Mass-Spectrometry-Based Lipidome and Proteome Profiling of Hottentotta saulcyi (Scorpiones: Buthidae) Venom
Abstract
1. Introduction
2. Results
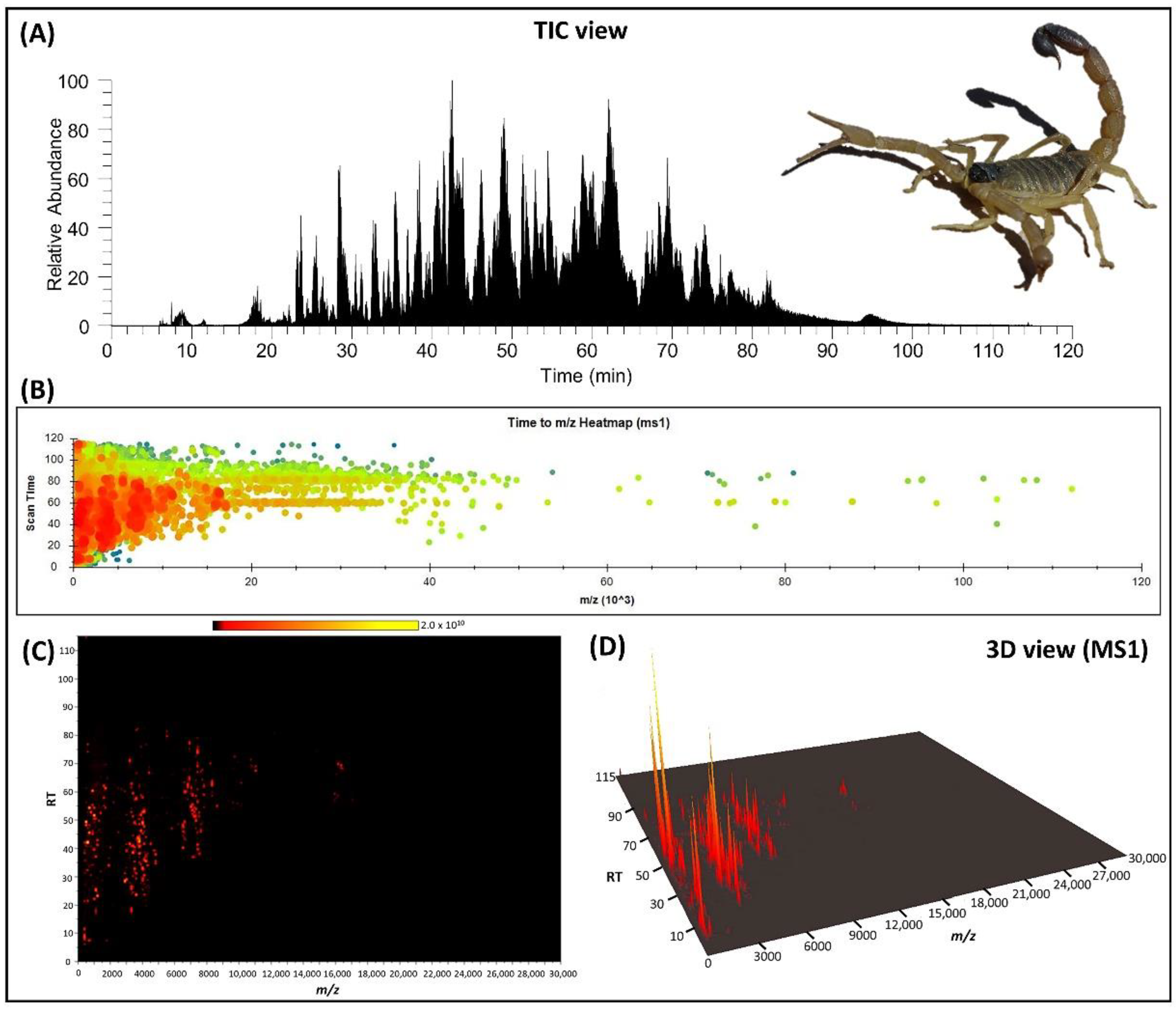
2.1. Mass Fingerprint of Crude Venom
2.2. Peptidomics
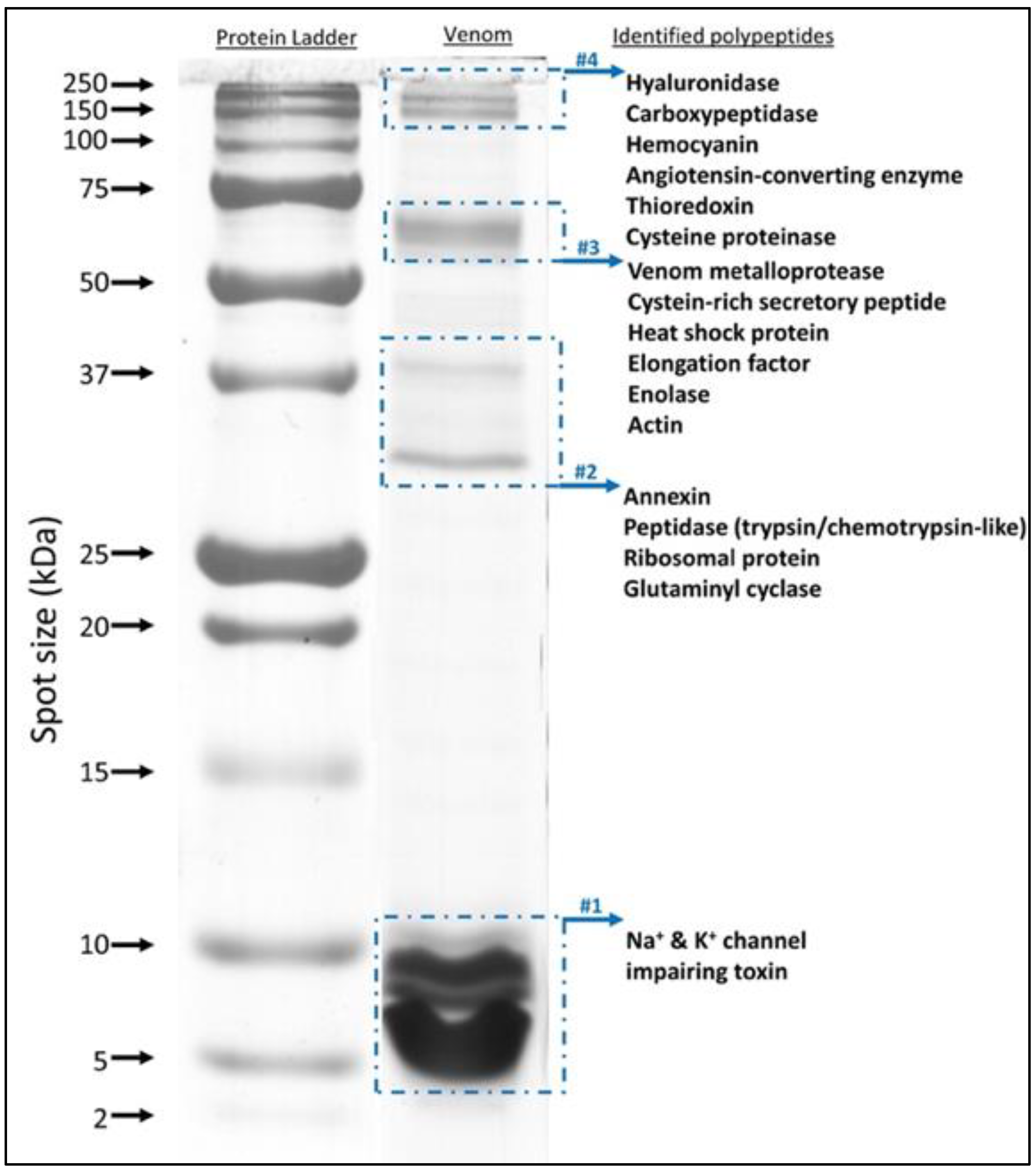
2.3. Protein Identification Using In-Gel and In-Solution Tryptic Digestion followed by LC-MS/MS
2.4. The Scorpion Venom Lipidome
3. Discussion
3.1. The Proteome of H. saulcyi Venom
3.2. The Lipidome of H. saulcyi Venom
4. Conclusions
5. Materials and Methods
5.1. Samples
5.2. Chemicals
5.3. Proteomics Sample Preparation
5.4. UHPLC-MS/MS Proteomics
5.5. Lipid Extraction and UHPLC-MS/MS Lipidomics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Santos, M.S.V.; Silva, C.G.L.; Neto, B.S.; Grangeiro Júnior, C.R.P.; Lopes, V.H.G.; Teixeira Júnior, A.G.; Bezerra, D.A.; Luna, J.V.C.P.; Cordeiro, J.B.; Júnior, J.G.; et al. Clinical and Epidemiological Aspects of Scorpionism in the World: A Systematic Review. Wilderness Environ. Med. 2016, 27, 504–518. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, M.A.; Quintero-Hernández, V.; Possani, L.D. Scorpion Venom Gland Transcriptomics and Proteomics: An Overview. In Venom Genomics and Proteomics; Gopalakrishnakone, P., Calvete, J.J., Eds.; Springer: Dordrecht, The Netherlands, 2016; pp. 105–124. [Google Scholar]
- Chippaux, J.P.; Goyffon, M. Epidemiology of Scorpionism. Acta Trop. 2008, 107, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, S.M.; Sabatier, J.-M. Venoms of Iranian Scorpions (Arachnida, Scorpiones) and Their Potential for Drug Discovery. Molecules 2019, 24, 2670. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Alewood, P.F. Modern Venom Profiling: Mining into Scorpion Venom Biodiversity. In Scorpion Venoms; Gopalakrishnakone, P., Possani, L.D., Schwartz, F.E., de la Vega, R.C.R., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 547–561. [Google Scholar]
- Zlotkin, E. Scorpion Venoms. In Comprehensive Molecular Insect Science; Gilbert, L.I., Ed.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 173–220. [Google Scholar]
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion Venom Components as Potential Candidates for Drug Development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef]
- Zeng, X.C.; Corzo, G.; Hahin, R. Scorpion Venom Peptides without Disulfide Bridges. IUBMB Life 2005, 57, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Albalas, Q. Scorpion Venom Peptides with No Disulfide Bridges: A Review. Peptides 2014, 51, 35–45. [Google Scholar] [CrossRef]
- de la Vega, R.C.R.; Schwartz, E.F.; Possani, L.D. Mining on Scorpion Venom Biodiversity. Toxicon 2010, 56, 1155–1161. [Google Scholar] [CrossRef]
- Quintero-Hernández, V.; Jiménez-Vargas, J.M.; Gurrola, G.B.; Valdivia, H.H.; Possani, L.D. Scorpion Venom Components That Affect Ion-Channels Function. Toxicon 2013, 76, 328–342. [Google Scholar] [CrossRef]
- King, G.F. Venoms as a Platform for Human Drugs: Translating Toxins into Therapeutics. Expert Opin. Biol. Ther. 2011, 11, 1469–1484. [Google Scholar] [CrossRef]
- Holford, M.; Daly, M.; King, G.F.; Norton, R.S. Venoms to the Rescue. Science 2018, 361, 842–844. [Google Scholar] [CrossRef]
- Akcan, M.; Stroud, M.R.; Hansen, S.J.; Clark, R.J.; Daly, N.L.; Craik, D.J.; Olson, J.M. Chemical Re-engineering of Chlorotoxin Improves Bioconjugation Properties for Tumor Imaging and Targeted Therapy. J. Med. Chem. 2011, 54, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Marie, Z.A.; Ibrahim, S.A. Lipid Content of Scorpion (Leiurus Quinquestriatus, H and E) Venom. Toxicon 1976, 14, 93–96. [Google Scholar] [CrossRef]
- Villar-Briones, A.; Aird, S. Organic and Peptidyl Constituents of Snake Venoms: The Picture is Vastly More Complex Than We Imagined. Toxins 2018, 10, 392. [Google Scholar] [CrossRef] [PubMed]
- Acunha, T.; Nardini, V.; Faccioli, L.H. A Lipidomics Approach Reveals New Insights into Crotalus Durissus Terrificus and Bothrops Moojeni Snake Venoms. Arch. Toxicol. 2021, 95, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Carcamo-Noriega, E.N.; Sathyamoorthi, S.; Banerjee, S.; Gnanamani, E.; Mendoza-Trujillo, M.; Mata-Espinosa, D.; Hernández-Pando, R.; Veytia-Bucheli, J.I.; Possani, L.D.; Zare, R.N. 1,4-Benzoquinone Antimicrobial Agents against Staphylococcus Aureus and Mycobacterium Tuberculosis Derived from Scorpion Venom. Proc. Natl. Acad. Sci. USA 2019, 116, 12642–12647. [Google Scholar] [CrossRef] [PubMed]
- Kovarik, F. A Revision of the genus Hottentotta (Scorpiones, Buthidae). Euscorpius 2007, 58, 1–107. [Google Scholar]
- Sanaei-Zadeh, H.; Marashi, S.M.; Dehghani, R. Epidemiological and Clinical Characteristics of Scorpionism in Shiraz (2012–2016); Development of a Clinical Severity Grading for Iranian Scorpion Envenomation. Med. J. Islam. Repub. Iran 2017, 31, 27. [Google Scholar] [CrossRef][Green Version]
- Dehghani, R.; Fathi, B. Scorpion Sting in Iran: A Review. Toxicon 2012, 60, 919–933. [Google Scholar] [CrossRef]
- Mille, B.G.; Peigneur, S.; Predel, R.; Tytgat, J. Trancriptomic Approach Reveals the Molecular Diversity of Hottentotta conspersus (Buthidae) Venom. Toxicon 2015, 99, 73–79. [Google Scholar] [CrossRef]
- Morgenstern, D.; Rohde, B.H.; King, G.F.; Tal, T.; Sher, D.; Zlotkin, E. The Tale of a Resting Gland: Transcriptome of a Replete Venom Gland from the Scorpion Hottentotta judaicus. Toxicon 2011, 57, 695–703. [Google Scholar] [CrossRef]
- Yağmur, E.A.; Özkan, Ö.; Karaer, K.Z. Determination of the Median Lethal Dose and Electrophoretic Pattern of Hottentotta saulcyi (Scorpiones, Buthidae) Scorpion Venom. J. Arthropod-Borne Dis. 2015, 9, 238–245. [Google Scholar] [PubMed]
- Ismael, B.N.; Abass, K.S.; Khalil, K.A.; Salih, K.A. Preparation of F(Ab’)2 Antivenom in Iraq against Scorpion (Hottentotta Saulcyi) Venom. Biologicals 2018, 56, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, H.R.; Nazari, A.; Noofeli, M.; Samiani, M. Bioassay of Derived Components from Venom of Iranian Medically Important Scorpions to Identify the Bradykinin Potentiating Factors. Arch. Razi Inst. 2019, 74, 385–394. [Google Scholar] [PubMed]
- Abdel-Rahman, M.A.; Omran, M.A.A.; Abdel-Nabi, I.M.; Ueda, H.; McVean, A. Intraspecific Variation in the Egyptian Scorpion Scorpio Maurus Palmatus Venom Collected from Different Biotopes. Toxicon 2009, 53, 349–359. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, Y.; He, Y.; Di, Z.; Wu, Y.; Cao, Z.; Li, W. Comparative Venom Gland Transcriptome Analysis of the Scorpion Lychas Mucronatus Reveals Intraspecific Toxic Gene Diversity and New Venomous Components. BMC Genom. 2010, 11, 452. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ravelo, R.; Coronas, F.I.V.; Zamudio, F.Z.; González-Morales, L.; López, G.E.; Urquiola, A.R.; Possani, L.D. The Cuban Scorpion Rhopalurus Junceus (Scorpiones, Buthidae): Component Variations in Venom Samples Collected in Different Geographical Areas. J. Venom. Anim. Toxins Incl. Trop. Dis. 2013, 19, 13. [Google Scholar] [CrossRef] [PubMed]
- Cid-Uribe, J.I.; Veytia-Bucheli, J.I.; Romero-Gutierrez, T.; Ortiz, E.; Possani, L.D. Scorpion Venomics: A 2019 Overview. Expert Rev. Proteom. 2020, 17, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Isbister, G.K.; Bawaskar, H.S. Scorpion Envenomation. N. Engl. J. Med. 2014, 371, 457–463. [Google Scholar] [CrossRef]
- Couraud, F.; Jover, E.; Dubois, J.M.; Rochat, H. Two Types of Scorpion Toxin Receptor Sites, One Related to the Activation, the Other to the Inactivation of the Action Potential Sodium Channel. Toxicon 1982, 20, 9–16. [Google Scholar] [CrossRef]
- Ma, Y.; Zhao, Y.; Zhao, R.; Zhang, W.; He, Y.; Wu, Y.; Cao, Z.; Guo, L.; Li, W. Molecular Diversity of Toxic Components from the Scorpion Heterometrus Petersii Venom Revealed by Proteomic and Transcriptome Analysis. Proteomics 2010, 10, 2471–2485. [Google Scholar] [CrossRef]
- Carmo, A.O.; Oliveira-Mendes, B.B.R.; Horta, C.C.R.; Magalhães, B.F.; Dantas, A.E.; Chaves, L.M.; Chávez-Olórtegui, C.; Kalapothakis, E. Molecular and Functional Characterization of Metalloserrulases, New Metalloproteases from the Tityus Serrulatus Venom Gland. Toxicon 2014, 90, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Chaim, O.M.; Trevisan-Silva, D.; Chaves-Moreira, D.; Wille, A.C.M.; Ferrer, V.P.; Matsubara, F.H.; Mangili, O.C.; da Silveira, R.B.; Gremski, L.H.; Gremski, W.; et al. Brown Spider (Loxosceles Genus) Venom Toxins: Tools for Biological Purposes. Toxins 2011, 3, 309–344. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.M.; Pimenta, A.M.C.; De Figueiredo, S.G.; Santoro, M.M.; Martin-Eauclaire, M.F.; Diniz, C.R.; De Lima, M.E. Enzymes with Gelatinolytic Activity Can Be Found in Tityus Bahiensis and Tityus Serrulatus Venoms. Toxicon 2002, 40, 1041–1045. [Google Scholar] [CrossRef]
- Brazón, J.; Guerrero, B.; D’Suze, G.; Sevcik, C.; Arocha-Piñango, C.L. Fibrin(Ogen)Olytic Enzymes in Scorpion (Tityus Discrepans) Venom. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2014, 168, 62–69. [Google Scholar] [CrossRef]
- Boldrini-França, J.; Pinheiro-Junior, E.L.; Peigneur, S.; Pucca, M.B.; Cerni, F.A.; Borges, R.J.; Costa, T.R.; Carone, S.E.I.; de Mattos Fontes, M.R.; Sampaio, S.V.; et al. Beyond Hemostasis: A Snake Venom Serine Protease with Potassium Channel Blocking and Potential Antitumor Activities. Sci. Rep. 2020, 10, 4476. [Google Scholar] [CrossRef]
- Bordon, K.C.F.; Wiezel, G.A.; Amorim, F.G.; Arantes, E.C. Arthropod Venom Hyaluronidases: Biochemical Properties and Potential Applications in Medicine and Biotechnology. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 43. [Google Scholar] [CrossRef]
- Cajado-Carvalho, D.; Kuniyoshi, A.K.; Duzzi, B.; Iwai, L.K.; de Oliveira, Ú.C.; de Azevedo, I.D.L.M.; Kodama, R.T.; Portaro, F.V. Insights into the Hypertensive Effects of Tityus Serrulatus Scorpion Venom: Purification of an Angiotensin-Converting Enzyme-Like Peptidase. Toxins 2016, 8, 348. [Google Scholar] [CrossRef]
- Gibbs, G.M.; Orta, G.; Reddy, T.; Koppers, A.J.; Martinez-López, P.; de la Vega-Beltràn, J.; Lo, J.C.Y.; Veldhuis, N.; Jamsai, D.; McIntyre, P.; et al. Cysteine-Rich Secretory Protein 4 Is an Inhibitor of Transient Receptor Potential M8 with a Role in Establishing Sperm Function. Proc. Natl. Acad. Sci. USA 2011, 108, 7034–7039. [Google Scholar] [CrossRef]
- Wang, J.; Shen, B.; Guo, M.; Lou, X.; Duan, Y.; Cheng, X.P.; Teng, M.; Niu, L.; Liu, Q.; Huang, Q.; et al. Blocking Effect and Crystal Structure of Natrin Toxin, a Cysteine-Rich Secretory Protein from Naja Atra Venom That Targets the BKCa Channel. Biochemistry 2005, 44, 10145–10152. [Google Scholar] [CrossRef]
- Deka, A.; Sharma, M.; Mukhopadhyay, R.; Devi, A.; Doley, R. Naja Kaouthia Venom Protein, Nk-CRISP, Upregulates Inflammatory Gene Expression in Human Macrophages. Int. J. Biol. Macromol. 2020, 160, 602–611. [Google Scholar] [CrossRef]
- Evans, E.R.J.; McIntyre, L.; Northfield, T.D.; Daly, N.L.; Wilson, D.T. Small Molecules in the Venom of the Scorpion Hormurus waigiensis. Biomedicines 2020, 8, 259. [Google Scholar] [CrossRef] [PubMed]
- Forster, Y.M.; Reusser, S.; Forster, F.; Bienz, S.; Bigler, L. VenoMS—A Website for the Low Molecular Mass Compounds in Spider Venoms. Metabolites 2020, 10, 327. [Google Scholar] [CrossRef] [PubMed]
- Stafforini, D.M.; McIntyre, T.M.; Zimmerman, G.A.; Prescott, S.M. Platelet-Activating Factor, a Pleiotrophic Mediator of Physiological and Pathological Processes. Crit. Rev. Clin. Lab. Sci. 2003, 40, 643–672. [Google Scholar] [CrossRef] [PubMed]
- Zoccal, K.F.; Sorgi, C.A.; Hori, J.I.; Paula-Silva, F.W.G.; Arantes, E.C.; Serezani, C.H.; Zamboni, D.S.; Faccioli, L.H. Opposing Roles of LTB4 and PGE2 in Regulating the Inflammasome-Dependent Scorpion Venom-Induced Mortality. Nat. Commun. 2016, 7, 10760. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Larrauri, A.; Presa, N.; Dominguez-Herrera, A.; Ouro, A.; Trueba, M.; Gomez-Muñoz, A. Role of Bioactive Sphingolipids in Physiology and Pathology. Essays Biochem. 2020, 64, 579–589. [Google Scholar] [PubMed]
- Rudd, A.K.; Devaraj, N.K. Traceless Synthesis of Ceramides in Living Cells Reveals Saturation-Dependent Apoptotic Effects. Proc. Natl. Acad. Sci. USA 2018, 115, 7485–7490. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Fong, Y.; Tsai, E.-M.; Chang, Y.-G.; Chou, H.L.; Wu, C.-Y.; Teng, Y.-N.; Liu, T.-C.; Yuan, S.-S.; Chiu, C.-C. Exogenous C8-Ceramide Induces Apoptosis by Overproduction of ROS and the Switch of Superoxide Dismutases SOD1 to SOD2 in Human Lung Cancer Cells. Int. J. Mol. Sci. 2018, 19, 3010. [Google Scholar] [CrossRef]
- Pizzuto, M.; Pelegrin, P. Cardiolipin in Immune Signaling and Cell Death. Trends Cell Biol. 2020, 30, 892–903. [Google Scholar] [CrossRef]
- Planas-Iglesias, J.; Dwarakanath, H.; Mohammadyani, D.; Yanamala, N.; Kagan, V.E.; Klein-Seetharaman, J. Cardiolipin Interactions with Proteins. Biophys. J. 2015, 109, 1282–1294. [Google Scholar] [CrossRef]
- Castoldi, A.; Monteiro, L.B.; van Teijlingen Bakker, N.; Sanin, D.E.; Rana, N.; Corrado, M.; Cameron, A.M.; Hässler, F.; Matsushita, M.; Caputa, G.; et al. Triacylglycerol Synthesis Enhances Macrophage Inflammatory Function. Nat. Commun. 2020, 11, 4107. [Google Scholar] [CrossRef]
- Lu, J.; Xu, Y.; Wang, J.; Singer, S.D.; Chen, G. The Role of Triacylglycerol in Plant Stress Response. Plants 2020, 9, 472. [Google Scholar] [CrossRef] [PubMed]
- Inceoglu, B.; Lango, J.; Jing, J.; Chen, L.; Doymaz, F.; Pessah, I.N.; Hammock, B.D. One Scorpion, Two Venoms: Prevenom of Parabuthus Transvaalicus Acts as an Alternative Type of Venom with Distinct Mechanism of Action. Proc. Natl. Acad. Sci. USA 2003, 100, 922–927. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-dye Binding. Anal. Biochem. 1976, 72, 248–255. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-Supervised Learning for Peptide Identification from Shotgun Proteomics Datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid Extraction by Methyl-Tert-Butyl Ether for High-Throughput Lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Kroeger, N.M.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Cochran, J.A.; Beecher, C.W.W.; Garrett, T.J.; Yost, R.A. LipidMatch: An Automated Workflow for Rule-Based Lipid Identification Using Untargeted High-Resolution Tandem Mass Spectrometry Data. BMC Bioinform. 2017, 18, 331. [Google Scholar] [CrossRef]

| RT * (min) | MW ** (Da) |
|---|---|
| 0–10 | 366.10, 393.01, 419.31, 441.29, 527.15, 610.25, 689.21, 735.04, 830.53, 896.48, 945.58, 1114.58, 1760.88, 1848.55 |
| 10–20 | 360.16, 389.22, 516.24, 741.47, 1121.62, 1210.48, 2200.16, 3276.27, 3357.40, 3588.60, 3904.57, 4059.84, 4186.87 |
| 20–30 | 374.22, 479.29, 534.31, 562.26, 842.39, 1032.63, 1138.61, 1186.50, 1212.58, 2138.07, 2201.16, 2254.55, 2950.13, 3006.15, 3313.41, 3359.40, 3387.41, 3434.58, 3462.50, 3590.60, 3874.56, 3974.65, 4083.90, 4174.96, 6802.07, 7177.19, 8843.36 |
| 30–40 | 429.27, 649.36, 804.47, 872.53, 1036.43, 1114.59, 1361.71, 1872.76, 1899.79, 3189.31, 3283.53, 3422.45, 3487.63, 3453.42, 3501.65, 3633.60, 3771.46, 3908.63, 3965.63, 4002.74, 4130.68, 4261.81, 4297.83, 4684.11, 4773.18, 4819.21, 6165.57, 6947.85, 7099.04, 7255.22, 7312.23, 7481.41, 7927.26, 8004.45, 8333.30 |
| 40–50 | 465.27, 522.29, 594.36, 643.40, 664.34, 709.28, 1162.52, 1176.54, 1300.67, 1349.60, 1998.96, 3189.28, 3411.84, 3445.66, 3544.52, 3738.45, 3797.50, 3824.54, 3853.80, 4050.58, 6488.80, 6536.80, 6563.86, 7308.22, 7668.39 |
| 50–60 | 765.35, 1002.40, 1575.65, 2146.16, 3375.45, 3547.46, 3632.32, 3820.60, 4063.62, 3968.71, 6633.89, 7026.07, 7074.05, 7109.11, 7121.10, 7135.16, 7264.21, 7325.25, 7339.23, 7371.39, 7405.41, 7462.21, 7866.45, 8589.02, 9970.49 |
| 60–70 | 621.37, 939.38, 1325.60, 1616.75, 1685.74, 2708.25, 4397.92, 4408.93, 4568.81, 5787.46, 6781.88, 6847.90, 6852.89, 6907.91, 6917.29, 7414.32, 7423.20, 7428.17, 7588.38, 7718.27, 8238.59, 8282.66, 8294.63, 10,162.66, 11,088.19, 11,737.29, 16,339.35, 16,348.28, 16,476.37, 18,298.81, 30,419.97, 37,947.32, 40,855.16 |
| 70–80 | 1098.77, 1149.31, 1307.75, 1420.94, 3242.44, 3941.52, 5524.45, 5555.44, 6932.92, 7407.20, 9761.33, 10,766.28, 14,957.42, 16,195.19, 24,694.55, 40,034.10 |
| 80–90 | 1331.50, 1576.64, 1864.05, 3617.49, 3676.49, 4128.9358, 5525.40, 5596.94, 11,839.56 |
| 90–100 | 386.98, 415.21, 558.92 |
| 100–110 | 468.30, 2268.11, 3111.71 |
| Peptide | m/z | z | Mass | Protein Name | Organism | Sequence ID | E-Value * | Description |
|---|---|---|---|---|---|---|---|---|
| SLENEVFWDVMKKLDFEGP | 761.7128 | 3 | 2282.1292 | putative toxin, partial | Hottentotta judaicus | F1CJ05 | 1 × 10−2 | - |
| WGELDFWDVMKKFFPDLP (−0.98) | 1135.0576 | 2 | 2268.1150 | putative toxin, partial | Hottentotta judaicus | F1CJ05 | 3 × 10−2 | - |
| FDEDLNVGFNDFGAPSRSH (+15.99) | 1070.4878 | 2 | 2138.9292 | venom neuropeptide-2 | Mesobuthus eupeus | E4VP42 | 2 × 10−5 | Neuropeptide signaling |
| DFDELDNVGFNDFGPASGVLQ (−0.98) | 1128.0040 | 2 | 2254.0066 | venom neuropeptide-3 | Mesobuthus eupeus | E4VP55 | 7 × 10−10 | Neuropeptide signaling |
| RSQPSGCNVGFNDFGPASRGPS (−0.98) | 1118.5049 | 2 | 2235.0015 | venom neuropeptide-2 | Mesobuthus eupeus | E4VP42 | 6 × 10−6 | Neuropeptide signaling |
| MLLDNVGFNDFGPASRHC | 996.9552 | 2 | 1991.8982 | venom neuropeptide-3 | Mesobuthus eupeus | E4VP55 | 2 × 10−7 | Neuropeptide signaling |
| QPQDLELDKSGFGGFH | 831.3739 | 2 | 1660.7480 | Putative orcokinin | Hottentotta judaicus | E4VP55 | 3 × 10−5 | Neuropeptide signaling |
| DLELDKSGFGGFH | 711.3378 | 2 | 1420.6624 | Putative orcokinin | Hottentotta judaicus | F8THJ9 | 3 × 10−4 | Neuropeptide signaling |
| RGGKELMNSLKEKLSEAKE | 537.5423 | 4 | 2146.1191 | U9-buthitoxin-Hja1 | Hottentotta judaicus | F1CIW9 | 4 × 10−9 | K+ channel impairing toxin |
| FAANTVLNGPEEEAAVENF | 1011.4805 | 2 | 2020.9377 | Putative toxin Tx297 | Mesobuthus martensii | B8XH54 | 4 × 10−5 | Bradykinin-potentiating peptide |
| EPDVLNGLLEEAAVPAAE | 918.9564 | 2 | 1835.9153 | Putative toxin Tx297 | Buthus occitanus israelis | B8XH54 | 4 × 10−2 | Bradykinin-potentiating peptide |
| PAALNHLNGPEEEAAVPAAE | 1000.4883 | 2 | 1998.9646 | Putative toxin Tx297 | Buthus occitanus israelis | B8XH54 | 1 × 10−5 | Bradykinin-potentiating peptide |
| HAPLKEKLSNMLETAHA | 945.5087 | 2 | 1888.9829 | Elongation factor | Leptotrombidium deliense | A0A443ST94 | 5 × 10−2 | GTP-binding protein |
| KNRELMNSLKEKLSE | 455.4981 | 4 | 1817.9668 | Potassium channel toxin | Mesobuthus eupeus | P0CH57 | 2 × 10−4 | K+ channel impairing toxin |
| WVPGNYPGVLSY | 676.3341 | 2 | 1350.6721 | Toxin b subunit beta | Androctonus crassicauda | P0C2A3 | 4 × 10−3 | Na+ channel impairing toxin |
| KKDGYPVDSGNCKYECLKDDYCNDLCLERKADKGYCYWGKVSCYCYGLPDNSPTKTSGKCNPA | 1180.5389 | 6 | 7071.1187 | Alpha-toxin CsE5 | Centruroides sculpturatus | P46066 | 2 × 10−68 | Na+ channel impairing toxin |
| ARDGYIANDRNCVYTCALNPYCDSECKKNGADSGYCQW (+15.99) FGRFGNACW (+15.99) CKNLPDKVPIRIPGECRG | 1220.3782 | 6 | 7312.2598 | MeuNaTxalpha-9 | Mesobuthus eupeus | D8UWD8 | 1 × 10−71 | Na+ channel impairing toxin |
| LKDGYIVDDRNCTYFCGTNAYCNEECVKLKGESGYCQWVGRYGNACWCYKLPDHVRTVQAGRCRS (−0.98) | 1245.5728 | 6 | 7462.3755 | Alpha-toxin Bot11 | Buthus occitanus tunetanus | P01486 | 5 × 10−72 | Na+ channel impairing toxin |
| GRDAYIADSENCTYTCALNPYCNDLCTKNGAKSGYCQW (+15.99) AGRYGNACW (+15.99) CIDLPDKVPIRISGSCR | 1012.1568 | 7 | 7074.1265 | Makatoxin-1 | Mesobuthus martensii | P56569 | 1 × 10−69 | Na+ channel impairing toxin |
| Accession | Description | Organism | Coverage (%) | #Peptides | #Unique | Avg. Mass | -10lgP |
|---|---|---|---|---|---|---|---|
| Na+- channel toxins | |||||||
| P0DJH8 | Alpha-toxin Bu1 (α-NaTx) | Buthacus macrocentrus | 60 | 6 | 3 | 7485 | 85.69 |
| D5HR49 | Neurotoxin 9 (Fragment) | Androctonus bicolor | 86 | 6 | 3 | 7750 | 82.98 |
| D5HR55 | Neurotoxin 2 (Fragment) | Hottentotta judaicus | 35 | 3 | 2 | 7421 | 48.05 |
| F1CJ50 | U1-buthitoxin-Hj1b | Hottentotta judaicus | 14 | 2 | 2 | 10,792 | 50.24 |
| B8XGY7 | Putative alpha toxin Tx405 (α-NaTx) | Buthus occitanus israelis | 32 | 4 | 3 | 9324 | 48.05 |
| B8XGY1 | Putative alpha toxin Tx93 (α-NaTx) | Buthus occitanus israelis | 31 | 4 | 3 | 9625 | 48.05 |
| F0V3W0 | Alpha neurotoxin precusor (α-NaTx) | Hottentotta judaicus | 61 | 6 | 4 | 9312 | 48.05 |
| Q56TT9 | Alpha-insect toxin BjaIT (α-NaTx) | Hottentotta judaicus | 61 | 6 | 4 | 9270 | 48.05 |
| F1CJ53 | Alpha-insect toxin BjaIT (Fragment) (α-NaTx) | Hottentotta judaicus | 65 | 4 | 3 | 4367 | 48.05 |
| Q86SE0 | Toxin Aam2 | Androctonus amoreuxi | 29 | 4 | 2 | 9283 | 112.38 |
| P45668 | Neurotoxin-2 (Fragment) | Hottentotta tamulus | 54 | 2 | 2 | 2686 | 106.55 |
| K+- channel toxins | |||||||
| A0A0K0LC05 | Potassium channel blocker AbKTx-2 (β-KTx) | Androctonus bicolor | 16 | 2 | 2 | 10,307 | 62.92 |
| A0A0K0LC09 | Potassium channel blocker AbKTx-7 (α-KTx) | Androctonus bicolor | 16 | 2 | 2 | 10,308 | 62.92 |
| A0A088D9U2 | Potassium channel blocker pMeKTx28-2 (β-KTx) | Mesobuthus eupeus | 16 | 2 | 2 | 10,264 | 62.92 |
| A0A0K0LC11 | Potassium channel blocker AbKTx-3 (α-KTx) | Androctonus bicolor | 19 | 2 | 2 | 8507 | 62.92 |
| A0A0K0LCJ0 | Potassium channel blocker AbKTx-5 (α-KTx) | Androctonus bicolor | 16 | 2 | 2 | 10,380 | 62.92 |
| A0A088DB26 | Potassium channel blocker pMeKTx28-3 (β-KTx) | Mesobuthus eupeus | 16 | 2 | 2 | 10,250 | 62.92 |
| A0A0K0LC08 | Potassium channel blocker AbKTx-4 (α-KTx) | Androctonus bicolor | 31 | 2 | 2 | 5163 | 62.92 |
| B8XH36 | Putative potassium channel toxin Tx633 (β-KTx) | Buthus occitanus israelis | 18 | 2 | 2 | 8688 | 62.92 |
| A0A143MGJ8 | Potassium channel toxin meuK28-2 (β-KTx) | Mesobuthus eupeus | 16 | 2 | 2 | 10,408 | 62.92 |
| A0A0U4GZ05 | Potassium channel toxin KTx3 (β-KTx) | Odontobuthus doriae | 16 | 2 | 2 | 10,313 | 62.92 |
| A9XE60 | Potassium channel toxin MeuTXK-beta-1 (β-KTx) | Mesobuthus eupeus | 10 | 3 | 3 | 10,338 | 46.55 |
| A0A0K0LC02 | Potassium channel blocker AbKTx-10 (α-KTx) | Androctonus bicolor | 10 | 2 | 2 | 10,110 | 46.55 |
| A9XE59 | Potassium channel toxin MeuTXK-beta-2 (β-KTx) | Mesobuthus eupeus | 10 | 3 | 3 | 10,328 | 46.55 |
| A0A0K0LBZ4 | Potassium channel blocker AbKTx-6 (α-KTx) | Androctonus bicolor | 10 | 2 | 2 | 10,103 | 46.55 |
| A0A0K0LCI9 | Potassium channel blocker AbKTx-11 (α-KTx) | Androctonus bicolor | 21 | 5 | 5 | 10,086 | 46.55 |
| A0A0K0LC06 | Potassium channel blocker AbKTx-9 (α-KTx) | Androctonus bicolor | 10 | 2 | 2 | 10,102 | 46.55 |
| E4VP56 | Putative bifunctional venom peptide-5 (β-KTx) | Mesobuthus eupeus | 14 | 3 | 3 | 7076 | 46.55 |
| E4VP14 | Putative bi-functional venom peptide (β-KTx) | Mesobuthus eupeus | 10 | 3 | 3 | 10,338 | 46.55 |
| Enzymes | |||||||
| P86100 | Hyaluronidase-1 | Mesobuthus martensii | 54 | 32 | 25 | 47,433 | 248.11 |
| A0A0C9RFM5 | Hyaluronidase | Tityus bahiensis | 11 | 4 | 3 | 46,533 | 85.94 |
| A0A1E1WWG5 | Hyaluronidase | Tityus obscurus | 21 | 5 | 4 | 46,678 | 94.81 |
| F1CIW6 | Hyaluronidase (Fragment) | Hottentotta judaicus | 52 | 15 | 9 | 20,715 | 182.73 |
| E4VNZ7 | Venom metalloprotease-1 | Mesobuthus eupeus | 35 | 32 | 24 | 44,842 | 239.74 |
| A0A0U4HEU8 | Venom protein VP4 | Odontobuthus doriae | 28 | 8 | 2 | 15,973 | 157.24 |
| A0A1E1WW02 | Putative metalloproteinase (Fragment) | Tityus obscurus | 6 | 7 | 4 | 41,037 | 89.28 |
| E4VNZ8 | Venom metalloprotease-2 (Fragment) | Mesobuthus eupeus | 32 | 29 | 21 | 35,602 | 239.74 |
| F1CIU8 | Putative M12B metalloprotease (Fragment) | Hottentotta judaicus | 16 | 7 | 7 | 38,519 | 192.13 |
| A0A0U1SF04 | Peptidase_M14 domain-containing protein (Fragment) | Isometrus maculatus | 31 | 5 | 2 | 23,077 | 119.46 |
| E4VP21 | Chymotrypsin-like protease-1 | Mesobuthus eupeus | 26 | 6 | 4 | 29,641 | 143.14 |
| F1CIY2 | Putative transmembranal serine protease (Fragment) | Hottentotta judaicus | 35 | 6 | 4 | 25,193 | 181.63 |
| F1CJ26 | M12B metalloprotease (Fragment) | Hottentotta judaicus | 37 | 12 | 12 | 27,664 | 125.9 |
| A0A2I9LNS6 | Acid phosphatase | Centruroides hentzi | 20 | 6 | 6 | 43,244 | 91.97 |
| A0A4Y2BUR0 | Carboxypeptidase E | Araneus ventricosus | 5 | 2 | 2 | 50,168 | 67.35 |
| A0A1S5QN46 | Carboxypeptidase E | Tityus serrulatus | 15 | 7 | 4 | 53,838 | 141.45 |
| A0A1W7RAV1 | Carboxypeptidase | Hadrurus spadix | 6 | 2 | 2 | 49,002 | 75.85 |
| A0A1E1WVT7 | Angiotensin-converting enzyme | Tityus obscurus | 8 | 4 | 4 | 72,883 | 65.33 |
| F1CJ87 | Putative angiotensin-converting enzyme (Fragment) | Hottentotta judaicus | 41 | 3 | 3 | 4575 | 103.37 |
| F1CJ25 | Putative angiotensin-converting enzyme (Fragment) | Hottentotta judaicus | 22 | 7 | 7 | 30,499 | 147.9 |
| Other components | |||||||
| A0A2I9LPW9 | Venom factor | Centruroides hentzi | 2 | 3 | 3 | 200,889 | 68.23 |
| F8THJ4 | CRISP3 (Fragment) | Hottentotta judaicus | 73 | 29 | 28 | 21,201 | 259.16 |
| T1E6Y3 | CAP-Iso-2 (Fragment) | Isometroides vescus | 8 | 2 | 2 | 44,650 | 53.16 |
| F1CJ75 | Putative cysteine-rich secretory peptide (Fragment) | Hottentotta judaicus | 40 | 14 | 11 | 23,920 | 219.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghezellou, P.; Jakob, K.; Atashi, J.; Ghassempour, A.; Spengler, B. Mass-Spectrometry-Based Lipidome and Proteome Profiling of Hottentotta saulcyi (Scorpiones: Buthidae) Venom. Toxins 2022, 14, 370. https://doi.org/10.3390/toxins14060370
Ghezellou P, Jakob K, Atashi J, Ghassempour A, Spengler B. Mass-Spectrometry-Based Lipidome and Proteome Profiling of Hottentotta saulcyi (Scorpiones: Buthidae) Venom. Toxins. 2022; 14(6):370. https://doi.org/10.3390/toxins14060370
Chicago/Turabian StyleGhezellou, Parviz, Kevin Jakob, Javad Atashi, Alireza Ghassempour, and Bernhard Spengler. 2022. "Mass-Spectrometry-Based Lipidome and Proteome Profiling of Hottentotta saulcyi (Scorpiones: Buthidae) Venom" Toxins 14, no. 6: 370. https://doi.org/10.3390/toxins14060370
APA StyleGhezellou, P., Jakob, K., Atashi, J., Ghassempour, A., & Spengler, B. (2022). Mass-Spectrometry-Based Lipidome and Proteome Profiling of Hottentotta saulcyi (Scorpiones: Buthidae) Venom. Toxins, 14(6), 370. https://doi.org/10.3390/toxins14060370






