDETARGCNRL NKKCNSDGDCCRYGERCISTGVNYYCRPDFGP |
| Bm12-b | 100 | 3.14E-36 | Chloride channel toxin | MKFLYGIVFIALFLTVMFATQTDGCGPCFTTD ANMARKCRECCGGNGKCFGPQCLCNRE |
| Marcin-18 | 100 | 9.09E-48 | Antimicrobial peptide | MQFKKQLMVIFLAYFLVVNESEAFFGH LFKLATKIIPSLFRRKNQRSRSIMKRDLE NLFDPYQRNLELDRLLKQLP |
| BmKn1 | 100 | 8.51E-43 | Antimicrobial peptide | MKSQTFFLLFLVVLLLAISQSEAFIGAVAGLLSKIFGK RSMRDMDTMKYLYDPSLSAADLKTLQKLMENY |
| BmOp-1 | 95.38 | 3.48E-34 | Orphan peptide | MKPRTFLVLFLVCILVMDAVVARRCGGNRRRKVI KMVRKLRPLVKVIRIIKKIRGRAPCGGSRLT |
| BmOp-2 | 86.59 | 9.63E-46 | Orphan peptide | MNKSLIILIVAIVVLSLWSAAEARQRRCQVYC SNSPKGCQVRCSWSFGKREIADTVEDFPGKL DEDDEAILKKLAIEELKNE |
| BmOp-3 | 83.76 | 5.15E-59 | Orphan peptide | MKTTGIILFVCAIIYSLYLEAESASIHSSEL QSNFHKLRKRQIHGRFGDYIPRGDLAQG KVSTGVEMFFPVDDDGEETRDTDDDGEQ TSRSEDDDDGLTSRRAAPRERHVNCNKRHY |
| BmLVP2-alpha | 85.71 | 8.46E-59 | Lipolysis-activating peptide | MMTLALFGIIFTLFSLIGSIHGDDEPGDYPTNVYGNK YYCTILGENEYCRKICKLHGVSYGYCYNSRCWCEY LEDKDVTIWNAVKNHCTNTLLYPNGK |
| Bpp BmK3 | 100 | 1.06E-44 | Bradykinin-potentiating peptide | MNKKTLLVIFFVTMLIVDEVNSFRFGSFLKKVWKSKL AKKLRSKGKQLLKDYANKVLNGPEEEAAAPAERRR |
| Makatoxin-2-1 | 90.91 | 6.3329E-39 | Inhibitor of nitric oxide (NO) synthase | MNYLIVISFALLLMTGVESGRDAYIADSENCTYFCGA NPYCNDLCTKNGAKSGYCQWVGKYGNGCW |
| BmPt2 | 91.25 | 1.62E-47 | Venom protein | MKSAVIVAITLCCLFNLYANAQKDCSL PVDTGRGKGWFLRYYYNKNSKTCESFI YGGVGGNKNNFLNIENCCKICKAKNC |
| BmPt3 | 93.68 | 3.94E-59 | Putative toxin-like peptide | MVKMQVIFIAFIAVIACSMVYGDSLSPWNEG DTYYGCQRQTDEFCNKICKLHLATGGGSC YQPFPFVKLCRCIGIDYDNSFFFGALEKQCPKLRE |
| BmPt4 | 78.75 | 1.77E-31 | Venom peptide | AGRFDPVSAAGAGRFDPVLEAGAGRFDP ASAAGAGRFDPVSAAGAGRFDPTLEAG AGRFNPALEAGAGRFDPAAEAGAGRF |
| BmPt5 | 66.67 | 4.99E-33 | Venom toxin | MFFLFLILLALIPMIKLEKECAMESLQGE FPRKDKGYLYECGEDIKCKKICKNHGRS ENDGKCCYGNCFCTGLHGKNIIKHQ |
| Name of Toxin | Sequnce |
|---|---|
| BmVt1 | KIVVKYVKQRIADHIPRRNFKIHKRKFANNIYDSCII |
| BmVt2 | AGRFDPALAAGARGYDPYYVGGPLGETKERIYIDE |
| BmVt3 | MRALTIFGVIFACLLLFRTAVADHDGDDHNEELEEDVKQEKKILGPYSNMGRRTSCKIWIWQMEEAQRPS |
| BmVt4 | IIHQIFQILLAALEKLNLMTKFLYLMIKAMIFLVEKLVVVEQNNVTWIHVRIMAIVMICGINTFVNVNDHFLE |
| BmVt5 | MEKILLNQFIVFIVGIFGMRDVNSEYVSENFRKFPNYISENKDIQNKNNARNLIPDIKSSLKETETAESKKRNV DLINFSEKVKLSDESKKSFERTAVIHVLNNKENYDKNSKNTLNENLQPSSSNINKLKVTEISNNMNL |
| BmVt6 | KVLEEEAGEGEEEEEENWQLMLQISVLFLYHKRDKIVYIIYDL |
| BmVt7 | ILSNSKPLKSSRNNDELHKLFLNKNDNQPIKEFLGNVEKNNILSFPQNILPVYHPDFANMFSDEISFQTNTVP |
| BmVt8 | KQEQDVLIQHRQQEQDVLTQYQQQERDVLIQH |
| BmVt9 | MKVLVLLLVVALVAAALADKRDNIKRDYGGVGGGYGGGGRGGFGGGRGGFGG GYGGGRGGGIGGGRGGFGGGRGGFGGGRGGFGGGYGGGKYGGKYGGK |
| BmVt10 | TMTFNRNYSLHHFRPCYVFIFLMSTIFWTLTNADEDNIKSTIDRDKRAPQLYSFGLGKKSYNIPVDSDNIES |
| BmVt11 | TRSSTPERFSTRDLISSFSSASSVSSRCSAMSASEVCCCSRAMSWANWVRRRSSSPTRSLARCSSRSSI |
| BmVt12 | MKCILINFLSLMLLANYSFGNEEQKKNMRALYPRKFYVERLRNDIVEEPFKKKNAYVTGG |
| BmVt13 | MRVDNSAVLLVLIYLAKLSLSAPVDHGKEENPDPSFNIDVEYK NYLEEVARLLENDEDFINQIKDVTHSKDFQPEDISF |
| BmVt14 | MRVVMLIFFLLCQALNLALVLQSEINFRYDSVETDLRSAELNNLDIWNED RKISRFLIEEEEISREISKYENAREISKRFLILVGTKWCGAGNKAE |
| BmVt15 | RRGTAEIREGRFGISRRAEEADLRREPRYQPVYQHEVQQDSYGQNQDSLRQNNAAQQRSHYQTYDGQLGR |
| BmVt16 | MKILSFIPVFLCLGIILNISCAENDLKRILNTSDEDLEKRWPDK |
| BmVt18 | DPPECTCPHGKKFDHGLMKCIAGPEINCDSEIEGKGKTWTAFCEEENMVPCKHNIG DPPECTCPHGKKFDHGLMKCIAGPEINCDSEIEGKGKTWTAFCEEENMVPC |
| BmVt21 | KDVNNLTYLIKKKLLEIIPKYKSPSIFSKSKMPIFPHENKESLFPKYKLQY |
| BmVt22 | IILVGYCVKVMEDLVIAIFGNWHVGVMIFIIGFQLGLERPTNVVQNNTSIYVTF |
| BmVt23 | MIKENMKKWLIYLLIPILSHSVVSSREGYRLNEQETYAADITPPHPQKHPYWYRRLWENQPPEQKDL YQNSYKNPFYLLDRKQLKNDDDEFELPNQAYLPLKEVGTNNRGADKIHSKSKIPEGHITVK |
| BmVt25 | MKVVLVTFLCILLLLSNQNLGNDAGKKEIQAVYRRKAYADPAKNDDVEIVDHNFFRFRRSEELN |
| BmVt26 | MKQFMFYFGIVIFATLLANRDVMAQNNKVILPGFIPEITQK |
| BmVt27 | NSGLSLFGMKIIGYIFGFALICSLPNQNVCHENEGQKRSKIDAENDDLKQRVFPRVRFSLSDEEKRLQQR LSPAVLASLTGLEKQRLSRETLEDFSWNTEKRFDPAIFGSLPDDTKERIDSAALASLAENEEERFYSTT |
| BmVt29 | LLIWSSLLPASPPSTKCVVFFFMPPLGEDSLKGQRKLFATLKFLPTV |
| BmVt30 | VPSPPTPDAILELNEKLRDGRIWEFVHQRLVESPLLRAMVFNELTPRSSELNLDISNMRRRRSLQDYELK |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luan, N.; Shen, W.; Liu, J.; Wen, B.; Lin, Z.; Yang, S.; Lai, R.; Liu, S.; Rong, M. A Combinational Strategy upon RNA Sequencing and Peptidomics Unravels a Set of Novel Toxin Peptides in Scorpion Mesobuthus martensii. Toxins 2016, 8, 286. https://doi.org/10.3390/toxins8100286
Luan N, Shen W, Liu J, Wen B, Lin Z, Yang S, Lai R, Liu S, Rong M. A Combinational Strategy upon RNA Sequencing and Peptidomics Unravels a Set of Novel Toxin Peptides in Scorpion Mesobuthus martensii. Toxins. 2016; 8(10):286. https://doi.org/10.3390/toxins8100286
Chicago/Turabian StyleLuan, Ning, Wang Shen, Jie Liu, Bo Wen, Zhilong Lin, Shilong Yang, Ren Lai, Siqi Liu, and Mingqiang Rong. 2016. "A Combinational Strategy upon RNA Sequencing and Peptidomics Unravels a Set of Novel Toxin Peptides in Scorpion Mesobuthus martensii" Toxins 8, no. 10: 286. https://doi.org/10.3390/toxins8100286
APA StyleLuan, N., Shen, W., Liu, J., Wen, B., Lin, Z., Yang, S., Lai, R., Liu, S., & Rong, M. (2016). A Combinational Strategy upon RNA Sequencing and Peptidomics Unravels a Set of Novel Toxin Peptides in Scorpion Mesobuthus martensii. Toxins, 8(10), 286. https://doi.org/10.3390/toxins8100286





