Isolation and Aflatoxin B1-Degradation Characteristics of a Microbacterium proteolyticum B204 Strain from Bovine Faeces
Abstract
:1. Introduction
2. Results
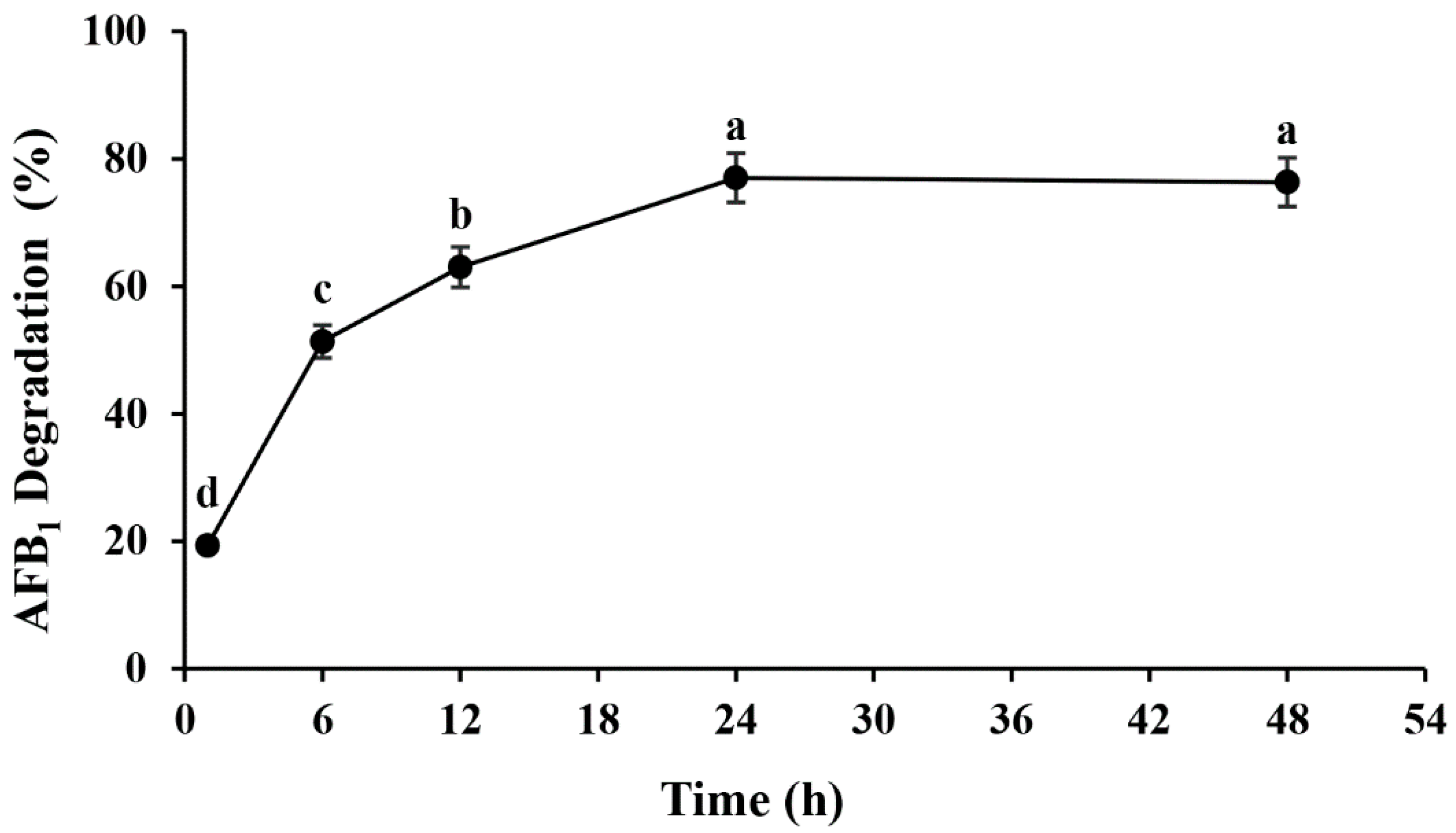
2.1. Isolation, Identification and Characterization of AFB1 Degradation Bacterial Strains
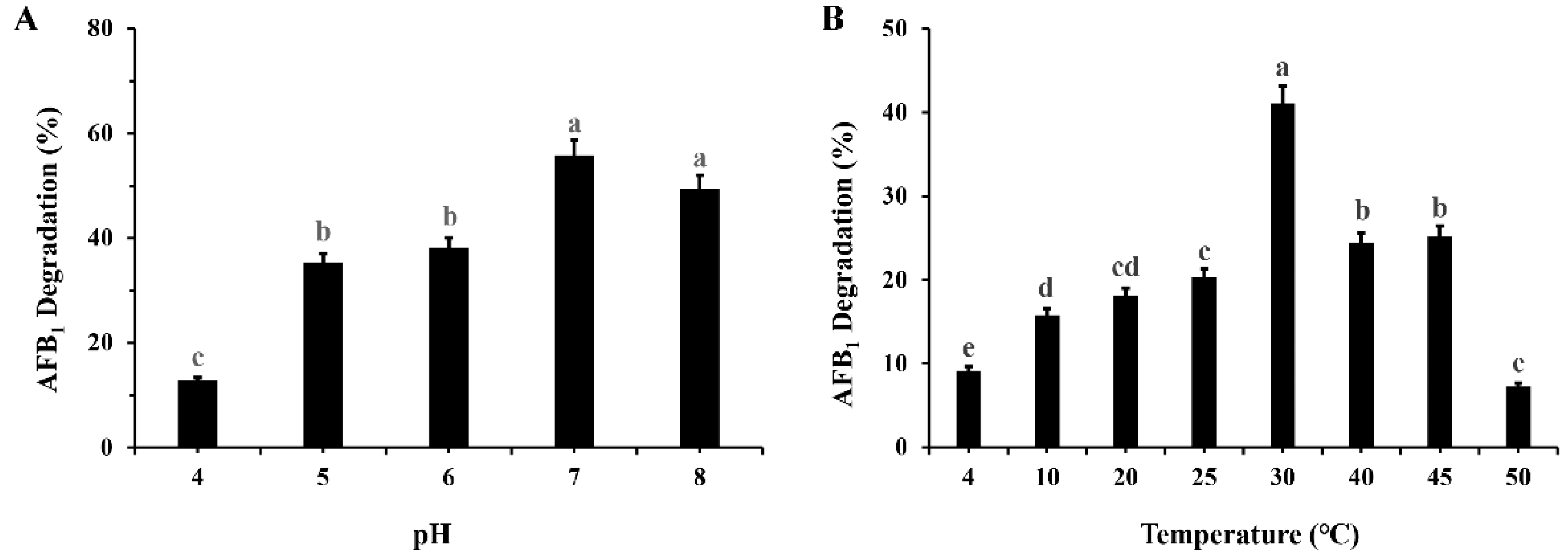
2.2. Effects of Incubation pH and Temperature on AFB1 Degradation by M. proteolyticum B204
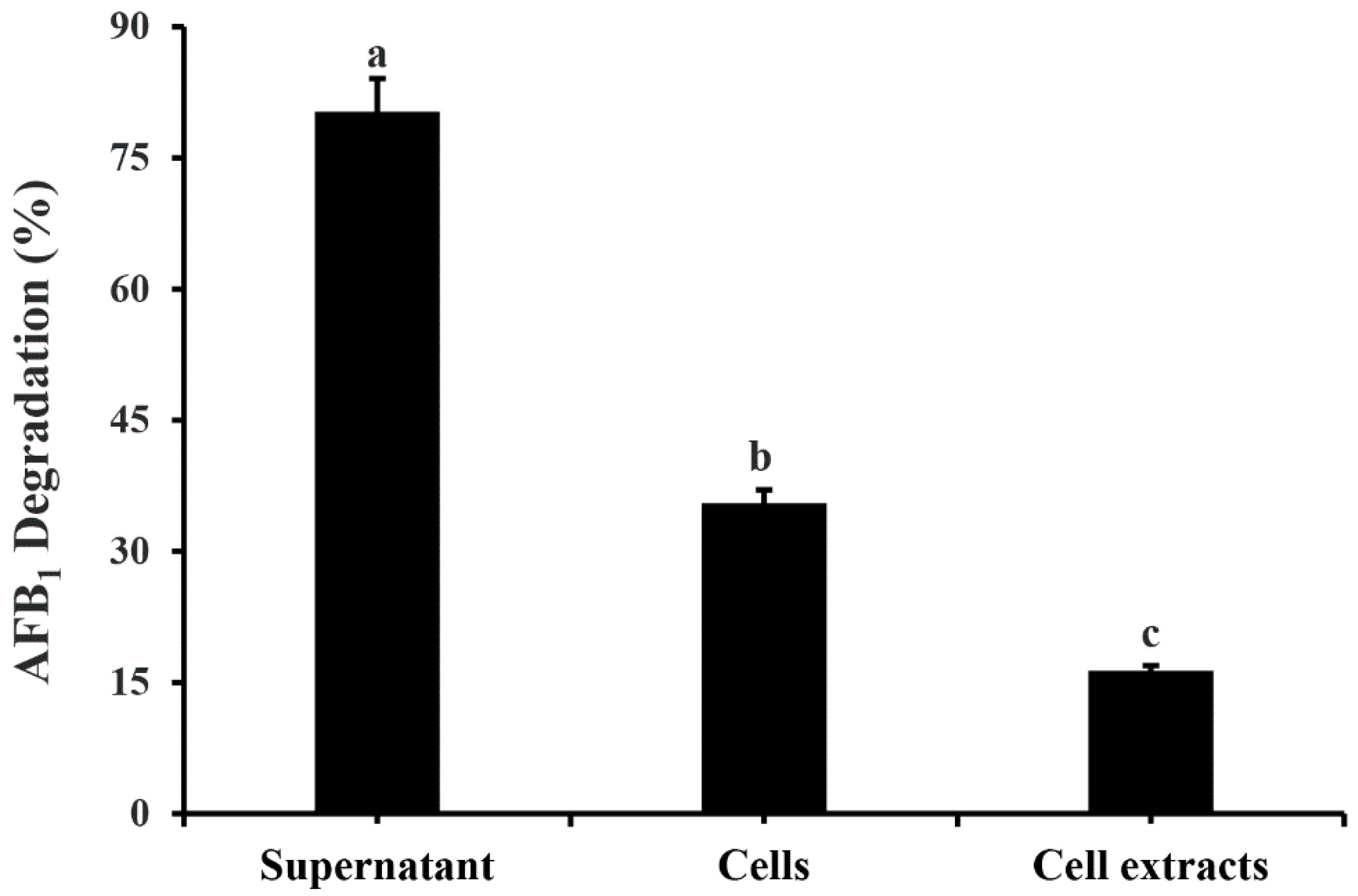
2.3. AFB1 Degradation by Cell-Free Culture Supernatant and Cell Extracts of M. proteolyticum B204
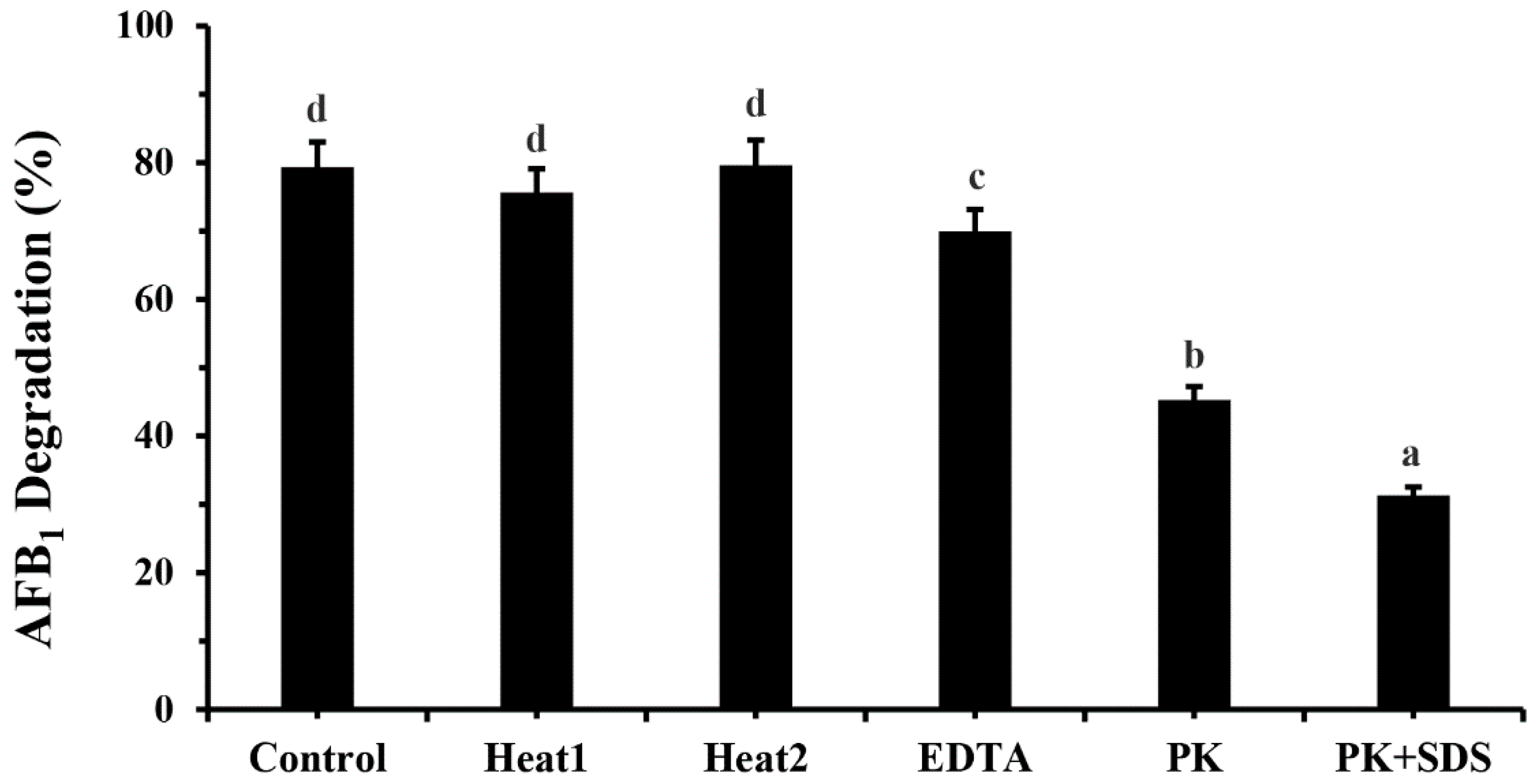
2.4. Effects of Heat Treatment, SDS, Proteinase K and EDTA on AFB1 Degradation by M. proteolyticum B204 Cell Culture Supernatant
2.5. Cytotoxicity Analysis of M. proteolyticum B204
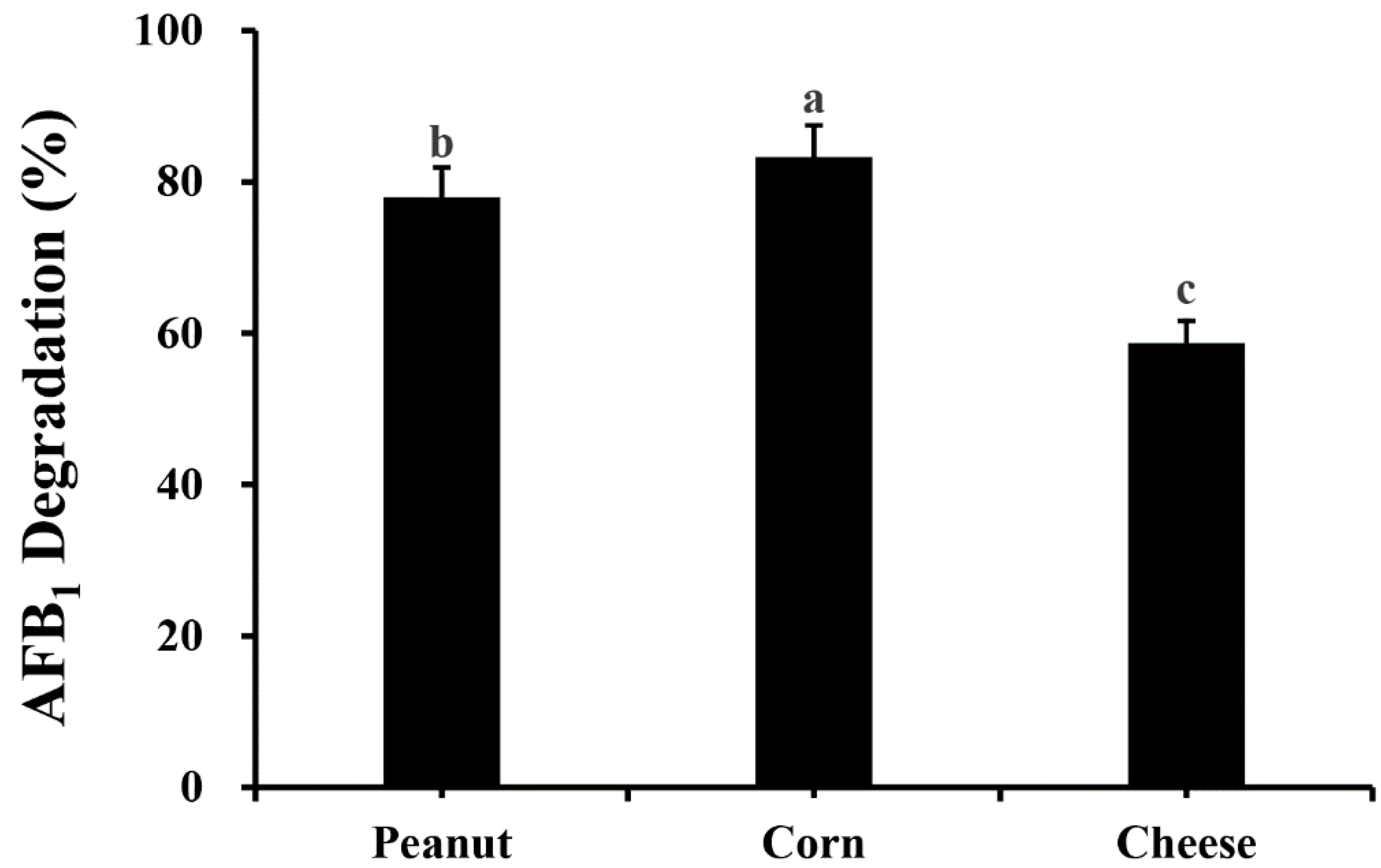
2.6. Application on Food Matrices of AFB1 Detoxification by M. proteolyticum B204
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemical and Medium
5.2. Two-Round Screening and Isolation of AFB1 Degradation Bacterium
5.3. Analysis of AFB1 Degradation Products Using HPLC
area in experimental group)/AFB1 peak area in control group × 100%
5.4. Identification of AFB1 Degradation Bacterium
5.5. Exploration of Culture Conditions by M. proteolyticum B204 on AFB1 Degradation
5.6. AFB1 Degradation by Cell-Free Culture Supernatant, Cell-Free Extracts and Cells
5.7. Effects of Heat Treatment, SDS, Proteinase and EDTA on AFB1 Degradation by Cell-Free Culture Supernatant
5.8. Cytotoxicity Analysis by MTT Assay
5.9. Application of AFB1 Detoxification by M. proteolyticum B204 to Food Matrices
5.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guan, Y.; Chen, J.; Nepovimova, E.; Long, M.; Wu, W.D.; Kuca, K. Aflatoxin Detoxification Using Microorganisms and Enzymes. Toxins 2021, 13, 46. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Q.; Li, Z.Y.; Wang, H.; Qiu, H.Y.; Zhang, M.H.; Li, S.; Luo, X.G.; Song, Y.J.; Zhou, H.; Ma, W.J.; et al. Rapid biodegradation of aflatoxin B1 by metabolites of Fusarium sp. WCQ3361 with broad working temperature range and excellent thermostability. J. Sci. Food Agirc. 2017, 97, 1342–1348. [Google Scholar] [CrossRef] [PubMed]
- Adebo, O.A.; Njobeh, P.B.; Gbashi, S.; Nwinyi, O.C.; Mavumengwana, V. Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. Nutr. 2017, 57, 3208–3217. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.R.N.; Salleh, B.; Saad, B.; Abbas, H.K.; Abel, C.A.; Shier, W.T. An overview of mycotoxin contamination in foods and its implications for human health. Toxin Rev. 2010, 29, 3–26. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, W.; Zhang, S. Purification and identification of an aflatoxin B1 degradation enzyme from Pantoea sp. T6. Toxicon 2019, 157, 35–42. [Google Scholar] [CrossRef]
- Cherkani-Hassani, A.; Ghanname, I.; Zinedine, A.; Sefrioui, H.; Qmichou, Z.; Mouane, N. Aflatoxin M1 prevalence in breast milk in Morocco: Associated factors and health risk assessment of newborns “CONTAMILK study”. Toxicon 2020, 187, 203–208. [Google Scholar] [CrossRef]
- Fang, L.; Chen, H.; Ying, X.; Lin, J.M. Micro-plate chemiluminescence enzyme immunoassay for aflatoxin B1 in agricultural products. Talanta 2011, 84, 216–222. [Google Scholar] [CrossRef]
- El-Nezami, H.; Kankaanpaa, P.; Salminen, S.; Ahokas, J. Ability of dairy strains of lactic acid bacteria to bind a common food carcinogen, aflatoxin B1. Food Chem. Toxicol. 1998, 36, 321–326. [Google Scholar] [CrossRef]
- Arzandeh, S.; Jinap, S. Effect of initial aflatoxin concentration, heating time and roasting temperature on aflatoxin reduction in contaminated peanuts and process optimisation using response surface modelling. Int. J. Food Sci. Technol. 2011, 46, 485–491. [Google Scholar] [CrossRef]
- Yu, Y.; Shi, J.; Xie, B.; He, Y.; Qin, Y.; Wang, D.; Shi, H.; Ke, Y.; Sun, Q. Detoxification of aflatoxin B1 in corn by chlorine dioxide gas. Food Chem. 2020, 328, 127121. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, F.; Liu, M.; Zhou, X.; Wang, M.; Cao, K.; Jin, S.; Shan, A.; Feng, X. Curcumin mitigates aflatoxin B1-induced liver injury via regulating the NLRP3 inflammasome and Nrf2 signaling pathway. Food Chem. Toxicol. 2022, 161, 112823. [Google Scholar] [CrossRef]
- Yang, H.; Wang, Y.; Yu, C.; Jiao, Y.; Zhang, R.; Jin, S.; Feng, X. Dietary Resveratrol Alleviates AFB1-Induced Ileum Damage in Ducks via the Nrf2 and NF-κB/NLRP3 Signaling Pathways and CYP1A1/2 Expressions. Agriculture 2022, 12, 54. [Google Scholar] [CrossRef]
- Aziz, N.H.; Moussa, L.A.A. Influence of gamma-radiation on mycotoxin producing moulds and mycotoxins in fruits. Food Control 2002, 13, 281–288. [Google Scholar] [CrossRef]
- Di Gregorio, M.C.; de Neeff, D.V.; Jager, A.V.; Corassin, C.H.; Carao, A.C.D.; de Albuquerque, R.; de Azevedo, A.C.; Oliveira, C.A.F. Mineral adsorbents for prevention of mycotoxins in animal feeds. Toxin Rev. 2014, 33, 125–135. [Google Scholar] [CrossRef]
- Kabak, B.; Dobson, A.D.; Var, I. Strategies to prevent mycotoxin contamination of food and animal feed: A review. Crit. Rev. Food Sci. Nutr. 2006, 46, 593–619. [Google Scholar] [CrossRef]
- Colovic, R.; Puvaca, N.; Cheli, F.; Avantaggiato, G.; Greco, D.; Duragic, O.; Kos, J.; Pinotti, L. Decontamination of Mycotoxin-Contaminated Feedstuffs and Compound Feed. Toxins 2019, 11, 617. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Chen, Y.; Kong, Q.; Ma, Y.; Liu, Y. Detoxification of Aflatoxin B1 by Zygosaccharomyces rouxii with Solid State Fermentation in Peanut Meal. Toxins 2017, 9, 42. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Duan, C.C.; Zhao, Y.J.; Gao, L.; Niu, C.H.; Xu, J.B.; Li, S.Y. Reduction of Aflatoxin B-1 Toxicity by Lactobacillus plantarum C88: A Potential Probiotic Strain Isolated from Chinese Traditional Fermented Food “Tofu”. PLoS ONE 2017, 12, e0170109. [Google Scholar] [CrossRef]
- Jebali, R.; Abbes, S.; Salah-Abbes, J.B.; Younes, R.B.; Haous, Z.; Oueslati, R. Ability of Lactobacillus plantarum MON03 to mitigate aflatoxins (B1 and M1) immunotoxicities in mice. J. Immunotoxicol. 2015, 12, 290–299. [Google Scholar] [CrossRef] [Green Version]
- Kumara, S.S.; Gayathri, D.; Hariprasad, P.; Venkateswaran, G.; Swamy, C.T. In vivo AFB1 detoxification by Lactobacillus fermentum LC5/a with chlorophyll and immunopotentiating activity in albino mice. Toxicon 2020, 187, 214–222. [Google Scholar] [CrossRef]
- Lahtinen, S.J.; Haskard, C.A.; Ouwehand, A.C.; Salminen, S.J.; Ahokas, J.T. Binding of aflatoxin B-1 to cell wall components of Lactobacillus rhamnosus strain GG. Food Addit. Contam. A 2004, 21, 158–164. [Google Scholar] [CrossRef]
- Chlebicz, A.; Slizewska, K. In Vitro Detoxification of Aflatoxin B1, Deoxynivalenol, Fumonisins, T-2 Toxin and Zearalenone by Probiotic Bacteria from Genus Lactobacillus and Saccharomyces cerevisiae Yeast. Probiotics Antimicrob. Proteins 2020, 12, 289–301. [Google Scholar] [CrossRef] [Green Version]
- Taheur, F.B.; Fedhila, K.; Chaieb, K.; Kouidhi, B.; Bakhrouf, A.; Abrunhosa, L. Adsorption of aflatoxin B1, zearalenone and ochratoxin A by microorganisms isolated from Kefir grains. Int. J. Food Microbiol. 2017, 251, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Ciegler, A.; Lillehoj, E.B.; Peterson, R.E.; Hall, H.H. Microbial detoxification of aflatoxin. Appl. Microbiol. 1966, 14, 934–939. [Google Scholar] [CrossRef]
- Adeniji, A.A.; Loots, D.T.; Babalola, O.O. Bacillus velezensis: Phylogeny, useful applications, and avenues for exploitation. Appl. Microbiol. Biotechnol. 2019, 103, 3669–3682. [Google Scholar] [CrossRef]
- Fan, Y.; Zhao, L.; Ma, Q.; Li, X.; Shi, H.; Zhou, T.; Zhang, J.; Ji, C. Effects of Bacillus subtilis ANSB060 on growth performance, meat quality and aflatoxin residues in broilers fed moldy peanut meal naturally contaminated with aflatoxins. Food Chem. Toxicol. 2013, 59, 748–753. [Google Scholar] [CrossRef]
- Farzaneh, M.; Shi, Z.Q.; Ghassempour, A.; Sedaghat, N.; Ahmadzadeh, M.; Mirabolfathy, M.; Javan-Nikkhah, M. Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control 2012, 23, 100–106. [Google Scholar] [CrossRef]
- Adebo, O.A.; Njobeh, P.B.; Mavumengwana, V. Degradation and detoxification of AFB(1) by Staphylocococcus warneri, Sporosarcina sp. and Lysinibacillus fusiformis. Food Control 2016, 68, 92–96. [Google Scholar] [CrossRef]
- Topcu, A.; Bulat, T.; Wishah, R.; Boyaci, I.H. Detoxification of aflatoxin B1 and patulin by Enterococcus faecium strains. Int. J. Food Microbiol. 2010, 139, 202–205. [Google Scholar] [CrossRef]
- Sangare, L.; Zhao, Y.J.; Folly, Y.M.E.; Chang, J.H.; Li, J.H.; Selvaraj, J.N.; Xing, F.G.; Zhou, L.; Wang, Y.; Liu, Y. Aflatoxin B-1 Degradation by a Pseudomonas Strain. Toxins 2015, 7, 3538–3539. [Google Scholar] [CrossRef] [Green Version]
- Singh, J.; Mehta, A. Protein-mediated degradation of aflatoxin B1 by Pseudomonas putida. Braz. J. Microbiol. 2019, 50, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.; Abdul Khalil, K.; Zahri, K.N.M.; Gomez-Fuentes, C.; Convey, P.; Zulkharnain, A.; Sabri, S.; Alias, S.A.; Gonzalez-Rocha, G.; Ahmad, S.A. Biosurfactant Production and Growth Kinetics Studies of the Waste Canola Oil-Degrading Bacterium Rhodococcus erythropolis AQ5-07 from Antarctica. Molecules 2020, 25, 3878. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Liu, D.; Mo, X.; Xie, C.; Yao, D. A fungal enzyme with the ability of aflatoxin B(1) conversion: Purification and ESI-MS/MS identification. Microbiol. Res. 2011, 166, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.-L.; Yao, D.-S.; Liang, R.; Ma, L.; Cheng, W.-Q.; Gu, L.-Q. Detoxification of aflatoxin B1 by enzymes isolated from Armillariella tabescens. Food Chem. Toxicol. 1998, 36, 563–574. [Google Scholar] [CrossRef]
- Xu, T.; Xie, C.; Yao, D.; Zhou, C.Z.; Liu, J. Crystal structures of Aflatoxin-oxidase from Armillariella tabescens reveal a dual activity enzyme. Biochem. Biophys. Res. Commun. 2017, 494, 621–625. [Google Scholar] [CrossRef]
- Alberts, J.F.; Gelderblom, W.C.; Botha, A.; van Zyl, W.H. Degradation of aflatoxin B1 by fungal laccase enzymes. Int. J. Food Microbiol. 2009, 135, 47–52. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, R.; Ng, T.B.; Lai, Y.; Yang, J.; Ye, X. A New Laccase of Lac 2 from the White Rot Fungus Cerrena unicolor 6884 and Lac 2-Mediated Degradation of Aflatoxin B1. Toxins 2020, 12, 476. [Google Scholar] [CrossRef]
- Zaid, A.M.A. Biodegradation of aflatoxin by peroxidase enzyme produced by local isolate of Pseudomonas sp. Int. J. Sci. Res. Manag. 2017, 5, 7449–7455. [Google Scholar]
- Li, C.H.; Li, W.Y.; Hsu, I.N.; Liao, Y.Y.; Yang, C.Y.; Taylor, M.C.; Liu, Y.F.; Huang, W.H.; Chang, H.H.; Huang, H.L.; et al. Recombinant Aflatoxin-Degrading F420H2-Dependent Reductase from Mycobacterium smegmatis Protects Mammalian Cells from Aflatoxin Toxicity. Toxins 2019, 11, 259. [Google Scholar] [CrossRef] [Green Version]
- Guan, S.; Ji, C.; Zhou, T.; Li, J.; Ma, Q.; Niu, T. Aflatoxin B1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 2008, 9, 1489–1503. [Google Scholar] [CrossRef] [Green Version]
- Verheecke, C.; Liboz, T.; Mathieu, F. Microbial degradation of aflatoxin B1: Current status and future advances. Int. J. Food Microbiol. 2016, 237, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wu, J.; Liu, Z.; Shi, Y.; Liu, J.; Xu, X.; Hao, S.; Mu, P.; Deng, F.; Deng, Y. Aflatoxin B1 Degradation and Detoxification by Escherichia coli CG1061 Isolated From Chicken Cecum. Front. Pharmacol. 2018, 9, 1548. [Google Scholar] [CrossRef]
- Caceres, I.; Snini, S.P.; Puel, O.; Mathieu, F. Streptomyces roseolus, A Promising Biocontrol Agent Against Aspergillus flavus, the Main Aflatoxin B-1 Producer. Toxins 2018, 10, 442. [Google Scholar] [CrossRef] [Green Version]
- D’Souza, D.H.; Brackett, R.E. The role of trace metal ions in aflatoxin B1 degradation by Flavobacterium aurantiacum. J. Food Prot. 1998, 61, 1666–1669. [Google Scholar] [CrossRef]
- Farzaneh, M.; Shi, Z.Q.; Ahmadzadeh, M.; Hu, L.B.; Ghassempour, A. Inhibition of the Aspergillus flavus Growth and Aflatoxin B1 Contamination on Pistachio Nut by Fengycin and Surfactin-Producing Bacillus subtilis UTBSP1. Plant Pathol. J. 2016, 32, 209–215. [Google Scholar] [CrossRef] [Green Version]
- Borsodi, A.K.; Pollak, B.; Keki, Z.; Rusznyak, A.; Kovacs, A.L.; Sproer, C.; Schumann, P.; Marialigeti, K.; Toth, E.M. Bacillus alkalisediminis sp. nov., an alkaliphilic and moderately halophilic bacterium isolated from sediment of extremely shallow soda ponds. Int. J. Syst. Evol. Microbiol. 2011, 61, 1880–1886. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Zhang, X.; Chen, H.; Huang, W.; Jiang, H.; Wang, C.; Xiao, Z.; Zhang, Y.; Xu, J. Isolation and Aflatoxin B1-Degradation Characteristics of a Microbacterium proteolyticum B204 Strain from Bovine Faeces. Toxins 2022, 14, 525. https://doi.org/10.3390/toxins14080525
Yan Y, Zhang X, Chen H, Huang W, Jiang H, Wang C, Xiao Z, Zhang Y, Xu J. Isolation and Aflatoxin B1-Degradation Characteristics of a Microbacterium proteolyticum B204 Strain from Bovine Faeces. Toxins. 2022; 14(8):525. https://doi.org/10.3390/toxins14080525
Chicago/Turabian StyleYan, Yi, Xinyue Zhang, Haiyan Chen, Wenmin Huang, Hongnian Jiang, Chulun Wang, Zhuang Xiao, Yuyu Zhang, and Jialiang Xu. 2022. "Isolation and Aflatoxin B1-Degradation Characteristics of a Microbacterium proteolyticum B204 Strain from Bovine Faeces" Toxins 14, no. 8: 525. https://doi.org/10.3390/toxins14080525
APA StyleYan, Y., Zhang, X., Chen, H., Huang, W., Jiang, H., Wang, C., Xiao, Z., Zhang, Y., & Xu, J. (2022). Isolation and Aflatoxin B1-Degradation Characteristics of a Microbacterium proteolyticum B204 Strain from Bovine Faeces. Toxins, 14(8), 525. https://doi.org/10.3390/toxins14080525




