The Ambivalent Nature of Bacteroides fragilis and the Interaction with Clostridioides difficile: Benefits and Disadvantages for the Human Host
Abstract
1. Introduction
2. The B. fragilis Toxin (BFT)
3. B. fragilis: Commensal and Pathogen
3.1. Commensal Role
3.1.1. Non-Toxigenic B. fragilis (NTBF)
3.1.2. Enterotoxigenic B. fragilis (ETBF)
3.2. Pathogenic Role
3.2.1. Non-Toxigenic B. fragilis (NTBF)
3.2.2. Enterotoxigenic B. fragilis (ETBF)
3.2.3. Antibiotic Resistance of B. fragilis
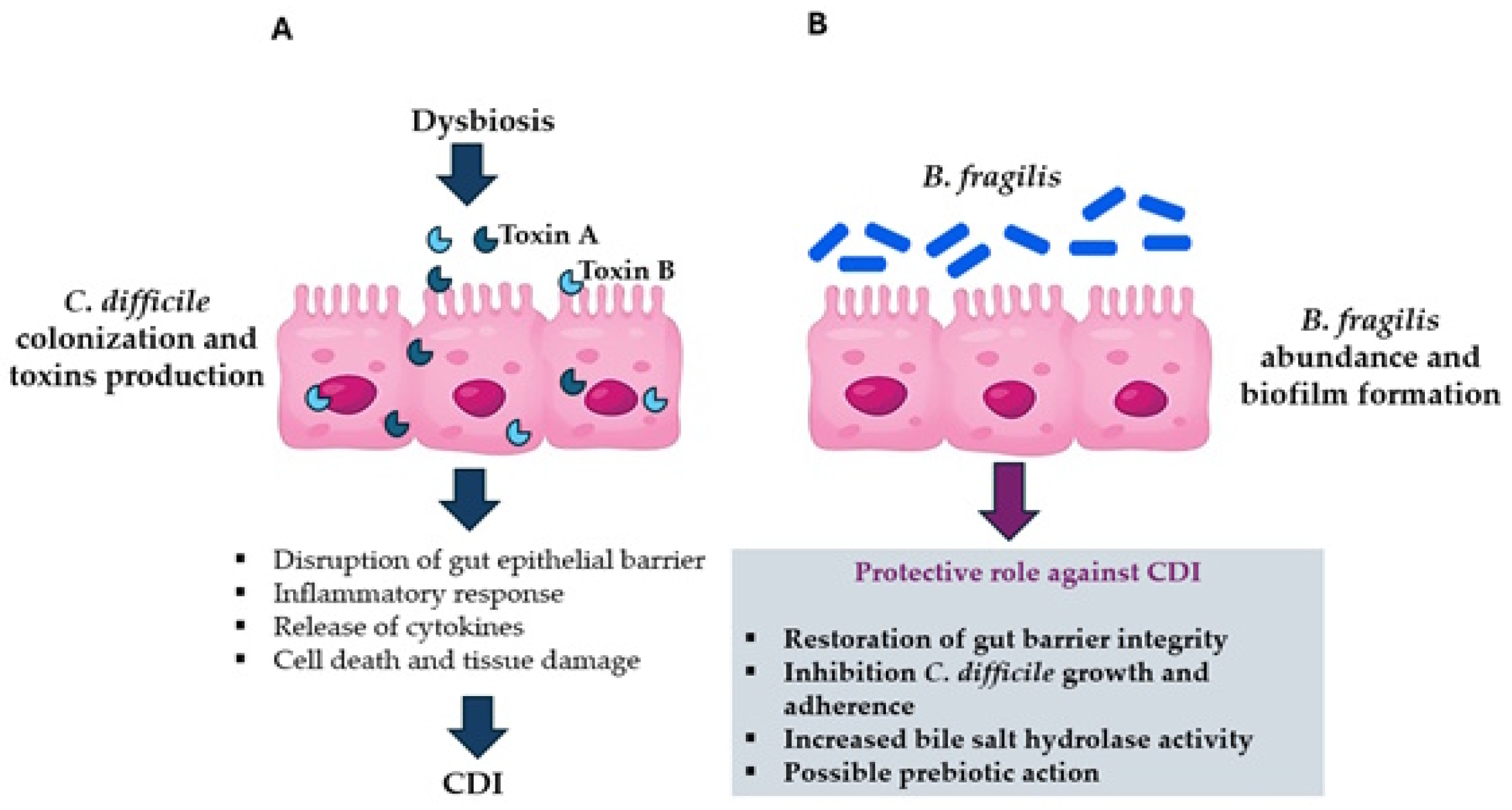
4. B. fragilis Interaction with C. difficile
4.1. Clostridioides Difficile Infection (CDI)
4.2. B. fragilis Protective Role Against CDI
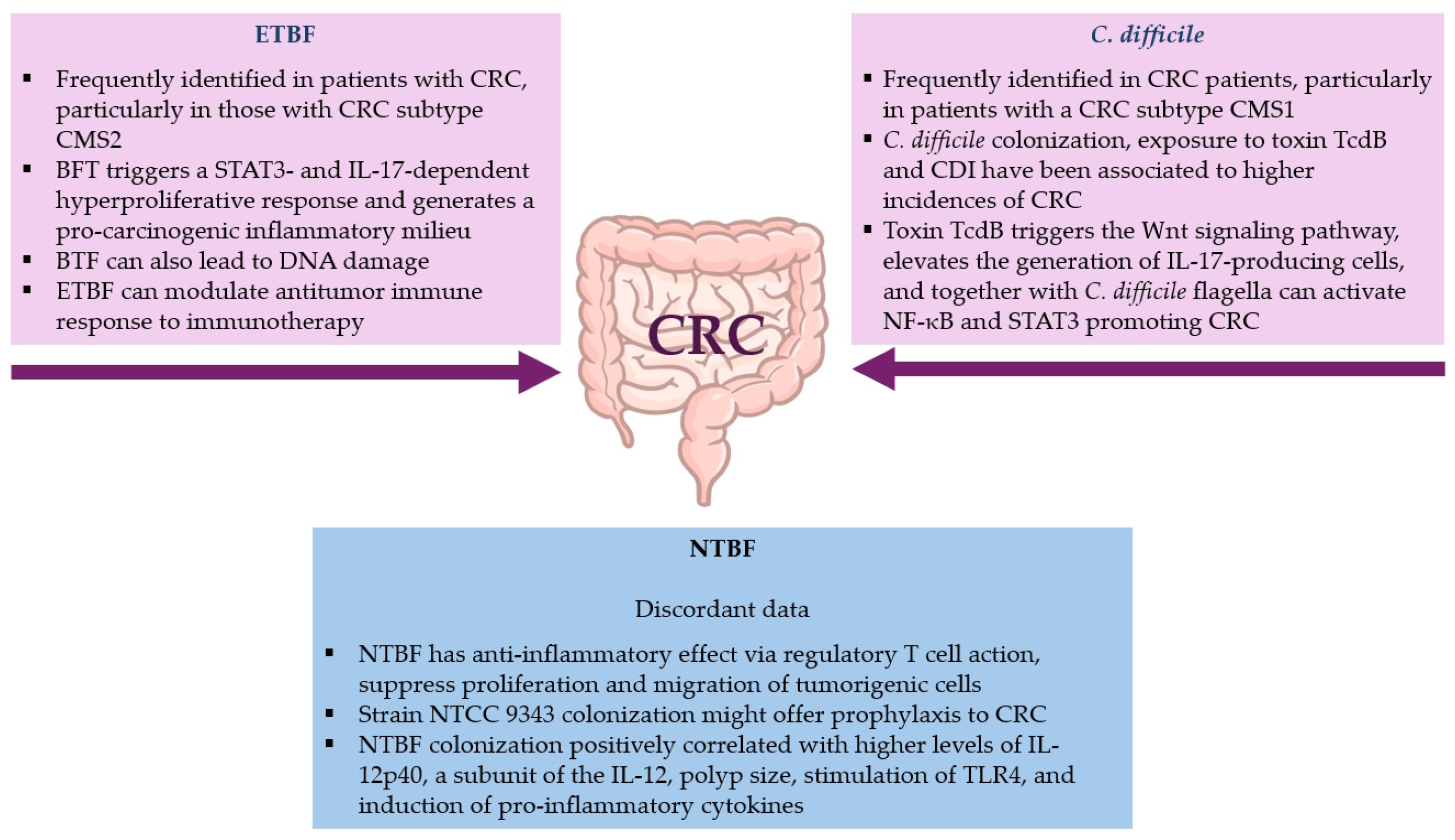
4.3. Involvement of B. fragilis and C. difficile in Colorectal Cancer (CRC)
4.3.1. B. fragilis Role in CRC
Enterotoxigenic B. fragilis (ETBF)
Non-Toxigenic B. fragilis (NTBF)
4.3.2. C. difficile’s Role in CRC
5. Conclusions and Future Research
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef]
- Gibson, G.R.; Macfarlane, G.T. Intestinal bacteria and disease. Human Health: The Contribution of Microorganisms; Gibson, S.A.W., Ed.; Springer: Berlin/Heidelberg, Germany, 1994; pp. 53–62. [Google Scholar]
- Zafar, H.; Saier, M.H., Jr. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Oles, R.E.; Terrazas, M.C.; Loomis, L.R.; Neal, M.J.; Paulchakrabarti, M.; Zuffa, S.; Hsu, C.Y.; Vasquez Ayala, A.; Lee, M.H.; Tribelhorn, C.; et al. Pathogenic Bacteroides fragilis strains can emerge from gut-resident commensals. bioRxiv 2024. [Google Scholar] [CrossRef]
- Patrick, S. A tale of two habitats: Bacteroides fragilis, a lethal pathogen and resident in the human gastrointestinal microbiome. Microbiology 2022, 168, 001156. [Google Scholar] [CrossRef]
- Patrick, S.; Duerden, B. Gram- negative non- spore forming obligate anaerobes. In Principles and Practice of Clinical Bacteriology, 2nd ed.; Gillespie, S.H., Hawkey, P., Eds.; Wiley: Hoboken, NJ, USA, 2006; pp. 541–556. [Google Scholar]
- Huang, Y.; Cao, J.; Zhu, M.; Wang, Z.; Jin, Z.; Xiong, Z. Nontoxigenic Bacteroides fragilis: A double-edged sword. Microbiol. Res. 2024, 286, 127796. [Google Scholar] [CrossRef]
- Wang, Y.; Begum-Haque, S.; Telesford, K.M.; Ochoa-Repáraz, J.; Christy, M.; Kasper, E.J.; Kasper, D.L.; Robson, S.C.; Kasper, L.H. A commensal bacterial product elicits and modulates migratory capacity of CD39(+) CD4 T regulatory subsets in the suppression of neuroinflammation. Gut Microbes 2014, 5, 552–561. [Google Scholar] [CrossRef]
- Pagliuca, C.; Cicatiello, A.G.; Colicchio, R.; Greco, A.; Cerciello, R.; Auletta, L.; Albanese, S.; Scaglione, E.; Pagliarulo, C.; Pastore, G.; et al. Novel approach for evaluation of Bacteroides fragilis protective role against Bartonella henselae liver damage in immunocompromised murine model. Front. Microbiol. 2016, 7, 1750. [Google Scholar] [CrossRef]
- Johnson, J.L.; Jones, M.B.; Cobb, B.A. Polysaccharide-experienced effector T cells induce IL-10 in FoxP3+ regulatory T cells to prevent pulmonary inflammation. Glycobiology 2018, 28, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Gopalakrishnan, V.; Daillère, R.; Zitvogel, L.; Wargo, J.A.; Kroemer, G. The gut microbiota influences anticancer immunosurveillance and general health. Nat. Rev. Clin. Oncol. 2018, 15, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.L.; Wu, S.; Geis, A.L.; Chan, G.V.; Gomes, T.A.M.; Beck, S.E.; Wu, X.; Fan, H.; Tam, A.J.; Chung, L.; et al. Non-toxigenic Bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide A. Mucosal Immunol. 2019, 12, 164–177. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, Z.; Yan, Y.; Ji, L.; He, J.; Xuan, B.; Shen, C.; Ma, Y.; Jiang, S.; Ma, D.; et al. Enterotoxigenic Bacteroides fragilis promotes intestinal inflammation and malignancy by inhibiting exosome-packaged miR-149-3p. Gastroenterology 2021, 161, 1552–1566.e12. [Google Scholar] [CrossRef] [PubMed]
- Parida, S.; Wu, S.; Siddharth, S.; Wang, G.; Muniraj, N.; Nagalingam, A.; Hum, C.; Mistriotis, P.; Hao, H.; Talbot, C.C., Jr.; et al. Procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and β-catenin axes. Cancer Discov. 2021, 1, 1138–1157. [Google Scholar] [CrossRef]
- Jimenez-Alesanco, A.; Eckhard, U.; Asencio Del Rio, M.; Vega, S.; Guevara, T.; Velazquez-Campoy, A.; Gomis-Rüth, F.X.; Abian, O. Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis. Protein Sci. 2022, 31, e4427. [Google Scholar] [CrossRef]
- Wilson, N.G.; Hernandez-Leyva, A.; Rosen, A.L.; Jaeger, N.; McDonough, R.T.; Santiago-Borges, J.; Lint, M.A.; Rosen, T.R.; Tomera, C.P.; Bacharier, L.B.; et al. The gut microbiota of people with asthma influences lung inflammation in gnotobiotic mice. iScience 2023, 26, 105991. [Google Scholar] [CrossRef]
- Sánchez, E.; Laparra, J.M.; Sanz, Y. Discerning the role of Bacteroides fragilis in celiac disease pathogenesis. Appl. Environ. Microbiol. 2012, 78, 6507–6515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, Q.; Hou, Y.; Zhang, X.; Yin, Z.; Cai, X.; Wei, W.; Wang, J.; He, D.; Wang, G.; et al. Bacteroides species differentially modulate depression-like behavior via gut-brain metabolic signaling. Brain Behav. Immun. 2022, 102, 11–22. [Google Scholar] [CrossRef]
- Xia, Y.; Xiao, Y.; Wang, Z.H.; Liu, X.; Alam, A.M.; Haran, J.P.; McCormick, B.A.; Shu, X.; Wang, X.; Ye, K. Bacteroides fragilis in the gut microbiomes of Alzheimer’s disease activates microglia and triggers pathogenesis in neuronal C/EBPβ transgenic mice. Nat. Commun. 2023, 6, 5471. [Google Scholar] [CrossRef]
- Tzianabos, A.O.; Onderdonk, A.B.; Rosner, B.; Cisneros, R.L.; Kasper, D.L. Structural features of polysaccharides that induce intra-abdominal abscesses. Science 1993, 262, 416–419. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, Y.; Cai, J.; Rimal, B.; Rocha, E.R.; Coleman, J.P.; Zhang, C.; Nichols, R.G.; Luo, Y.; Kim, B.; et al. Bile salt hydrolase in non-enterotoxigenic Bacteroides potentiates colorectal cancer. Nat. Commun. 2023, 14, 755. [Google Scholar] [CrossRef] [PubMed]
- Czepiel, J.; Dróżdż, M.; Pituch, H.; Kuijper, E.J.; Perucki, W.; Mielimonka, A.; Goldman, S.; Wultańska, D.; Garlicki, A.; Biesiada, G. Clostridium difficile infection: Review. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1211–1221. [Google Scholar] [CrossRef]
- Kelly, C.P.; LaMont, J.T. Clostridium difficile–more difficult than ever. N. Engl. J. Med. 2008, 359, 1932–1940. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.S.; Yang, Y.; Zhang, K.; Qian, Z.; Zhang, Y.; Liu, Y.; Wang, Y.; Bai, Y.; Fan, H.; Zhao, X.; et al. Bacteroides fragilis prevents Clostridium difficile infection in a mouse model by restoring gut barrier and microbiome regulation. Front. Microbiol. 2018, 9, 2976. [Google Scholar] [CrossRef]
- Zamani, S.; Taslimi, R.; Sarabi, A.; Jasemi, S.; Sechi, L.A.; Feizabadi, M.M. Enterotoxigenic Bacteroides fragilis: A possible etiological candidate for bacterially-induced colorectal precancerous and cancerous lesions. Front. Cell Infect. Microbiol. 2020, 9, 449. [Google Scholar] [CrossRef]
- Nezhadi, J.; Lahouty, M.; Rezaee, M.A.; Fadaee, M. Clostridium difficile as a potent trigger of colorectal carcinogenesis. Discov. Oncol. 2025, 16, 910. [Google Scholar] [CrossRef]
- Myers, L.L.; Firehammer, B.D.; Shoop, D.S.; Border, M.M. Bacteroides fragilis: A possible cause of acute diarrheal disease in newborn lambs. Infect. Immun. 1984, 44, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Myers, L.L.; Shoop, D.S.; Firehammer, B.D.; Border, M.M. Association of enterotoxigenic Bacteroides fragilis with diarrheal disease in calves. J. Infect. Dis. 1985, 152, 1344–1347. [Google Scholar] [CrossRef]
- Sears, C.L. The toxins of Bacteroides fragilis. Toxicon 2001, 39, 1737–1746. [Google Scholar] [CrossRef]
- Franco, A.A.; Buckwold, S.; Shin, J.W.; Ascon, M.; Sears, C.L. Mutation of the zinc-binding metalloprotease motif affects Bacteroides fragilis toxin activity without affecting propeptide processing. Infect. Immun. 2005, 73, 5273–5277. [Google Scholar] [CrossRef]
- Chambers, F.G.; Koshy, S.S.; Saidi, R.F.; Clark, D.P.; Moore, R.D.; Sears, C.L. Bacteroides fragilis toxin exhibits polar activity on monolayers of human intestinal epithelial cells (T84 cells) in vitro. Infect. Immun. 1997, 65, 3561–3570. [Google Scholar] [CrossRef] [PubMed]
- Obiso, R.J., Jr.; Azghani, A.O.; Wilkins, T.D. The Bacteroides fragilis toxin fragilysin disrupts the paracellular barrier of epithelial cells. Infect. Immun. 1997, 65, 1431–1439. [Google Scholar] [CrossRef]
- Kim, J.M.; Oh, Y.K.; Kim, Y.J.; Oh, H.B.; Cho, Y.J. Polarized secretion of CXC chemokines by human intestinal epithelial cells in response to Bacteroides fragilis enterotoxin: NF-kappa B plays a major role in the regulation of IL-8 expression. Clin. Exp. Immunol. 2001, 123, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Powell, J.; Mathioudakis, N.; Kane, S.; Fernandez, E.; Sears, C.L. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-κB pathway. Infect. Immun. 2004, 72, 5832–5839. [Google Scholar] [CrossRef] [PubMed]
- Moncrief, J.S.; Duncan, A.J.; Wright, R.L.; Barroso, L.A.; Wilkins, T.D. Molecular characterization of the fragilysin pathogenicity islet of enterotoxigenic Bacteroides fragilis. Infect. Immun. 1998, 66, 1735–1739. [Google Scholar] [CrossRef]
- Franco, A.A.; Cheng, R.K.; Chung, G.T.; Wu, S.; Oh, H.B.; Sears, C.L. Molecular evolution of the pathogenicity island of enterotoxigenic Bacteroides fragilis strains. J. Bacteriol. 1999, 181, 6623–6633. [Google Scholar] [CrossRef]
- Buckwold, S.L.; Shoemaker, N.B.; Sears, C.L.; Franco, A.A. Identification and characterization of conjugative transposons CTn86 and CTn9343 in Bacteroides fragilis strains. Appl. Environ. Microbiol. 2007, 73, 53–63. [Google Scholar] [CrossRef]
- Franco, A.A.; Cheng, R.K.; Goodman, A.; Sears, C.L. Modulation of bft expression by the Bacteroides fragilis pathogenicity island and its flanking region. Mol. Microbiol. 2002, 45, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Liu, C.X.; Kato, H.; Watanabe, K.; Tanaka, Y.; Yamamoto, T.; Suzuki, K.; Ueno, K. A new subtype of the metalloprotease toxin gene and the incidence of the three bft subtypes among Bacteroides fragilis isolates in Japan. FEMS Microbiol. Lett. 2000, 185, 171–176. [Google Scholar] [CrossRef][Green Version]
- Wu, S.; Dreyfus, L.A.; Tzianabos, A.O.; Hayashi, C.; Sears, C.L. Diversity of the metalloprotease toxin produced by enterotoxigenic Bacteroides fragilis. Infect. Immun. 2002, 70, 2463–2471. [Google Scholar] [CrossRef]
- Avila-Campos, M.J.; Liu, C.; Song, Y.; Rowlinson, M.C.; Finegold, S.M. Determination of bft gene subtypes in Bacteroides fragilis clinical isolates. J. Clin. Microbiol. 2007, 45, 1336–1338. [Google Scholar] [CrossRef]
- Akpinar, M.; Aktaş, E.; Cömert, F.; Külah, C.; Sümbüloĝlu, V. Evaluation of the prevalence of enterotoxigenic Bacteroides fragilis and the distribution bft gene subtypes in patients with diarrhea. Anaerobe 2010, 16, 505–509. [Google Scholar] [CrossRef]
- Ulger, N.; Rajendram, D.; Yagci, A.; Gharbia, S.; Shah, H.N.; Gulluoglu, B.M.; Akin, L.M.; Demirkalem, P.; Celenk, T.; Soyletir, G. The distribution of the bft alleles among enterotoxigenic Bacteroides fragilis strains from stool specimens and extraintestinal sites. Anaerobe 2006, 12, 71–74. [Google Scholar] [CrossRef]
- Akhi, M.T.; Jedari Seifi, S.; Asgharzadeh, M.; Ahangarzadeh Rezaee, M.; Abdoli Oskuei, S.; Pirzadeh, T.; Memar, M.Y.; Alizadeh, N.; Seifi Yarijan Sofla, H. Role of enterotoxigenic Bacteroides fragilis in children less than 5 years of age with diarrhea in Tabriz, Iran. Jundishapur J. Microbiol. 2016, 9, e32163. [Google Scholar] [CrossRef]
- Chung, G.T.; Franco, A.A.; Wu, S.; Rhie, G.E.; Cheng, R.; Oh, H.B.; Sears, C.L. Identification of a third metalloprotease toxin gene in extraintestinal isolates of Bacteroides fragilis. Infect. Immun. 1999, 67, 4945–4949. [Google Scholar] [CrossRef]
- Valguarnera, E.; Wardenburg, J.B. Good gone bad: One toxin away from disease for Bacteroides fragilis. J. Mol. Biol. 2020, 432, 765–785. [Google Scholar] [CrossRef]
- Nakano, V.; Gomes, D.A.; Arantes, R.M.; Nicoli, J.R.; Avila-Campos, M.J. Evaluation of the pathogenicity of the Bacteroides fragilis toxin gene subtypes in gnotobiotic mice. Curr. Microbiol. 2006, 53, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Shin, J.; Zhang, G.; Cohen, M.; Franco, A.; Sears, C.L. The Bacteroides fragilis toxin binds to a specific intestinal epithelial cell receptor. Infect. Immun. 2006, 74, 5382–5390. [Google Scholar] [CrossRef] [PubMed]
- Yekani, M.; Baghi, H.B.; Naghili, B.; Vahed, S.Z.; Sóki, J.; Memar, M.Y. To resist and persist: Important factors in the pathogenesis of Bacteroides fragilis. Microb. Pathog. 2020, 149, 104506. [Google Scholar] [CrossRef]
- Wu, S.; Morin, P.J.; Maouyo, D.; Sears, C.L. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 2003, 124, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Gwon, S.Y.; Kim, M.S.; Lee, S.; Rhee, K.J. Bacteroides fragilis toxin induces IL-8 secretion in HT29/C1 cells through disruption of E-cadherin junctions. Immune Netw. 2013, 13, 213–217. [Google Scholar] [CrossRef]
- Kim, J.M.; Cho, Y.K.; Oh, S.J.; Jung, H.Y.; Kim, Y.J.; Kim, N. Nuclear factor-kappa B activation pathway in intestinal epithelial cells is a major regulator of chemokine gene expression and neutrophil migration induced by Bacteroides fragilis enterotoxin. Clin. Exp. Immunol. 2002, 130, 59–66. [Google Scholar] [CrossRef]
- Kim, J.M.; Jung, H.J.; Lee, J.Y.; Youn, J.; Lee, C.H.; Kim, K.H. Mitogen-activated protein kinase and activator protein-1 dependent signals are essential for Bacteroides fragilis enterotoxin-induced enteritis. Eur. J. Immunol. 2005, 35, 2648e2657. [Google Scholar] [CrossRef]
- Wick, E.C.; Rabizadeh, S.; Albesiano, E.; Wu, X.; Wu, S.; Chan, J.; Rhee, K.J.; Ortega, G.; Huso, D.L.; Pardoll, D.; et al. Stat3 activation in murine colitis induced by enterotoxigenic Bacteroides fragilis. Inflamm. Bowel Dis. 2014, 20, 821–834. [Google Scholar] [CrossRef]
- Rhee, K.J.; Wu, S.; Wu, X.; Huso, D.L.; Karim, B.; Franco, A.A.; Rabizadeh, S.; Golub, J.E.; Mathews, L.E.; Shin, J.; et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect. Immun. 2009, 77, 1708–1718. [Google Scholar] [CrossRef] [PubMed]
- Casterline, B.W.; Hecht, A.L.; Choi, V.M.; Wardenburg, J.B. The Bacteroides fragilis pathogenicity island links virulence and strain competition. Gut Microbes 2017, 8, 374–383. [Google Scholar] [CrossRef]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef]
- Browne, H.P.; Neville, B.A.; Forster, S.C.; Lawley, T.D. Transmission of the gut microbiota: Spreading of health. Nat. Rev. Microbiol. 2017, 15, 531–543. [Google Scholar] [CrossRef]
- Nagpal, R.; Tsuji, H.; Takahashi, T.; Nomoto, K.; Kawashima, K.; Nagata, S.; Yamashiro, Y. Ontogenesis of the gut microbiota composition in healthy, full-Term, vaginally born and breast-fed infants over the first 3 years of life: A quantitative bird’s-eye view. Front. Microbiol. 2017, 8, 1388. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Tillotson, G.; MacKenzie, T.N.; Warren, C.A.; Wexler, H.M.; Goldstein, E.J.C. Bacteroides and related species: The keystone taxa of the human gut microbiota. Anaerobe 2024, 85, 102819. [Google Scholar] [CrossRef] [PubMed]
- Tufail, M.A.; Schmitz, R.A. Exploring the probiotic potential of Bacteroides spp. within One Health paradigm. Probiotics Antimicrob. Proteins 2025, 17, 681–704. [Google Scholar] [CrossRef]
- Alvarez, C.A.; Jones, M.B.; Hambor, J.; Cobb, B.A. Characterization of polysaccharide A response reveals interferon responsive gene signature and immunomodulatory marker expression. Front. Immunol. 2020, 11, 556813. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qie, J.; Zhu, D.; Zhang, X.; Zhang, Q.; Xu, Y.; Wang, Y.; Mi, K.; Pei, Y.; Liu, Y.; et al. The landscape in the gut microbiome of long-lived families reveals new insights on longevity and aging—relevant neural and immune function. Gut Microbes 2022, 14, 2107288. [Google Scholar] [CrossRef]
- La-Ongkham, O.; Nakphaichit, M.; Leelavatcharamas, V.; Keawsompong, S.; Nitisinprasert, S. Distinct gut microbiota of healthy children from two different geographic regions of Thailand. Arch. Microbiol. 2015, 197, 561–573. [Google Scholar] [CrossRef]
- Kahleova, H.; Rembert, E.; Alwarith, J.; Yonas, W.N.; Tura, A.; Holubkov, R.; Agnello, M.; Chutkan, R.; Barnard, N.D. Effects of a low-fat vegan diet on gut microbiota in overweight individuals and relationships with body weight, body composition, and insulin sensitivity. A randomized clinical trial. Nutrients 2020, 12, 2917. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, D.; Zhou, G.; Li, C. Dietary pattern, gut microbiota, and Alzheimer’s disease. J. Agric. Food Chem. 2020, 68, 12800–12809. [Google Scholar] [CrossRef]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef]
- Seicaru, E.M.; Popa Ilie, I.R.; Cătinean, A.; Crăciun, A.M.; Ghervan, C. Enhancing metformin effects by adding gut microbiota modulators to ameliorate the metabolic status of obese, insulin-resistant hosts. J. Gastrointestin Liver Dis. 2022, 31, 344–354. [Google Scholar] [CrossRef]
- Ochoa-Repáraz, J.; Mielcarz, D.W.; Ditrio, L.E.; Burroughs, A.R.; Begum-Haque, S.; Dasgupta, S.; Kasper, D.L.; Kasper, L.H. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J. Immunol. 2010, 185, 4101–4108. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L.; Jones, M.B.; Cobb, B.A. Bacterial capsular polysaccharide prevents the onset of asthma through T-cell activation. Glycobiology 2015, 25, 368–375. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, D.; Zhao, X.; Luo, Y.; Yu, H.; Zhou, Y.; Gao, Y.; Han, X.; Duan, Y.; Fang, N.; et al. Bacteroides fragilis prevents aging-related atrial fibrillation in rats via regulatory T cells-mediated regulation of inflammation. Pharm. Res. 2022, 177, 10614. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Pan, Y.; Xia, X.; Liang, J.; Liu, F.; Dou, H.; Hou, Y. Bacteroides fragilis alleviates the symptoms of lupus nephritis via regulating CD1d and CD86 expressions in B cells. Eur. J. Pharm. 2020, 884, 173421. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Vernay, T.; Cannie, I.; Gaboriau, F.; Gall, F.D.L.; Tamanai-Shacoori, Z.; Burel, A.; Jolivet-Gougeon, A.; Loréal, O.; Bousarghin, L. Bacteroides fragilis prevents Salmonella Heidelberg translocation in co-culture model mimicking intestinal epithelium. Benef. Microbes 2020, 11, 391–401. [Google Scholar] [CrossRef]
- Li, Z.; Deng, H.; Zhou, Y.; Tan, Y.; Wang, X.; Han, Y.; Liu, Y.; Wang, Y.; Yang, R.; Bi, Y.; et al. Bioluminescence imaging to track Bacteroides fragilis Inhibition of Vibrio parahaemolyticus infection in mice. Front. Cell Infect. Microbiol. 2017, 7, 170. [Google Scholar] [CrossRef]
- Fan, H.; Chen, Z.; Lin, R.; Liu, Y.; Wu, X.; Puthiyakunnon, S.; Wang, Y.; Zhu, B.; Zhang, Q.; Bai, Y.; et al. Bacteroides fragilis strain ZY-312 defense against cronobacter sakazakii-induced necrotizing enterocolitis in vitro and in a neonatal rat model. mSystems 2019, 4, e00305-19. [Google Scholar] [CrossRef]
- Sack, R.B.; Myers, L.L.; Almeido-Hill, J.; Shoop, D.S.; Bradbury, W.C.; Reid, R.; Santosham, M. Enterotoxigenic Bacteroides fragilis: Epidemiologic studies of its role as a human diarrhoeal pathogen. J. Diarrhoeal Dis. Res. 1992, 10, 4–9. [Google Scholar] [PubMed]
- Sack, R.B.; Albert, M.J.; Alam, K.; Neogi, P.K.; Akbar, M.S. Isolation of enterotoxigenic Bacteroides fragilis from Bangladeshi children with diarrhea: A controlled study J. Clin. Microbiol. 1994, 32, 960–963. [Google Scholar] [CrossRef]
- Zhang, G.; Svenungsson, B.; Karnell, A.; Weintraub, A. Prevalence of enterotoxigenic Bacteroides fragilis in adult patients with diarrhea and healthy controls. Clin. Infect. Dis. 1999, 29, 590–594. [Google Scholar] [CrossRef]
- San Joaquin, V.H.; Griffis, J.C.; Lee, C.; Sears, C.L. Association of Bacteroides fragilis with childhood diarrhea. Scand. J. Infect. Dis. 1995, 27, 211–215. [Google Scholar] [CrossRef]
- Pathela, P.; Hasan, K.Z.; Roy, E.; Alam, K.; Huq, F.; Siddique, A.K.; Sack, R.B. Enterotoxigenic Bacteroides fragilis-associated diarrhea in children 0-2 years of age in rural Bangladesh. J. Infect. Dis. 2005, 191, 1245–1252. [Google Scholar] [CrossRef][Green Version]
- Vu Nguyen, T.; Le Van, P.; Le Huy, C.; Weintraub, A. Diarrhea caused by enterotoxigenic Bacteroides fragilis in children less than 5 years of age in Hanoi Vietnam. Anaerobe 2005, 11, 109–114. [Google Scholar] [CrossRef]
- Durmaz, B.; Dalgalar, M.; Durmaz, R. Prevalence of enterotoxigenic Bacteroides fragilis in patients with diarrhea: A controlled study. Anaerobe 2005, 11, 318–321. [Google Scholar] [CrossRef]
- Rivera-Chavez, F.; Mekalanos, J.J. Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 2019, 572, 244–248. [Google Scholar] [CrossRef]
- Russell, A.B.; Wexler, A.G.; Harding, B.N.; Whitney, J.C.; Bohn, A.J.; Goo, Y.A.; Tran, B.Q.; Barry, N.A.; Zheng, H.; Peterson, S.B.; et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 2014, 16, 227–236. [Google Scholar] [CrossRef]
- Chatzidaki-Livanis, M.; Geva-Zatorsky, N.; Comstock, L.E. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl. Acad. Sci. USA 2016, 113, 3627–3632. [Google Scholar] [CrossRef] [PubMed]
- Coyne, M.J.; Roelofs, K.G.; Comstock, L.E. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genom. 2016, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Pierce, J.V.; Bernstein, H.D. Genomic diversity of enterotoxigenic strains of Bacteroides fragilis. PLoS ONE 2016, 11, e0158171. [Google Scholar] [CrossRef] [PubMed]
- Gemmell, C.G.; Peterson, P.K.; Schmeling, D.; Mathews, J.; Quie, P.G. Antibiotic-induced modification of Bacteroides fragilis and its susceptibility to phagocytosis by human polymorphonuclear leukocytes. Eur. J. Clin. Microbiol. 1983, 2, 327–334. [Google Scholar] [CrossRef]
- Reid, J.H.; Patrick, S. Phagocytic and serum killing of capsulate and non-capsulate Bacteroides fragilis. J. Med. Microbiol. 1984, 17, 247–257. [Google Scholar] [CrossRef]
- Simon, G.L.; Klempner, M.S.; Kasper, D.L.; Gorbach, S.L. Alterations in opsonophagocytic killing by neutrophils of Bacteroides fragilis associated with animal and laboratory passage: Effect of capsular polysaccharide. J. Infect. Dis. 1982, 145, 72–77. [Google Scholar] [CrossRef]
- Krinos, C.M.; Coyne, M.J.; Weinacht, K.G.; Tzianabos, A.O.; Kasper, D.L.; Comstock, L.E. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 2001, 414, 555–558. [Google Scholar] [CrossRef]
- Coyne, M.J.; Kalka-Moll, W.; Tzianabos, A.O.; Kasper, D.L.; Comstock, L.E. Bacteroides fragilis NCTC9343 produces at least three distinct capsular polysaccharides: Cloning, characterization, and reassignment of polysaccharide B and C biosynthesis loci. Infect. Immun. 2000, 68, 6176–6181. [Google Scholar] [CrossRef] [PubMed]
- Vieira, J.M.; Vallim, D.C.; Ferreira, E.O.; Seabra, S.H.; Vommaro, R.C.; Avelar, K.E.; De, S.W.; Ferreira, M.C.; Domingues, R.M. Bacteroides fragilis interferes with iNOS activity and leads to pore formation in macrophage surface. Biochem. Biophys. Res. Commun. 2005, 326, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Coyne, M.J.; Tzianabos, A.O.; Mallory, B.C.; Carey, V.J.; Kasper, D.L.; Comstock, L.E. Polysaccharide biosynthesis locus required for virulence of Bacteroides fragilis. Infect. Immun. 2001, 69, 4342–4350. [Google Scholar] [CrossRef]
- Kalka-Moll, W.M.; Wang, Y.; Comstock, L.E.; Gonzalez, S.E.; Tzianabos, A.O.; Kasper, D.L. Immunochemical and biological characterization of three capsular polysaccharides from a single Bacteroides fragilis strain. Infect. Immun. 2001, 69, 2339–2344. [Google Scholar] [CrossRef]
- Ramírez-Pérez, O.; Cruz-Ramón, V.; Chinchilla-López, P.; Méndez-Sánchez, N. The role of the gut microbiota in bile acid metabolism. Ann. Hepatol. 2017, 16 (Suppl. S1), s15–s20. [Google Scholar] [CrossRef]
- Huang, Y.; Cao, J.; Zhu, M.; Wang, Z.; Jin, Z.; Xiong, Z. Bacteroides fragilis aggravates high-fat diet-induced non-alcoholic fatty liver disease by regulating lipid metabolism and remodeling gut microbiota. Microbiol. Spectr. 2024, 12, e0339323. [Google Scholar] [CrossRef]
- Long, S.L.; Gahan, C.G.M.; Joyce, S.A. Interactions between gut bacteria and bile in health and disease. Mol. Aspects Med. 2017, 56, 54–65. [Google Scholar] [CrossRef]
- Cai, J.; Rimal, B.; Jiang, C.; Chiang, J.Y.L.; Patterson, A.D. Bile acid metabolism and signaling, the microbiota, and metabolic disease. Pharmacol. Ther. 2022, 237, 108238. [Google Scholar] [CrossRef]
- Pumbwe, L.; Skilbeck, C.A.; Nakano, V.; Avila-Campos, M.J.; Piazza, R.M.; Wexler, H.M. Bile salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb. Pathog. 2007, 43, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Tang, L.; Li, S.; Liu, S.; He, J.; Li, P.; Wang, S.; Yang, M.; Zhang, L.; Lei, Y.; et al. Gut microbiota alters host bile acid metabolism to contribute to intrahepatic cholestasis of pregnancy. Nat. Commun. 2023, 14, 1305. [Google Scholar] [CrossRef] [PubMed]
- Sofi, M.H.; Johnson, B.M.; Gudi, R.R.; Jolly, A.; Gaudreau, M.C.; Vasu, C. Polysaccharide A-dependent opposing effects of mucosal and systemic exposures to human gut commensal Bacteroides fragilis in type 1 diabetes. Diabetes 2019, 68, 1975–1989. [Google Scholar] [CrossRef]
- Shi, G.; Lin, Y.; Wu, Y.; Zhou, J.; Cao, L.; Chen, J.; Li, Y.; Tan, N.; Zhong, S. Bacteroides fragilis supplementation deteriorated metabolic dysfunction, inflammation, and aorta atherosclerosis by inducing gut microbiota dysbiosis in animal model. Nutrients 2022, 14, 2199. [Google Scholar] [CrossRef]
- Myers, L.L.; Shoop, D.S.; Stackhouse, L.L.; Newman, F.S.; Flaherty, R.J.; Letson, G.W.; Sack, R.B. Isolation of enterotoxigenic Bacteroides fragilis from humans with diarrhea. J. Clin. Microbiol. 1987, 25, 2330–2333. [Google Scholar] [CrossRef]
- Caceres, M.; Zhang, G.; Weintraub, A.; Nord, C.E. Prevalence and antimicrobial susceptibility of enterotoxigenic Bacteroides fragilis in children with diarrhoea in Nicaragua. Anaerobe 2000, 6, 143–148. [Google Scholar] [CrossRef]
- Sears, C.L. Enterotoxigenic Bacteroides fragilis: A rogue among symbiotes. Clin. Microbiol. Rev. 2009, 22, 349–369. [Google Scholar] [CrossRef]
- Saidi, R.F.; Sears, C.L. Bacteroides fragilis toxin rapidly intoxicates human intestinal epithelial cells (HT29/C1) in vitro. Infect. Immun. 1996, 64, 5029–5034. [Google Scholar] [CrossRef]
- Cohen, S.H.; Shetab, R.; Tang-Feldman, Y.G.; Sarma, P.; Silva, J., Jr.; Prindiville, T.P. Prevalence of enterotoxigenic Bacteroides fragilis in hospital-acquired diarrhea. Diagn. Microbiol. Infect. Dis. 2006, 55, 251–254. [Google Scholar] [CrossRef]
- Sears, C.L.; Islam, S.; Saha, A.; Arjumand, M.; Alam, N.H.; Faruque, A.S.; Salam, M.A.; Shin, J.; Hecht, D.; Weintraub, A.; et al. Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clin. Infect. Dis. 2008, 47, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Patrick, S.; Stewart, L.D.; Damani, N.; Wilson, K.G.; Lutton, D.A.; Larkin, M.J.; Poxton, I.; Brown, R. Immunological detection of Bacteroides fragilis in clinical samples. J. Med. Microbiol. 1995, 43, 99–109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choi, V.M.; Herrou, J.; Hecht, A.L.; Teoh, W.P.; Turner, J.R.; Crosson, S.; Bubeck Wardenburg, J. Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice. Nat. Med. 2016, 22, 563–567. [Google Scholar] [CrossRef]
- Lukiw, W.J.; Arceneaux, L.; Li, W.; Bond, T.; Zhao, Y. Gastrointestinal (GI)-tract microbiome derived neurotoxins and their potential contribution to inflammatory neurodegeneration in Alzheimer’s Disease (AD). J. Alzheimers Dis. Parkinsonism 2021, 11, 525. [Google Scholar]
- Lukiw, W.J. Bacteroides fragilis lipopolysaccharide and inflammatory signaling in Alzheimer’s disease. Front. Microbiol. 2016, 7, 1544. [Google Scholar] [CrossRef]
- Xia, S.; Ma, L.; Li, H.; Li, Y.; Yu, L. Prevalence of enterotoxigenic Bacteroides fragilis in patients with colorectal cancer: A systematic review and meta-analysis. Front. Cell Infect. Microbiol. 2025, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Scott, N.; Whittle, E.; Jeraldo, P.; Chia, N. A systemic review of the role of enterotoxic Bacteroides fragilis in colorectal cancer. Neoplasia 2022, 29, 100797. [Google Scholar] [CrossRef] [PubMed]
- Ogane, K.; Tarumoto, N.; Kodana, M.; Onodera, A.; Imai, K.; Sakai, J.; Kawamura, T.; Takeuchi, S.; Murakami, T.; Mitsutake, K.; et al. Antimicrobial susceptibility and prevalence of resistance genes in Bacteroides fragilis isolated from blood culture bottles in two tertiary care hospitals in Japan. Anaerobe 2020, 64, 102215. [Google Scholar] [CrossRef]
- Jasemi, S.; Emaneini, M.; Ahmadinejad, Z.; Fazeli, M.S.; Sechi, L.A.; Sadeghpour Heravi, F.; Feizabadi, M.M. Antibiotic resistance pattern of Bacteroides fragilis isolated from clinical and colorectal specimens. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 27. [Google Scholar] [CrossRef]
- Pumbwe, L.; Wareham, D.W.; Aduse-Opoku, J.; Brazier, J.S.; Wexler, H.M. Genetic analysis of mechanisms of multidrug resistance in a clinical isolate of Bacteroides fragilis. Clin. Microbiol. Infect. 2007, 13, 183–189. [Google Scholar] [CrossRef]
- Veloo, A.C.M.; Baas, W.H.; Haan, F.J.; Coco, J.; Rossen, J.W. Prevalence of antimicrobial resistance genes in Bacteroides spp. and Prevotella spp. Dutch clinical isolates. Clin. Microbiol. Infect. 2019, 25, 1156.e9–1156.e13. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, N.B.; Vlamakis, H.; Hayes, K.; Salyers, A.A. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 2001, 67, 561–568. [Google Scholar] [CrossRef]
- Herin, O.; Hedberg, M.; Edlund, C. Efflux-mediated fluoroquinolone resistance in the Bacteroides fragilis group. Anaerobe 2002, 8, 277–282. [Google Scholar] [CrossRef]
- Yekani, M.; Rezaee, M.A.; Beheshtirouy, S.; Baghi, H.B.; Bazmani, A.; Farzinazar, A.; Memar, M.Y.; Sóki, J. Carbapenem resistance in Bacteroides fragilis: A review of molecular mechanisms. Anaerobe 2022, 76, 102606. [Google Scholar] [CrossRef]
- Ghotaslou, R.; Bannazadeh Baghi, H.; Alizadeh, N.; Yekani, M.; Arbabi, S.; Memar, M.Y. Mechanisms of Bacteroides fragilis resistance to metronidazole. Infect. Genet. Evol. 2018, 64, 156–163. [Google Scholar] [CrossRef]
- Roh, K.H.; Kim, S.; Kim, C.K.; Yum, J.H.; Kim, M.S.; Yong, D.; Jeong, S.H.; Lee, K.; Kim, J.M.; Chong, Y. New cfiA variant and novel insertion sequence elements in carbapenem-resistant Bacteroides fragilis isolates from Korea. Diagn. Microbiol. Infect. Dis. 2010, 66, 343–348. [Google Scholar] [CrossRef]
- Steffens, L.S.; Nicholson, S.; Paul, L.V.; Nord, C.E.; Patrick, S.; Abratt, V.R. Bacteroides fragilis RecA protein overexpression causes resistance to metronidazole. Res. Microbiol. 2010, 161, 346–354. [Google Scholar] [CrossRef]
- Ghotaslou, R.; Yekani, M.; Memar, M.Y. The role of efflux pumps in Bacteroides fragilis resistance to antibiotics. Microbiol. Res. 2018, 210, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Veeranagouda, Y.; Husain, F.; Boente, R.; Moore, J.; Smith, C.J.; Rocha, E.R.; Patrick, S.; Wexler, H.M. Deficiency of the ferrous iron transporter FeoAB is linked with metronidazole resistance in Bacteroides fragilis. J. Antimicrob. Chemother. 2014, 69, 2634–2643. [Google Scholar] [CrossRef] [PubMed]
- Nagy, E.; Földes, J. Inactivation of metronidazole by Enterococcus faecalis J. Antimicrob. Chemother. 1991, 27, 63–70. [Google Scholar] [CrossRef]
- Boyanova, L.; Markovska, R.; Mitov, I. Multidrug resistance in anaerobes. Future Microbiol. 2019, 14, 1055–1064. [Google Scholar] [CrossRef]
- Kierzkowska, M.; Majewska, A.; Szymanek-Majchrzak, K.; Sawicka-Grzelak, A.; Mlynarczyk, A.; Mlynarczyk, G. The presence of antibiotic resistance genes and bft genes as well as antibiotic susceptibility testing of Bacteroides fragilis strains isolated from inpatients of the Infant Jesus Teaching Hospital, Warsaw during 2007–2012. Anaerobe 2019, 56, 109–115. [Google Scholar] [CrossRef]
- Jamal, W.; Khodakhast, F.B.; AlAzmi, A.; Sόki, J.; AlHashem, G.; Rotimi, V.O. Prevalence and antimicrobial susceptibility of enterotoxigenic extra-intestinal Bacteroides fragilis among 13-year collection of isolates in Kuwait. BMC Microbiol. 2020, 20, 14. [Google Scholar] [CrossRef]
- Wallace, M.J.; Jean, S.; Wallace, M.A.; Burnham, C.D.; Dantas, G. Comparative genomics of Bacteroides fragilis group isolates reveals species-dependent resistance mechanisms and validates clinical tools for resistance prediction. mBio 2022, 13, e0360321. [Google Scholar] [CrossRef]
- Rotstein, O.D.; Kao, J.; Houston, K. Reciprocal synergy between Escherichia coli and Bacteroides fragilis in an intra-abdominal infection model. J. Med. Microbiol. 1989, 29, 269–276. [Google Scholar] [CrossRef]
- Sommese, L.; Pagliuca, C.; Avallone, B.; Ippolito, R.; Casamassimi, A.; Costa, V.; Colicchio, R.; Cerciello, R.; D’Armiento, M.; Scarpato, M.; et al. Evidence of Bacteroides fragilis protection from Bartonella henselae-induced damage. PLoS ONE 2012, 7, e49653. [Google Scholar] [CrossRef]
- Hassall, J.; Cheng, J.K.J.; Unnikrishnan, M. Dissecting individual interactions between pathogenic and commensal bacteria within a multispecies gut microbial community. mSphere 2021, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Anonye, B.O.; Hassall, J.; Patient, J.; Detamornrat, U.; Aladdad, A.M.; Schuller, S.; Rose, F.; Unnikrishnan, M. Probing Clostridium difficile infection in complex human gut cellular models. Front. Microbiol. 2019, 10, 879. [Google Scholar] [CrossRef]
- Jasemi, S.; Molicotti, P.; Fais, M.; Cossu, I.; Simula, E.R.; Sechi, L.A. Biological mechanisms of enterotoxigenic Bacteroides fragilis toxin: Linking inflammation, colorectal cancer, and clinical implications. Toxins 2025, 17, 305. [Google Scholar] [CrossRef] [PubMed]
- Elliott, B.; Androga, G.O.; Knight, D.R.; Riley, T.V. Clostridium difficile infection: Evolution, phylogeny and molecular epidemiology. Infect. Genet. Evol. 2017, 49, 1–11. [Google Scholar] [CrossRef]
- Longtin, Y.; Gilca, R.; Loo, V.G. Effect of detecting and isolating asymptomatic Clostridium difficile carriers-reply. JAMA Intern. Med. 2016, 176, 1573. [Google Scholar] [CrossRef] [PubMed]
- Le Monnier, A.; Candela, T.; Mizrahi, A.; Bille, E.; Bourgeois-Nicolaos, N.; Cattoir, V.; Farfour, E.; Grall, I.; Lecointe, D.; Limelette, A.; et al. One-day prevalence of asymptomatic carriage of toxigenic and non-toxigenic Clostridioides difficile in 10 French hospitals. J. Hosp. Infect. 2022, 129, 65–74. [Google Scholar] [CrossRef]
- Schubert, A.M.; Sinani, H.; Schloss, P.D. Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. mBio 2015, 6, e00974-15. [Google Scholar] [CrossRef]
- Haak, B.W.; Lankelma, J.M.; Hugenholtz, F.; Belzer, C.; de Vos, W.M.; Wiersinga, W.J. Long-term impact of oral vancomycin, ciprofloxacin and metronidazole on the gut microbiota in healthy humans. J. Antimicrob. Chemother. 2019, 74, 782–786. [Google Scholar] [CrossRef]
- Mougiou, D.; Gioula, G.; Skoura, L.; Anastassopoulou, C.; Kachrimanidou, M. Insights into the interaction between Clostridioides difficile and the gut microbiome. J. Pers. Med. 2025, 15, 94. [Google Scholar] [CrossRef]
- Hopkins, M.J.; Macfarlane, G.T. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J. Med. Microbiol. 2002, 51, 448–454. [Google Scholar] [CrossRef]
- Theriot, C.M.; Bowman, A.A.; Young, V.B. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 2016, 1, 16. [Google Scholar] [CrossRef] [PubMed]
- Winston, J.A.; Theriot, C.M. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 2016, 41, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Thanissery, R.; Winston, J.A.; Theriot, C.M. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile cids. Anaerobe 2017, 45, 86–100. [Google Scholar] [CrossRef]
- Leffler, D.A.; Lamont, J.T. Clostridium difficile infection. N. Engl. J. Med. 2015, 373, 287–288. [Google Scholar] [PubMed]
- Eeuwijk, J.; Ferreira, G.; Yarzabal, J.P.; Robert-Du Ry van Beest Holle, M. A systematic literature review on risk factors for and timing of Clostridioides difficile infection in the United States. Infect. Dis. Ther. 2024, 13, 273–298. [Google Scholar] [CrossRef]
- Braun, V.; Hundsberger, T.; Leukel, P.; Sauerborn, M.; von Eichel-Streiber, C. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 1996, 81, 29–38. [Google Scholar] [CrossRef]
- Cohen, S.H.; Tang, Y.J.; Silva, J. Analysis of the pathogenicity locus in Clostridium difficile strains. J. Infect. Dis. 2000, 181, 659–663. [Google Scholar] [CrossRef]
- Chandra, H.; Sorg, J.A.; Hassett, D.J.; Sun, X. Regulatory transcription factors of Clostridioides difficile pathogenesis with a focus on toxin regulation. Crit. Rev. Microbiol. 2023, 49, 334–349. [Google Scholar] [CrossRef]
- Aktories, K.; Schwan, C.; Jank, T. Clostridium difficile toxin biology. Annu. Rev. Microbiol. 2017, 71, 281–307. [Google Scholar] [CrossRef]
- Gerding, D.N.; Johnson, S.; Rupnik, M.; Aktories, K. Clostridium difficile binary toxin CDT: Mechanism, epidemiology, and potential clinical importance. Gut Microbes 2014, 5, 15–27. [Google Scholar] [CrossRef]
- Valiente, E.; Cairns, M.D.; Wren, B.W. The Clostridium difficile PCR ribotype 027 lineage: A pathogen on the move. Clin. Microbiol. Infect. 2014, 20, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Smits, W.K. Hype or hypervirulence: A reflection on problematic C. difficile strains. Virulence 2013, 4, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Villalobos, N.A.; Ruiz-Hernandez, F.G.; Méndez-Arellano, A.C.; Azamar-Márquez, J.M.; Camacho-Ortiz, A. Epidemiologic profile of community-acquired Clostridioides difficile infections: A systematic review and meta-analysis. Epidemiol. Infect. 2025, 153, e46. [Google Scholar] [CrossRef]
- Buddle, J.E.; Fagan, R.P. Pathogenicity and virulence of Clostridioides difficile. Virulence 2023, 14, 2150452. [Google Scholar] [CrossRef]
- Markovska, R.; Dimitrov, G.; Gergova, R.; Boyanova, L. Clostridioides difficile, a New “Superbug”. Microorganisms 2023, 11, 845. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.A.; Anderson, D.J. Hospital Infection Control: Clostridioides difficile. Clin. Colon. Rectal Surg. 2020, 33, 98–108. [Google Scholar] [CrossRef]
- Chilton, C.H.; Pickering, D.S.; Freeman, J. Microbiologic factors affecting Clostridium difficile recurrence. Clin. Microbiol. Infect. 2018, 24, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Porcari, S.; Baunwall, S.M.D.; Occhionero, A.S.; Ingrosso, M.R.; Ford, A.C.; Hvas, C.L.; Gasbarrini, A.; Cammarota, G.; Ianiro, G. Fecal microbiota transplantation for recurrent C. difficile infection in patients with inflammatory bowel disease: A systematic review and meta-analysis. J. Autoimmun. 2023, 141, 103036. [Google Scholar] [CrossRef]
- Ressler, A.M.; Rao, K.; Young, V.B. Current approaches to treat and prevent recurrence of Clostridioides difficile. Gastroenterol. Clin. N. Am. 2025, 54, 259–275. [Google Scholar] [CrossRef]
- Gupta, A.; Ananthakrishnan, A.N. Economic burden and cost-effectiveness of therapies for Clostridiodes difficile infection: A narrative review. Therap. Adv. Gastroenterol. 2021, 14, 17562848211018654. [Google Scholar] [CrossRef]
- Kartalidis, P.; Skoulakis, A.; Tsilipounidaki, K.; Florou, Z.; Petinaki, E.; Fthenakis, G.C. Clostridioides difficile as a dynamic vehicle for the dissemination of antimicrobial-resistance determinants: Review and in silico analysis. Microorganisms 2021, 25, 1383. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, E.; Amir, I.; Zafran, M.; Gophna, U.; Samra, Z.; Pitlik, S.; Bishara, J. The correlation between Clostridium-difficile infection and human gut concentrations of Bacteroidetes phylum and clostridial species. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 377–383. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, B.; Xu, J.; Liu, Y.; Qiu, E.; Li, Z.; Li, Z.; He, Y.; Zhou, H.; Bai, Y.; et al. Bacteroides fragilis protects against antibiotic-associated diarrhea in rats by modulating intestinal defenses. Front. Immunol. 2018, 9, 1040. [Google Scholar] [CrossRef]
- Frost, L.R.; Cheng, J.K.J.; Unnikrishnan, M. Clostridioides difficile biofilms: A mechanism of persistence in the gut? PLoS Pathog. 2021, 17, e1009348. [Google Scholar] [CrossRef]
- Slater, R.T.; Frost, L.R.; Jossi, S.E.; Millard, A.D.; Unnikrishnan, M. Clostridioides difficile LuxS mediates inter-bacterial interactions within biofilms. Sci. Rep. 2019, 9, 9903. [Google Scholar] [CrossRef]
- Hardie, K.R.; Heurlier, K. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer 2 in biofilm development. Nat. Rev. Microbiol. 2008, 6, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.M.; Sorg, J.A. Gut associated metabolites and their roles in Clostridioides difficile pathogenesis. Gut Microb. 2022, 14, 2094672. [Google Scholar] [CrossRef]
- Sorg, J.A.; Sonenshein, A.L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008, 190, 2505–2512. [Google Scholar] [CrossRef]
- Foley, M.H.; Walker, M.E.; Stewart, A.K.; O’Flaherty, S.; Gentry, E.C.; Patel, S.; Beaty, V.V.; Allen, G.; Pan, M.; Simpson, J.B.; et al. Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut. Nat. Microbiol. 2023, 8, 611–628. [Google Scholar] [CrossRef] [PubMed]
- Mullish, B.H.; McDonald, J.A.K.; Pechlivanis, A.; Allegretti, J.R.; Kao, D.; Barker, G.F.; Kapila, D.; Petrof, E.O. Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut 2019, 68, 1791–1800. [Google Scholar] [CrossRef]
- Gupta, S.; Allen-Vercoe, E.; Petrof, E.O. Fecal microbiota transplantation: In perspective. Therap. Adv. Gastroenterol. 2016, 9, 229–239. [Google Scholar] [CrossRef]
- Seekatz, A.M.; Theriot, C.M.; Rao, K.; Chang, Y.M.; Freeman, A.E.; Kao, J.Y.; Young, V.B. Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe 2018, 53, 64–73. [Google Scholar] [CrossRef]
- Imwattana, K.; Kiratisin, P.; Riley, T.V. Antimicrobial-resistant Bacteroides fragilis in Thailand and their inhibitory effect in vitro on the growth of Clostridioides difficile. Anaerobe 2022, 73, 102505. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.; Silver, H.J.; Hazleton, K.; Lozupone, C.; Nicholson, M.R. The impact of diet on Clostridioides difficile infection: A review. J. Infect. Dis. 2025, 231, e1010–e1018. [Google Scholar] [CrossRef] [PubMed]
- Singh, C.; Singh, A.; Singh, D.; Upadhyay, R. Potential therapeutic solution for Clostridioides difficile infection: Current scenario and future prospects. Med. Microecol. 2025, 24, 100121. [Google Scholar] [CrossRef]
- Lewis, S.; Burmeister, S.; Brazier, J. Effect of the prebiotic oligofructose on relapse of Clostridium difficile–associated diarrhea: A randomized, controlled study. Clin. Gastroenterol. Hepatol. 2005, 3, 442–448. [Google Scholar] [CrossRef]
- Piotrowski, M.; Wultańska, D.; Pituch, H. Effect of prebiotics on Bacteroides sp. adhesion and biofilm formation and synbiotic effect on Clostridioides difficile. Future Microbiol. 2022, 17, 363–375. [Google Scholar] [CrossRef]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Drewes, J.; Chen, J.; Markham, N.; Knippel, R.; Domingue, J.; Tam, A.; Chan, J.L.; Kim, L.; McMann, M.; Stevens, C.; et al. Human colon cancer-derived Clostridioides difficile strains drive colonic tumorigenesis in mice. Cancer Discov. 2022, 12, 1873–1885. [Google Scholar] [CrossRef]
- Permain, J.; Hock, B.; Eglinton, T.; Purcell, R. Functional links between the microbiome and the molecular pathways of colorectal carcinogenesis. Cancer Metastasis Rev. 2024, 43, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Morley-Bunker, A.; Walker, L.; Currie, M.; Pearson, J.; Eglinton, T. Translating colorectal cancer genetics into clinically useful biomarkers. Color. Dis. 2016, 18, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Conlin, A.; Smith, G.; Carey, F.A.; Wolf, C.R.; Steele, R.J. The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut 2005, 54, 1283–1286. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 2, 1350–1356. [Google Scholar] [CrossRef]
- Purcell, R.V.; Visnovska, M.; Biggs, P.J.; Schmeier, S.; Frizelle, F.A. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci. Rep. 2017, 7, 11590. [Google Scholar] [CrossRef]
- DeDecker, L.; Coppedge, B.; Avelar-Barragan, J.; Karnes, W.; Whiteson, K. Microbiome distinctions between the CRC carcinogenic pathways. Gut Microbes 2021, 13, 1–12. [Google Scholar] [CrossRef]
- Yang, H.; Gan, Y.; Jiang, S.; Zhu, X.; Xia, Y.; Gong, D.; Xie, X.; Gong, Y.; Zhang, Y.; Lei, Q.; et al. Genomic alterations in Bacteroides fragilis favor adaptation in colorectal cancer microenvironment. BMC Genom. 2025, 26, 269. [Google Scholar] [CrossRef]
- Toprak, N.U.; Yagci, A.; Gulluoglu, B.M.; Akin, M.L.; Demirkalem, P.; Celenk, T.; Soyletir, G. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. 2006, 12, 782–786. [Google Scholar] [CrossRef]
- Pandey, H.; Tang, D.W.T.; Wong, S.H.; Lal, D. Gut microbiota in colorectal Cancer: Biological role and therapeutic opportunities. Cancers 2023, 15, 866. [Google Scholar] [CrossRef]
- Khodaverdi, N.; Zeighami, H.; Jalilvand, A.; Haghi, F.; Hesami, N. High frequency of enterotoxigenic Bacteroides fragilis and Enterococcus faecalis in the paraffin-embedded tissues of Iranian colorectal cancer patients. BMC Cancer 2021, 21, 1353. [Google Scholar] [CrossRef]
- Purcell, R.V.; Pearson, J.; Aitchison, A.; Dixon, L.; Frizelle, F.A.; Keenan, J.I. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS ONE 2017, 12, e0171602. [Google Scholar] [CrossRef]
- Moore, W.; Moore, L.H. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 1995, 61, 3202–3207. [Google Scholar] [CrossRef]
- Kingston, D.G.I.; Van Tassell, R.L.; Wilkins, T.D. The fecapentaenes, potent mutagens from human feces. Chem. Res. Toxicol. 1990, 3, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Garvey, M. Intestinal dysbiosis: Microbial imbalance impacts on colorectal cancer initiation, progression and disease mitigation. Biomedicines 2024, 12, 740. [Google Scholar] [CrossRef] [PubMed]
- Haghi, F.; Goli, E.; Mirzaei, B.; Zeighami, H. The association between fecal enterotoxigenic B. fragilis with colorectal cancer. BMC Cancer 2019, 19, 879. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Rhee, K.J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, L.; Zhao, H.; Yan, Y.; Lu, J. The role of interleukins in colorectal cancer. Int. J. Biol. Sci. 2020, 16, 2323–2339. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Karin, M. Dangerous liaisons: STAT3 and NF-κB collaboration and crosstalk in cancer. Cytokine Growth Factor. Rev. 2010, 21, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; McLoughlin, R.M.; Cobb, B.A.; Charrel-Dennis, M.; Zaleski, K.J.; Golenbock, D.; Tzianabos, A.O.; Kasper, D.L. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J. Exp. Med. 2006, 203, 2853–2863. [Google Scholar] [CrossRef]
- Thiele Orberg, E.; Fan, H.; Tam, A.J.; Dejea, C.M.; Destefano Shields, C.E.; Wu, S.; Chung, L.; Finard, B.B.; Wu, X.; Fathi, P.; et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017, 10, 421–433. [Google Scholar] [CrossRef]
- Chung, L.; Thiele Orberg, E.; Geis, A.L.; Chan, J.L.; Fu, K.; DeStefano Shields, C.E.; Dejea, C.M.; Fathi, P.; Chen, J.; Finard, B.B.; et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 2018, 23, 203–214. [Google Scholar] [CrossRef]
- Sears, C.L.; Geis, A.L.; Housseau, F. Bacteroides fragilis subverts mucosal biology: From symbiont to colon carcinogenesis. J. Clin. Investig. 2014, 124, 4166–4172. [Google Scholar] [CrossRef]
- Zhang, L.; Shay, J.W. Multiple roles of APC and its therapeutic implications in colorectal cancer J. Natl. Cancer Inst. 2017, 109, djw332. [Google Scholar] [CrossRef]
- Cheng, W.T.; Kantilal, H.K.; Davamani, F. The mechanism of Bacteroides fragilis toxin contributes to colon cancer formation. Malays. J. Med. Sci. 2020, 27, 9–21. [Google Scholar] [CrossRef]
- Allen, J.; Hao, S.; Sears, C.L.; Timp, W. Epigenetic changes induced by Bacteroides fragilis toxin. Infect. Immun. 2019, 87, 12. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, A.; Eger, A.; Wolf, J.; Beug, H.; Foisner, R. E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J. Cell Biol. 2001, 154, 1185–1196. [Google Scholar] [CrossRef]
- Allen, J.; Sears, C.L. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: Contributions to colorectal cancer development. Genome Med. 2019, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef]
- Allen, J.; Rosendahl Huber, A.; Pleguezuelos-Manzano, C.; Puschhof, J.; Wu, S.; Wu, X.; Boot, C.; Saftien, A.; O’Hagan, H.M.; Wang, H.; et al. Colon tumors in enterotoxigenic Bacteroides fragilis (ETBF)-colonized mice do not display a unique mutational signature but instead possess host-dependent alterations in the APC gene. Microbiol. Spectr. 2022, 10, e0105522. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Ting, N.L.; Wong, C.C.; Huang, P.; Jiang, L.; Liu, C.; Lin, Y.; Li, S.; Liu, Y.; Xie, M.; et al. Bacteroides fragilis promotes chemoresistance in colorectal cancer, and its elimination by phage VA7 restores chemosensitivity. Cell Host Microbe 2025, 33, 941–956.e10. [Google Scholar] [CrossRef]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Sittipo, P.; Lobionda, S.; Choi, K.; Sari, I.N.; Kwon, H.Y.; Lee, Y.K. Toll-like receptor 2-mediated suppression of colorectal cancer pathogenesis by polysaccharide A from Bacteroides fragilis. Front. Microbiol. 2018, 9, 1588. [Google Scholar] [CrossRef]
- Lee, Y.K.; Mehrabian, P.; Boyajian, S.; Wu, W.L.; Selicha, J.; Vonderfecht, S.; Mazmanian, S.K. The protective role of Bacteroides fragilis in a murine model of colitis-associated colorectal cancer. mSphere 2018, 3, 11. [Google Scholar] [CrossRef]
- Yang, J.; Wang, X.; Hu, T.; Huang, H.; Chen, G.; Jin, B.; Zeng, G.; Liu, J. Entero-toxigenic Bacteroides fragilis contributes to intestinal barrier injury and colorectal cancer progression by mediating the BFT/STAT3/ZEB2 pathway. Cell Cycle 2024, 23, 70–82. [Google Scholar] [CrossRef]
- Kordahi, M.C.; Stanaway, I.B.; Avril, M.; Chac, D.; Blanc, M.P.; Ross, B.; Diener, C.; Jain, S.; McCleary, P.; Parker, A.; et al. Genomic and functional characterization of a mucosal symbiont involved in early-stage colorectal cancer. Cell Host Microbe 2021, 29, 1589–1598.e6. [Google Scholar] [CrossRef]
- Yang, Y.; Cao, X.; Kato, N.; Wang, Y. Gut Bacteroides fragilis in health and diseases: An updated review. J. Future Foods 2025, in press. [CrossRef]
- Allali, I.; Boukhatem, N.; Bouguenouch, L.; Hardi, H.; Boudouaya, H.A.; Cadenas, M.B.; Ouldim, K.; Amzazi, S.; Azcarate-Peril, M.A.; Ghazal, H. Gut microbiome of Moroccan colorectal cancer patients. Med. Microbiol. Immunol. 2018, 207, 211–225. [Google Scholar] [CrossRef]
- Jahani-Sherafat, S.; Azimirad, M.; Alebouyeh, M.; Amoli, H.A.; Hosseini, P.; Ghasemian-Safaei, H.; Moghim, S. The rate and importance of Clostridium difficile in colorectal cancer patients. Gastroenterol. Hepatol. Bed Bench 2019, 12, 358–363. [Google Scholar] [PubMed]
- Zheng, Y.; Luo, Y.; Lv, Y.; Huang, C.; Sheng, Q.; Zhao, P.; Ye, J.; Jiang, W.; Liu, L.; Song, X.; et al. Clostridium difficile colonization in preoperative colorectal cancer patients. Oncotarget 2017, 8, 11877–11886. [Google Scholar] [CrossRef]
- Armin, S.; Shamsian, S.; Drakhshanfar, H. Colonization with Clostridium difficile in children with cancer. Iran. J. Pediatr. 2013, 23, 473–476. [Google Scholar] [PubMed]
- Magat, E.M.; Balanag, G.A.; CariÑo, A.M.; Fellizar, A.; Ortin, T.S.; Guevarra, L., Jr.; Albano, P.M. Clostridioides difficile antibody response of colorectal cancer patients versus clinically healthy individuals. Biosci. Microbiota Food Health 2020, 39, 123–127. [Google Scholar] [CrossRef]
- Shahbazi, T.; Bakhshi, B.; Rasekhi, A.; Fazeli, M.S.; Fallah, F. Significant presence of Clostridioides difficile in colorectal cancer patients by TaqMan Real-Time PCR. Iran. J. Med. Microbiol. 2024, 18, 106–112. [Google Scholar] [CrossRef]
- Lugito, N.P.; Shin, A.; Kelly, C.P. A 21 year-old male colorectal cancer Clostridium difficile and intestinal amebiasis infection. Indones. J. Cancer 2014, 8, 71. [Google Scholar]
- Fang, W.J.; Jing, D.Z.; Luo, Y.; Fu, C.Y.; Zhao, P.; Qian, J.; Tian, B.R.; Chen, X.G.; Zheng, Y.L.; Zheng, Y.; et al. Clostridium difficile carriage in hospitalized cancer patients: A prospective investigation in eastern China. BMC Infect. Dis. 2014, 14, 523. [Google Scholar] [CrossRef]
- Geier, D.A.; Geier, M.R. Colon cancer risk following intestinal Clostridioides difficile infection: A longitudinal cohort study. J. Clin. Med. Res. 2023, 15, 310. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.K.; Cardeiro, M.; Frankel, L.; Kim, E.; Takabe, K.; Rashid, O.M. Incidence of colorectal cancer after intestinal infection due to Clostridioides difficile. World J. Oncol. 2024, 15, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.M.; Sears, C.L. The role of the gut microbiome in cancer: A review, with special focus on colorectal neoplasia and Clostridioides difficile. Clin. Infect. Dis. 2023, 77 (Suppl. S6), S471–S478. [Google Scholar] [CrossRef]
- Kyne, L.; Warny, M.; Qamar, A.; Kelly, C.P. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N. Engl. J. Med. 2000, 342, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Fellows, R.C.; Chun, S.K.; Larson, N.; Fortin, B.M.; Mahieu, A.L.; Song, W.A.; Seldin, M.M.; Pannunzio, N.R.; Masri, S. Disruption of the intestinal clock drives dysbiosis and impaired barrier function in colorectal cancer. Sci. Adv. 2024, 10, eado1458. [Google Scholar] [CrossRef]
- Kulecka, M.; Zeber-Lubecka, N.; Bałabas, A.; Czarnowski, P.; Bagińska, K.; Głowienka, M.; Kluska, A.; Piątkowska, M.; Dąbrowska, M.; Waker, E.; et al. Diarrheal-associated gut dysbiosis in cancer and inflammatory bowel disease patients is exacerbated by Clostridioides difficile infection. Front. Cell Infect. Microbiol. 2023, 13, 1190910. [Google Scholar] [CrossRef]
- Ozma, M.A.; Fadaee, M.; Hosseini, H.M.; Ataee, M.H.; Mirhosseini, S.A. A critical review of postbiotics as promising novel therapeutic agents for clostridial infections. Probiotics Antimicrob. Proteins 2024, 17, 656. [Google Scholar] [CrossRef]
- Miao, E.A.; Andersen-Nissen, E.; Warren, S.E.; Aderem, A. TLR5 and Ipaf: Dual sensors of bacterial flagellin in the innate immune system. Semin. Immunopathol. 2007, 29, 275–288. [Google Scholar] [CrossRef]
- Sokol, C.L.; Luster, A.D. The chemokine system in innate immunity. Cold Spring Harb. Perspect. Biol. 2015, 7, a016303. [Google Scholar] [CrossRef]
- Mantovani, A.; Romero, P.; Palucka, A.K.; Marincola, F.M. Tumour immunity: Effector response to tumour and role of the microenvironment. Lancet 2008, 371, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Batah, J.; Kobeissy, H.; Bui Pham, P.T.; Denève-Larrazet, C.; Kuehne, S.; Collignon, A.; Janoir-Jouveshomme, C.; Marvaud, J.C.; Kansau, I. Clostridium difficile flagella induce a pro-inflammatory response in intestinal epithelium of mice in cooperation with toxins. Sci. Rep. 2017, 7, 3256. [Google Scholar] [CrossRef]
- Sun, X.; Savidge, T.; Feng, H. The enterotoxicity of Clostridium difficile toxins. Toxins 2010, 2, 1848–1880. [Google Scholar] [CrossRef]
- Chandrasekaran, R.; Lacy, D.B. The role of toxins in Clostridium difficile infection. FEMS Microbiol. Rev. 2017, 41, 723–750. [Google Scholar] [CrossRef]
- Mola, S.; Pandolfo, C.; Sica, A.; Porta, C. The macrophages-microbiota interplay in colorectal cancer (CRC)-related inflammation: Prognostic and therapeutic significance. Int. J. Mol. Sci. 2020, 21, 6866. [Google Scholar] [CrossRef]
- Hu, Y.; Dong, Z.; Liu, K. Unraveling the complexity of STAT3 in cancer: Molecular understanding and drug discovery. J. Exp. Clin. Cancer Res. 2024, 43, 23. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Lalowski, P.; Zielińska, D. The most promising next-generation probiotic candidates-Impact on human health and potential application in food technology. Fermentation 2024, 10, 444. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, Q.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. A potential species of next-generation probiotics? The dark and light sides of Bacteroides fragilis in health. Food Res. Int. 2019, 126, 108590. [Google Scholar] [CrossRef] [PubMed]
- Yamin, D.; Uskoković, V.; Wakil, A.M.; Goni, M.D.; Shamsuddin, S.H.; Mustafa, F.H.; Alfouzan, W.A.; Alissa, M.; Alshengeti, A.; Almaghrabi, R.H.; et al. Current and future technologies for the detection of antibiotic-resistant bacteria. Diagnostics 2023, 13, 3246. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, X.; Xu, C.; Huang, J.; Zhang, L.; Qiu, P.; Zheng, D.; Chen, W.; Zhang, S. Pathogen virulence genes: Advances, challenges and future directions in infectious disease research. Int. J. Mol. Med. 2025, 56, 173. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Important Safety Alert Regarding Use of Fecal Microbiota for Transplantation and Risk of Serious Adverse Reactions Due to Transmission of Multi-Drug Resistant Organisms FDA. 2019. Available online: https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/safety-alert-regarding-use-fecal-microbiota-transplantation-and-risk-serious-adverse-events-likely (accessed on 15 October 2025).
- Ruszkowski, J.; Kachlik, Z.; Walaszek, M.; Storman, D.; Podkowa, K.; Garbarczuk, P.; Jemioło, P.; Łyzińska, W.; Nowakowska, K.; Grych, K.; et al. Fecal microbiota transplantation from patients into animals to establish human microbiota-associated animal models: A scoping review. J. Transl. Med. 2025, 23, 662. [Google Scholar] [CrossRef]
- Gong, D.; Adomako-Bonsu, A.G.; Wang, M.; Li, J. Three specific gut bacteria in the occurrence and development of colorectal cancer: A concerted effort. PeerJ 2023, 11, e15777. [Google Scholar] [CrossRef] [PubMed]
- Aneke-Nash, C.; Yoon, G.; Du, M.; Liang, P. Antibiotic use and colorectal neoplasia: A systematic review and meta-analysis. BMJ Open Gastroenterol. 2021, 8, e000601. [Google Scholar] [CrossRef] [PubMed]
| Commensal Role | |
| NTBF and ETBF strains | |
| Degrade polysaccharides of plants | |
| Produce short-chain fatty acids | |
| Show regulation properties | |
| Show anti-inflammatory properties | |
| Prevent gut dysbiosis | |
| Prevent bacterial infection | |
| Mitigate several diseases | |
| ETBF strains | |
| Abolish B. fragilis toxin (BFT) production or produce a nondamaging BFT in the intestine of carriers, favoring bacterial survival and transmission | |
| Pathogenic role | |
| NTBF and ETBF strains | |
| Show an expansive pangenome characterized by extensive genetic diversity | |
| Show the capability to evade the host immune response Show resistance/multi-resistance to antibiotics | |
| NTBF strains | |
| Contribute to the development and progression of metabolic disorders (obesity and diabetes) and atherosclerotic cardiovascular disease | |
| Induce intra-abdominal abscess development and intrahepatic cholestasis (ICP) | |
| ETBF strains | |
| Cause gastrointestinal infection and intestinal and extra-intestinal abscesses formation | |
| Promote chronic inflammation, neurodegeneration and carcinogenesis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spigaglia, P. The Ambivalent Nature of Bacteroides fragilis and the Interaction with Clostridioides difficile: Benefits and Disadvantages for the Human Host. Toxins 2025, 17, 513. https://doi.org/10.3390/toxins17100513
Spigaglia P. The Ambivalent Nature of Bacteroides fragilis and the Interaction with Clostridioides difficile: Benefits and Disadvantages for the Human Host. Toxins. 2025; 17(10):513. https://doi.org/10.3390/toxins17100513
Chicago/Turabian StyleSpigaglia, Patrizia. 2025. "The Ambivalent Nature of Bacteroides fragilis and the Interaction with Clostridioides difficile: Benefits and Disadvantages for the Human Host" Toxins 17, no. 10: 513. https://doi.org/10.3390/toxins17100513
APA StyleSpigaglia, P. (2025). The Ambivalent Nature of Bacteroides fragilis and the Interaction with Clostridioides difficile: Benefits and Disadvantages for the Human Host. Toxins, 17(10), 513. https://doi.org/10.3390/toxins17100513





