The Enzymatic Core of Scorpion Venoms
Abstract
:1. Introduction
2. Results
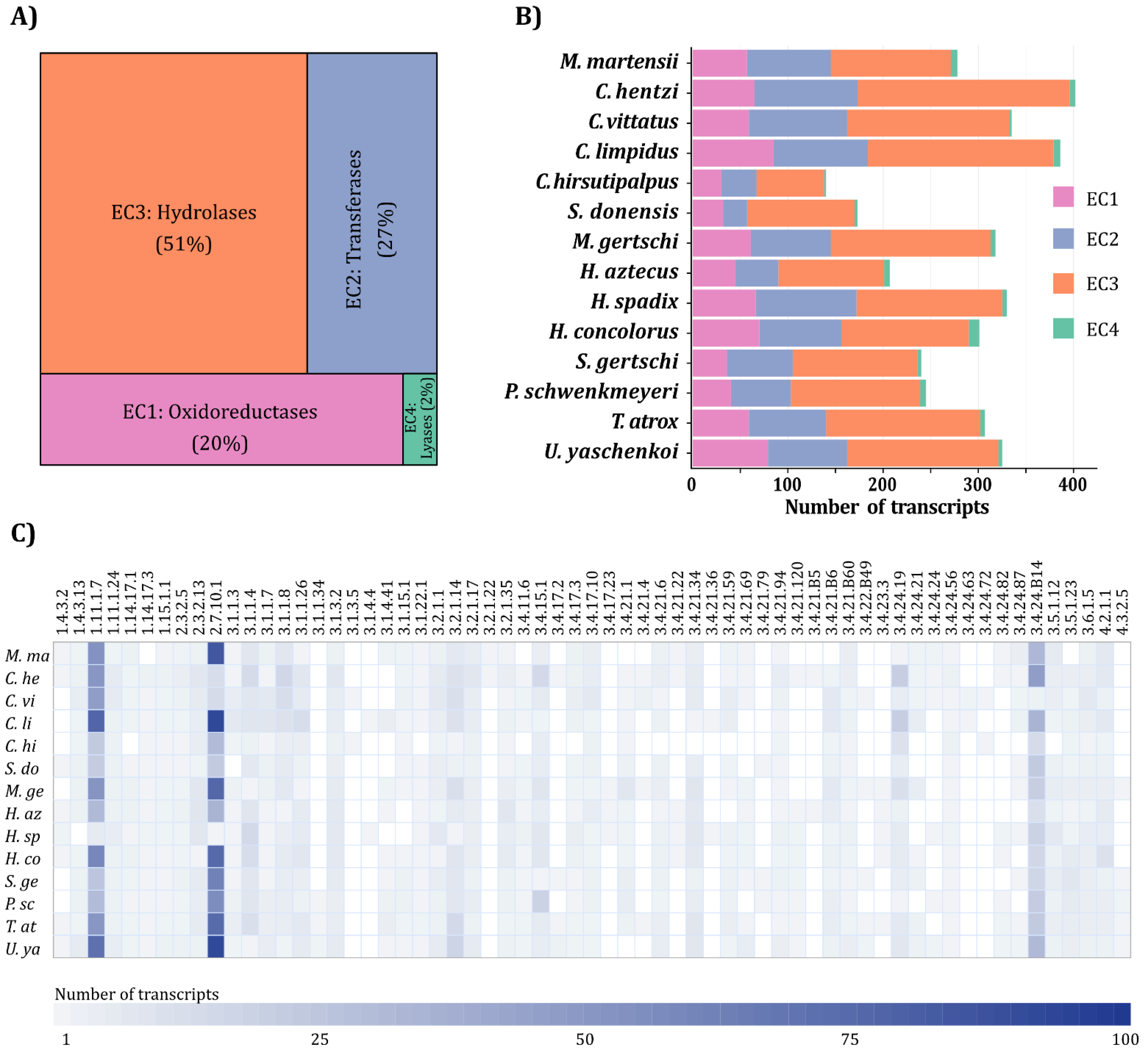
2.1. Transcriptomic Analysis
2.2. Putative Enzyme Identification, Orthogroup Assignment and Annotation
2.3. Transcript Sequence Validation by Proteomic Matching
2.4. Novel Enzymes
2.4.1. Toxic Enzymes
2.4.2. Enzymes Involved in Prey Pre-Digestion
2.4.3. Enzymes as Spreading Factors
2.4.4. Enzymes as Venom Component Activators
2.4.5. Enzymes with Preservative Function
2.4.6. Multifunctional Enzymes
2.4.7. Enzymes with Non-Elucidated Function in Venom
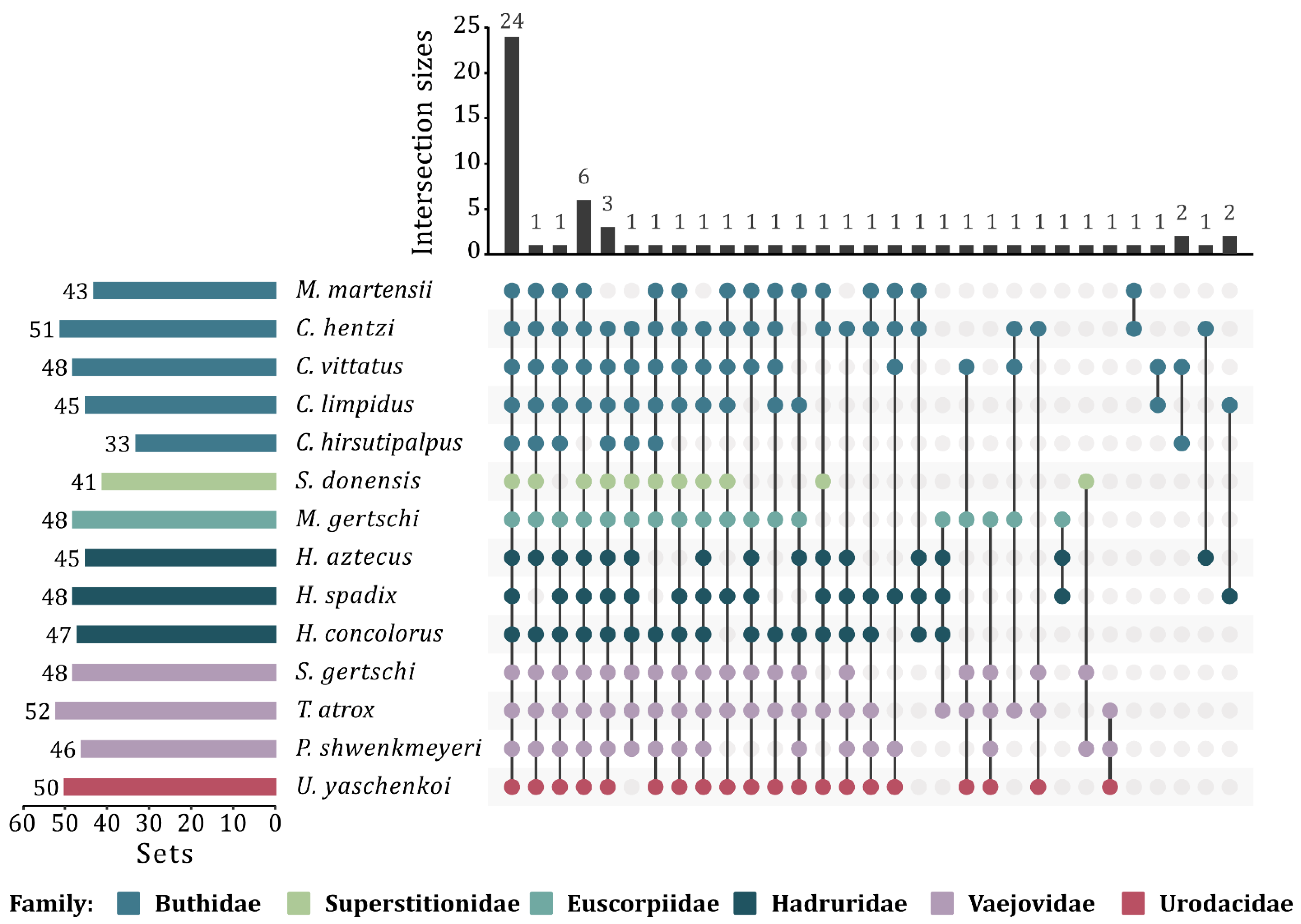
2.5. Determination of the Enzymatic Core
2.6. Venom Gland RNA-seq Quantification
3. Discussion
3.1. Transcriptomes Used in the Study
3.2. Putative Enzyme Identification, Assignment and Annotation of Orthogroups
3.3. Validation of Transcriptome-Derived Enzymes
3.4. Novel Enzymes
3.4.1. Toxic Enzymes
3.4.2. Enzymes Involved in Pre-Digestion of Prey
3.4.3. Spreading Enzymes
3.4.4. Venom Component Activators
3.4.5. Enzymes as Preservative Agents
3.4.6. Enzymes with Multiple Physiological Effects
3.4.7. Enzymes with Non-Elucidated Function in Venom
3.5. Scorpion Venom Enzymatic Core
3.6. RNA-seq Quantification
4. Conclusions
5. Materials and Methods
5.1. Transcriptomes Used in the Study
5.2. Read Filtering and De Novo Transcriptome Assembly
5.3. Identification of Transcript Sequences and Putative Coding for Enzymes
5.4. Identification, Assignment and Annotation of Orthogroups
5.5. Identification of Enzymes in the Venom
5.6. RNA-seq Quantification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion venom components as potential candidates for drug development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, S.; Knerr, J.M.; Argemi, L.; Bordon, K.C.F.; Pucca, M.B.; Cerni, F.A.; Arantes, E.C.; Caliskan, F.; Laustsen, A.H. Scorpion venom: Detriments and benefits. Biomedicines 2020, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Prudencio, G.; Possani, L.D.; Becerril, B.; Ortiz, E. The dual alpha-amidation system in scorpion venom glands. Toxins 2019, 11, 425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira-Mendes, B.B.R.; Miranda, S.E.M.; Sales-Medina, D.F.; Magalhaes, B.F.; Kalapothakis, Y.; Souza, R.P.; Cardoso, V.N.; de Barros, A.L.B.; Guerra-Duarte, C.; Kalapothakis, E.; et al. Inhibition of Tityus serrulatus venom hyaluronidase affects venom biodistribution. PLoS Negl. Trop. Dis. 2019, 13, e0007048. [Google Scholar] [CrossRef] [PubMed]
- Borchani, L.; Sassi, A.; Shahbazzadeh, D.; Strub, J.M.; Tounsi-Guetteti, H.; Boubaker, M.S.; Akbari, A.; Van Dorsselaer, A.; El Ayeb, M. Heminecrolysin, the first hemolytic dermonecrotic toxin purified from scorpion venom. Toxicon 2011, 58, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Duarte, C.; Rebello Horta, C.C.; Ribeiro Oliveira-Mendes, B.B.; de Freitas Magalhães, B.; Costal-Oliveira, F.; Stransky, S.; Fonseca de Freitas, C.; Campolina, D.; de Oliveira Pardal, P.P.; Lira-da-Silva, R.; et al. Determination of hyaluronidase activity in Tityus spp. scorpion venoms and its inhibition by Brazilian antivenoms. Toxicon 2019, 167, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W. A brief review of the scientific history of several lesser-known snake venom proteins: L-amino acid oxidases, hyaluronidases and phosphodiesterases. Toxicon 2013, 62, 75–82. [Google Scholar] [CrossRef]
- Gremski, L.H.; da Justa, H.C.; da Silva, T.P.; Polli, N.L.C.; Antunes, B.C.; Minozzo, J.C.; Wille, A.C.M.; Senff-Ribeiro, A.; Arni, R.K.; Veiga, S.S. Forty years of the description of brown spider venom phospholipases-D. Toxins 2020, 12, 164. [Google Scholar] [CrossRef] [Green Version]
- Valavi, E.; Ansari, M.J. Hemolytic uremic syndrome following Hemiscorpius lepturus (scorpion) sting. Indian J. Nephrol. 2008, 18, 166–168. [Google Scholar] [CrossRef]
- Ortiz, E.; Rendon-Anaya, M.; Rego, S.C.; Schwartz, E.F.; Possani, L.D. Antarease-like Zn-metalloproteases are ubiquitous in the venom of different scorpion genera. Biochim. Biophys. Acta 2014, 1840, 1738–1746. [Google Scholar] [CrossRef]
- Fletcher, P.L., Jr.; Fletcher, M.D.; Weninger, K.; Anderson, T.E.; Martin, B.M. Vesicle-associated membrane protein (VAMP) cleavage by a new metalloprotease from the Brazilian scorpion Tityus serrulatus. J. Biol. Chem. 2010, 285, 7405–7416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bordon, K.C.F.; Cologna, C.T.; Fornari-Baldo, E.C.; Pinheiro-Júnior, E.L.; Cerni, F.A.; Amorim, F.G.; Anjolette, F.A.P.; Cordeiro, F.A.; Wiezel, G.A.; Cardoso, I.A.; et al. From animal poisons and venoms to medicines: Achievements, challenges and perspectives in drug discovery. Front. Pharmacol. 2020, 11, 1132. [Google Scholar] [CrossRef] [PubMed]
- Mohamed Abd El-Aziz, T.; Garcia Soares, A.; Stockand, J.D. Snake venoms in drug discovery: Valuable therapeutic tools for life saving. Toxins 2019, 11, 564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karapetian, H. Reptilase time (RT). Methods Mol. Biol. 2013, 992, 273–277. [Google Scholar] [CrossRef]
- Gempeler-Messina, P.M.; Volz, K.; Buhler, B.; Muller, C. Protein C activators from snake venoms and their diagnostic use. Haemostasis 2001, 31, 266–272. [Google Scholar] [CrossRef]
- González-Santillán, E.; Possani, L.D. North American scorpion species of public health importance with a reappraisal of historical epidemiology. Acta Trop. 2018, 187, 264–274. [Google Scholar] [CrossRef]
- Soleglad, M.E.; Fet, V. High-level systematics and phylogeny of the extant scorpions (Scorpiones: Orthosterni). Euscorpius 2003, 2003, 1–56. [Google Scholar] [CrossRef]
- Santibáñez-López, C.E.; González-Santillán, E.; Monod, L.; Sharma, P.P. Phylogenomics facilitates stable scorpion systematics: Reassessing the relationships of Vaejovidae and a new higher-level classification of Scorpiones (Arachnida). Mol. Phylogenet. Evol. 2019, 135, 22–30. [Google Scholar] [CrossRef]
- Soleglad, M.E.; Sissom, W.E. Phylogeny of the family Euscorpiidae Laurie, 1896 (Scorpiones): A major revision. In Scorpions 2001. In memoriam Gary, A. Polis; Fet, V., Selden, P.A., Eds.; British Arachnological Society: Grays, UK, 2001; pp. 25–111. [Google Scholar]
- Santibáñez-López, C.E.; Graham, M.R.; Sharma, P.P.; Ortiz, E.; Possani, L.D. Hadrurid scorpion toxins: Evolutionary conservation and selective pressures. Toxins 2019, 11, 637. [Google Scholar] [CrossRef] [Green Version]
- Santibáñez-López, C.E.; Cid-Uribe, J.I.; Batista, C.V.; Ortiz, E.; Possani, L.D. Venom gland transcriptomic and proteomic analyses of the enigmatic scorpion Superstitionia donensis (Scorpiones: Superstitioniidae), with insights on the evolution of its venom components. Toxins 2016, 8, 367. [Google Scholar] [CrossRef] [Green Version]
- Ward, M.J.; Ellsworth, S.A.; Rokyta, D.R. Venom-gland transcriptomics and venom proteomics of the Hentz striped scorpion (Centruroides hentzi; Buthidae) reveal high toxin diversity in a harmless member of a lethal family. Toxicon 2018, 142, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Cid-Uribe, J.I.; Meneses, E.P.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Dissecting toxicity: The venom gland transcriptome and the venom proteome of the highly venomous scorpion Centruroides limpidus (Karsch, 1879). Toxins 2019, 11, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdez-Velázquez, L.L.; Cid-Uribe, J.; Romero-Gutierrez, M.T.; Olamendi-Portugal, T.; Jimenez-Vargas, J.M.; Possani, L.D. Transcriptomic and proteomic analyses of the venom and venom glands of Centruroides hirsutipalpus, a dangerous scorpion from Mexico. Toxicon 2020, 179, 21–32. [Google Scholar] [CrossRef] [PubMed]
- McElroy, T.; McReynolds, C.N.; Gulledge, A.; Knight, K.R.; Smith, W.E.; Albrecht, E.A. Differential toxicity and venom gland gene expression in Centruroides vittatus. PLoS ONE 2017, 12, e0184695. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Liang, H.; Shou, Z.; Yao, Y.; Lv, Y.; Shang, J.; Lu, W.; Jia, C.; Liu, Q.; Zhang, H.; et al. De novo transcriptomic and proteomic analysis and potential toxin screening of Mesobuthus martensii samples from four different provinces. J. Ethnopharmacol. 2021, 265, 113268. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Ward, M.J. Venom-gland transcriptomics and venom proteomics of the black-back scorpion (Hadrurus spadix) reveal detectability challenges and an unexplored realm of animal toxin diversity. Toxicon 2017, 128, 23–37. [Google Scholar] [CrossRef] [Green Version]
- Santibáñez-López, C.E.; Cid-Uribe, J.I.; Zamudio, F.Z.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Venom gland transcriptomic and venom proteomic analyses of the scorpion Megacormus gertschi Diaz-Najera, 1966 (Scorpiones: Euscorpiidae: Megacorminae). Toxicon 2017, 133, 95–109. [Google Scholar] [CrossRef]
- Cid-Uribe, J.I.; Santibáñez-Lopez, C.E.; Meneses, E.P.; Batista, C.V.F.; Jiménez-Vargas, J.M.; Ortiz, E.; Possani, L.D. The diversity of venom components of the scorpion species Paravaejovis schwenkmeyeri (Scorpiones: Vaejovidae) revealed by transcriptome and proteome analyses. Toxicon 2018, 151, 47–62. [Google Scholar] [CrossRef]
- Romero-Gutierrez, M.T.; Santibáñez-López, C.E.; Jiménez-Vargas, J.M.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Transcriptomic and proteomic analyses reveal the diversity of venom components from the vaejovid scorpion Serradigitus gertschi. Toxins 2018, 10, 359. [Google Scholar] [CrossRef] [Green Version]
- Romero-Gutierrez, T.; Peguero-Sanchez, E.; Cevallos, M.A.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. A deeper examination of Thorellius atrox scorpion venom components with omic techonologies. Toxins 2017, 9, 399. [Google Scholar] [CrossRef] [Green Version]
- Luna-Ramírez, K.; Quintero-Hernández, V.; Juárez-González, V.R.; Possani, L.D. Whole transcriptome of the venom gland from Urodacus yaschenkoi scorpion. PLoS ONE 2015, 10, e0127883. [Google Scholar] [CrossRef] [PubMed]
- Fuzita, F.J.; Pinkse, M.W.; Patane, J.S.; Juliano, M.A.; Verhaert, P.D.; Lopes, A.R. Biochemical, transcriptomic and proteomic analyses of digestion in the scorpion Tityus serrulatus: Insights into function and evolution of digestion in an ancient arthropod. PLoS ONE 2015, 10, e0123841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthukrishnan, S.; Merzendorfer, H.; Arakane, Y.; Yang, Q. Chitin metabolic pathways in insects and their regulation. In Extracellular Composite Matrices in Arthropods; Cohen, E., Moussian, B., Eds.; Springer International Publishing: New York, NY, USA, 2016; pp. 31–65. [Google Scholar]
- Colinet, D.; Cazes, D.; Belghazi, M.; Gatti, J.L.; Poirié, M. Extracellular superoxide dismutase in insects: Characterization, function, and interspecific variation in parasitoid wasp venom. J. Biol. Chem. 2011, 286, 40110–40121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peiren, N.; de Graaf, D.C.; Vanrobaeys, F.; Danneels, E.L.; Devreese, B.; Van Beeumen, J.; Jacobs, F.J. Proteomic analysis of the honey bee worker venom gland focusing on the mechanisms of protection against tissue damage. Toxicon 2008, 52, 72–83. [Google Scholar] [CrossRef]
- Valdez-Cruz, N.A.; Batista, C.V.; Possani, L.D. Phaiodactylipin, a glycosylated heterodimeric phospholipase A from the venom of the scorpion Anuroctonus phaiodactylus. Eur. J. Biochem. 2004, 271, 1453–1464. [Google Scholar] [CrossRef]
- Cajado-Carvalho, D.; Kuniyoshi, A.K.; Duzzi, B.; Iwai, L.K.; Oliveira, U.C.; Junqueira de Azevedo, I.L.; Kodama, R.T.; Portaro, F.V. Insights into the hypertensive effects of Tityus serrulatus scorpion venom: Purification of an angiotensin-converting enzyme-like peptidase. Toxins 2016, 8, 348. [Google Scholar] [CrossRef] [Green Version]
- Cid-Uribe, J.I.; Veytia-Bucheli, J.I.; Romero-Gutierrez, T.; Ortiz, E.; Possani, L.D. Scorpion venomics: A 2019 overview. Expert Rev. Proteom. 2020, 17, 67–83. [Google Scholar] [CrossRef]
- Jeske, L.; Placzek, S.; Schomburg, I.; Chang, A.; Schomburg, D. BRENDA in 2019: A European ELIXIR core data resource. Nucleic Acids Res. 2019, 47, D542–D549. [Google Scholar] [CrossRef]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef] [Green Version]
- Isbister, G.K.; Bawaskar, H.S. Scorpion envenomation. N. Engl. J. Med. 2014, 371, 457–463. [Google Scholar] [CrossRef]
- Laraba-Djebari, F.; Chérifi, F. Pathophysiological and pharmacological effects of snake venom components: Molecular targets. J. Clin. Toxicol. 2014, 4, 190. [Google Scholar] [CrossRef] [Green Version]
- Oldrati, V.; Arrell, M.; Violette, A.; Perret, F.; Sprungli, X.; Wolfender, J.L.; Stocklin, R. Advances in venomics. Mol. Biosyst. 2016, 12, 3530–3543. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.P.; Fernández, R.; Esposito, L.A.; González-Santillán, E.; Monod, L. Phylogenomic resolution of scorpions reveals multilevel discordance with morphological phylogenetic signal. Proc. Biol. Sci. 2015, 282, 20142953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, E.P.; Rautsaw, R.M.; Strickland, J.L.; Holding, M.L.; Hogan, M.P.; Mason, A.J.; Rokyta, D.R.; Parkinson, C.L. Comparative venom-gland transcriptomics and venom proteomics of four Sidewinder Rattlesnake (Crotalus cerastes) lineages reveal little differential expression despite individual variation. Sci. Rep. 2018, 8, 15534. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [Green Version]
- Cajado-Carvalho, D.; Galvão, J.; Kuniyoshi, A.K.; Carneiro, P.D.S.; Paes Leme, A.F.; Pauletti, B.A.; Marengo, E.B.; Portaro, F.V. Tityus serrulatus scorpion venom: In vitro tests and their correlation with in vivo lethal dose assay. Toxins 2017, 9, 380. [Google Scholar] [CrossRef] [Green Version]
- Feng, L.; Gao, R.; Gopalakrishnakone, P. Isolation and characterization of a hyaluronidase from the venom of Chinese red scorpion Buthus martensi. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2008, 148, 250–257. [Google Scholar] [CrossRef]
- Incamnoi, P.; Patramanon, R.; Thammasirirak, S.; Chaveerach, A.; Uawonggul, N.; Sukprasert, S.; Rungsa, P.; Daduang, J.; Daduang, S. Heteromtoxin (HmTx), a novel heterodimeric phospholipase A(2) from Heterometrus laoticus scorpion venom. Toxicon 2013, 61, 62–71. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Yin, X.; Guo, S.; Gao, J.; Luo, L.; Liao, X.; Li, M.; Su, H.; Huang, Z.; Xu, J.; Pei, J.; et al. Kinetic analysis of effects of temperature and time on the regulation of venom expression in Bungarus multicinctus. Sci. Rep. 2020, 10, 14142. [Google Scholar] [CrossRef] [PubMed]
- Watson, E.L. GTP-binding proteins and regulated exocytosis. Crit. Rev. Oral Biol. Med. 1999, 10, 284–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, T.S.; Georgieva, D.; Genov, N.; Murakami, M.T.; Sinha, M.; Kumar, R.P.; Kaur, P.; Kumar, S.; Dey, S.; Sharma, S.; et al. Enzymatic toxins from snake venom: Structural characterization and mechanism of catalysis. FEBS J. 2011, 278, 4544–4576. [Google Scholar] [CrossRef] [PubMed]
- Tambourgi, D.V.; Magnoli, F.C.; van den Berg, C.W.; Morgan, B.P.; de Araujo, P.S.; Alves, E.W.; Da Silva, W.D. Sphingomyelinases in the venom of the spider Loxosceles intermedia are responsible for both dermonecrosis and complement-dependent hemolysis. Biochem. Biophys. Res. Commun. 1998, 251, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, A.C.M.; de Santana, C.J.C.; Melani, R.D.; Domont, G.B.; Castro, M.S.; Fontes, W.; Roepstorff, P.; Júnior, O.R.P. Exploring the biological activities and proteome of Brazilian scorpion Rhopalurus agamemnon venom. J. Proteom. 2021, 237, 104119. [Google Scholar] [CrossRef]
- Kamio, M.; Ko, K.C.; Zheng, S.; Wang, B.; Collins, S.L.; Gadda, G.; Tai, P.C.; Derby, C.D. The chemistry of escapin: Identification and quantification of the components in the complex mixture generated by an L-amino acid oxidase in the defensive secretion of the sea snail Aplysia californica. Chemistry 2009, 15, 1597–1603. [Google Scholar] [CrossRef]
- Naumann, G.B.; Silva, L.F.; Silva, L.; Faria, G.; Richardson, M.; Evangelista, K.; Kohlhoff, M.; Gontijo, C.M.; Navdaev, A.; de Rezende, F.F.; et al. Cytotoxicity and inhibition of platelet aggregation caused by an L-amino acid oxidase from Bothrops leucurus venom. Biochim. Biophys. Acta 2011, 1810, 683–694. [Google Scholar] [CrossRef] [Green Version]
- Izidoro, L.F.; Sobrinho, J.C.; Mendes, M.M.; Costa, T.R.; Grabner, A.N.; Rodrigues, V.M.; da Silva, S.L.; Zanchi, F.B.; Zuliani, J.P.; Fernandes, C.F.; et al. Snake venom L-amino acid oxidases: Trends in pharmacology and biochemistry. Biomed Res. Int. 2014, 2014, 196754. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Primers. 2017, 3, 17063. [Google Scholar] [CrossRef]
- Amr, Z.S.; Abu Baker, M.A.; Al-Saraireh, M.; Warrell, D.A. Scorpions and scorpion sting envenoming (scorpionism) in the Arab countries of the Middle East. Toxicon 2021, 191, 83–103. [Google Scholar] [CrossRef]
- Smith, L.R.; Potgieter, P.D.; Chappell, W.A. Scorpion sting producing severe muscular paralysis. A case report. S. Afr. Med. J. 1983, 64, 69–70. [Google Scholar] [PubMed]
- Villa-Manzano, A.I.; Vazquéz-Solís, M.G.; Zamora-López, X.X.; Arias-Corona, F.; Palomera-Ávila, F.M.; Pulido-Galaviz, C.; Pacifuentes-Orozco, A. Scorpionism causing severe acute flaccid paralysis. Case report. Rev. Med. Inst. Mex. Seguro Soc. 2016, 54, 265–268. [Google Scholar] [PubMed]
- Thany, S.H.; Tricoire-Leignel, H.; Lapied, B. Identification of cholinergic synaptic transmission in the insect nervous system. Adv. Exp. Med. Biol. 2010, 683, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gepner, J.I.; Hall, L.M.; Sattelle, D.B. Insect acetylcholine receptors as a site of insecticide action. Nature 1978, 276, 188–190. [Google Scholar] [CrossRef]
- Walter, A.; Bechsgaard, J.; Scavenius, C.; Dyrlund, T.S.; Sanggaard, K.W.; Enghild, J.J.; Bilde, T. Characterisation of protein families in spider digestive fluids and their role in extra-oral digestion. BMC Genom. 2017, 18, 600. [Google Scholar] [CrossRef] [PubMed]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.; Kerkkamp, H.M.; Vos, R.A.; Guerreiro, I.; Calvete, J.J.; et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 2013, 110, 20651–20656. [Google Scholar] [CrossRef] [Green Version]
- Pedrini, N.; Ortiz-Urquiza, A.; Huarte-Bonnet, C.; Zhang, S.; Keyhani, N.O. Targeting of insect epicuticular lipids by the entomopathogenic fungus Beauveria bassiana: Hydrocarbon oxidation within the context of a host-pathogen interaction. Front. Microbiol. 2013, 4, 24. [Google Scholar] [CrossRef] [Green Version]
- McReynolds, C.N. Effect of seasons and scorpion size on the foraging and diet of the striped bark scorpion, Centruroides vittatus (Buthidae: Scorpiones) in blackbrush habitat of south Texas. Euscorpius Occas Publ. Scorpiol. 2020, 323, 1–18. [Google Scholar]
- Fuzita, F.J.; Pinkse, M.W.; Verhaert, P.D.; Lopes, A.R. Cysteine cathepsins as digestive enzymes in the spider Nephilengys cruentata. Insect Biochem. Mol. Biol. 2015, 60, 47–58. [Google Scholar] [CrossRef]
- Girish, K.S.; Jagadeesha, D.K.; Rajeev, K.B.; Kemparaju, K. Snake venom hyaluronidase: An evidence for isoforms and extracellular matrix degradation. Mol. Cell. Biochem. 2002, 240, 105–110. [Google Scholar] [CrossRef]
- Long-Rowe, K.O.; Burnett, J.W. Characteristics of hyaluronidase and hemolytic activity in fishing tentacle nematocyst venom of Chrysaora quinquecirrha. Toxicon 1994, 32, 165–174. [Google Scholar] [CrossRef]
- Tu, A.T.; Hendon, R.R. Characterization of lizard venom hyaluronidase and evidence for its action as a spreading factor. Comp. Biochem. Physiol. B 1983, 76, 377–383. [Google Scholar] [CrossRef]
- Trevisan-Silva, D.; Gremski, L.H.; Chaim, O.M.; da Silveira, R.B.; Meissner, G.O.; Mangili, O.C.; Barbaro, K.C.; Gremski, W.; Veiga, S.S.; Senff-Ribeiro, A. Astacin-like metalloproteases are a gene family of toxins present in the venom of different species of the brown spider (genus Loxosceles). Biochimie 2010, 92, 21–32. [Google Scholar] [CrossRef]
- Simpson, J.W. Distribution of collagenolytic enzyme activity among snake venoms. Comp. Biochem. Physiol Part B Comp. Biochem. 1975, 51, 425–431. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Escalante, T.; Rucavado, A.; Herrera, C. Hemorrhage caused by snake venom metalloproteinases: A journey of discovery and understanding. Toxins 2016, 8, 93. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Clemetson, J.M.; Clemetson, K.J. Snake venoms and hemostasis. J. Thromb. Haemost. 2005, 3, 1791–1799. [Google Scholar] [CrossRef]
- Dhananjaya, B.L.; D’Souza, C.J. The pharmacological role of nucleotidases in snake venoms. Cell Biochem. Funct. 2010, 28, 171–177. [Google Scholar] [CrossRef]
- Aird, S.D. Ophidian envenomation strategies and the role of purines. Toxicon 2002, 40, 335–393. [Google Scholar] [CrossRef]
- Katkar, G.D.; Sundaram, M.S.; NaveenKumar, S.K.; Swethakumar, B.; Sharma, R.D.; Paul, M.; Vishalakshi, G.J.; Devaraja, S.; Girish, K.S.; Kemparaju, K. NETosis and lack of DNase activity are key factors in Echis carinatus venom-induced tissue destruction. Nat. Commun. 2016, 7, 11361. [Google Scholar] [CrossRef] [Green Version]
- Possani, L.D.; Merino, E.; Corona, M.; Bolivar, F.; Becerril, B. Peptides and genes coding for scorpion toxins that affect ion-channels. Biochimie 2000, 82, 861–868. [Google Scholar] [CrossRef]
- Zheng, L.L.; Niu, S.; Hao, P.; Feng, K.; Cai, Y.D.; Li, Y. Prediction of protein modification sites of pyrrolidone carboxylic acid using mRMR feature selection and analysis. PLoS ONE 2011, 6, e28221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coelho, V.A.; Cremonez, C.M.; Anjolette, F.A.; Aguiar, J.F.; Varanda, W.A.; Arantes, E.C. Functional and structural study comparing the C-terminal amidated beta-neurotoxin Ts1 with its isoform Ts1-G isolated from Tityus serrulatus venom. Toxicon 2014, 83, 15–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estrada, G.; Restano-Cassulini, R.; Ortiz, E.; Possani, L.D.; Corzo, G. Addition of positive charges at the C-terminal peptide region of CssII, a mammalian scorpion peptide toxin, improves its affinity for sodium channels Nav1.6. Peptides 2011, 32, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, G.; Li, B.; Chi, C.; Wu, X. Cloning, co-expression with an amidating enzyme, and activity of the scorpion toxin BmK ITa1 cDNA in insect cells. Mol. Biotechnol. 2003, 24, 21–26. [Google Scholar] [CrossRef]
- Katopodis, A.G.; May, S.W. Novel substrates and inhibitors of peptidylglycine alpha-amidating monooxygenase. Biochemistry 1990, 29, 4541–4548. [Google Scholar] [CrossRef]
- Ullah, A.; Souza, T.A.; Abrego, J.R.; Betzel, C.; Murakami, M.T.; Arni, R.K. Structural insights into selectivity and cofactor binding in snake venom L-amino acid oxidases. Biochem. Biophys. Res. Commun. 2012, 421, 124–128. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.Y.; Wang, J.Q.; Zhang, Z.B.; Huang, J.M.; Zhu, J.Y. Unraveling the venom components of an encyrtid endoparasitoid wasp Diversinervus elegans. Toxicon 2017, 136, 15–26. [Google Scholar] [CrossRef]
- Liu, N.Y.; Xu, Z.W.; Yan, W.; Ren, X.M.; Zhang, Z.Q.; Zhu, J.Y. Venomics reveals novel ion transport peptide-likes (ITPLs) from the parasitoid wasp Tetrastichus brontispae. Toxicon 2018, 141, 88–93. [Google Scholar] [CrossRef]
- Ramanaiah, M.; Venkaiah, B. Characterization of superoxide dismutase from south Indian scorpion venom. Biochem. Int. 1992, 26, 113–123. [Google Scholar]
- Liu, N.Y.; Huang, J.M.; Ren, X.M.; Xu, Z.W.; Yan, N.S.; Zhu, J.Y. Superoxide dismutase from venom of the ectoparasitoid Scleroderma guani inhibits melanization of hemolymph. Arch. Insect Biochem. Physiol. 2018, 99, e21503. [Google Scholar] [CrossRef]
- Casewell, N.R.; Harrison, R.A.; Wüster, W.; Wagstaff, S.C. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genom. 2009, 10, 564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Riverol, A.; Lasa, A.M.; Dos Santos-Pinto, J.R.A.; Palma, M.S. Insect venom phospholipases A1 and A2: Roles in the envenoming process and allergy. Insect Biochem. Mol. Biol. 2019, 105, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Vardjan, N.; Mattiazzi, M.; Rowan, E.G.; Krizaj, I.; Petrovic, U.; Petan, T. Neurotoxic phospholipase A2 toxicity model: An insight from mammalian cells. Commun. Integr. Biol. 2013, 6, e23600. [Google Scholar] [CrossRef] [PubMed]
- Montecucco, C.; Gutierrez, J.M.; Lomonte, B. Cellular pathology induced by snake venom phospholipase A2 myotoxins and neurotoxins: Common aspects of their mechanisms of action. Cell. Mol. Life Sci. 2008, 65, 2897–2912. [Google Scholar] [CrossRef]
- Cintra-Francischinelli, M.; Caccin, P.; Chiavegato, A.; Pizzo, P.; Carmignoto, G.; Angulo, Y.; Lomonte, B.; Gutiérrez, J.M.; Montecucco, C. Bothrops snake myotoxins induce a large efflux of ATP and potassium with spreading of cell damage and pain. Proc. Natl. Acad. Sci. USA 2010, 107, 14140–14145. [Google Scholar] [CrossRef] [Green Version]
- Zambelli, V.O.; Picolo, G.; Fernandes, C.A.H.; Fontes, M.R.M.; Cury, Y. Secreted phospholipases A(2) from animal venoms in pain and analgesia. Toxins 2017, 9, 406. [Google Scholar] [CrossRef] [Green Version]
- Barboni, E.; Kemeny, D.M.; Campos, S.; Vernon, C.A. The purification of acid phosphatase from honey bee venom (Apis mellifica). Toxicon 1987, 25, 1097–1103. [Google Scholar] [CrossRef]
- Francischetti, I.M.; Mather, T.N.; Ribeiro, J.M. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochem. Biophys. Res. Commun. 2003, 305, 869–875. [Google Scholar] [CrossRef] [Green Version]
- Heidarpour, M.; Ennaifer, E.; Ahari, H.; Srairi-Abid, N.; Borchani, L.; Khalili, G.; Amini, H.; Anvar, A.A.; Boubaker, S.; El-Ayeb, M.; et al. Histopathological changes induced by Hemiscorpius lepturus scorpion venom in mice. Toxicon 2012, 59, 373–378. [Google Scholar] [CrossRef]
- Medina-Santos, R.; Guerra-Duarte, C.; de Almeida Lima, S.; Costal-Oliveira, F.; Alves de Aquino, P.; Oliveira do Carmo, A.; Ferreyra, C.B.; Gonzalez-Kozlova, E.E.; Kalapothakis, E.; Chávez-Olórtegui, C. Diversity of astacin-like metalloproteases identified by transcriptomic analysis in Peruvian Loxosceles laeta spider venom and in vitro activity characterization. Biochimie 2019, 167, 81–92. [Google Scholar] [CrossRef]
- Barbosa Pereira, P.J.; Segura-Martín, S.; Oliva, B.; Ferrer-Orta, C.; Avilés, F.X.; Coll, M.; Gomis-Rüth, F.X.; Vendrell, J. Human procarboxypeptidase B: Three-dimensional structure and implications for thrombin-activatable fibrinolysis inhibitor (TAFI). J. Mol. Biol. 2002, 321, 537–547. [Google Scholar] [CrossRef]
- Calvete, J.J.; Fasoli, E.; Sanz, L.; Boschetti, E.; Righetti, P.G. Exploring the venom proteome of the western diamondback rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteom. Res. 2009, 8, 3055–3067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junqueira-de-Azevedo, I.L.; Bastos, C.M.; Ho, P.L.; Luna, M.S.; Yamanouye, N.; Casewell, N.R. Venom-related transcripts from Bothrops jararaca tissues provide novel molecular insights into the production and evolution of snake venom. Mol. Biol. Evol. 2015, 32, 754–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Velasco, J.; Card, D.C.; Andrew, A.L.; Shaney, K.J.; Adams, R.H.; Schield, D.R.; Casewell, N.R.; Mackessy, S.P.; Castoe, T.A. Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom. Mol. Biol. Evol. 2015, 32, 173–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rendón-Anaya, M.; Delaye, L.; Possani, L.D.; Herrera-Estrella, A. Global transcriptome analysis of the scorpion Centruroides noxius: New toxin families and evolutionary insights from an ancestral scorpion species. PLoS ONE 2012, 7, e43331. [Google Scholar] [CrossRef] [Green Version]
- Simone, Y.; van der Meijden, A. Armed stem to stinger: A review of the ecological roles of scorpion weapons. J. Venom. Anim. Toxins Incl. Trop. Dis. 2021, 27, e20210002. [Google Scholar] [CrossRef]
- Nisani, Z.; Boskovic, D.S.; Dunbar, S.G.; Kelln, W.; Hayes, W.K. Investigating the chemical profile of regenerated scorpion (Parabuthus transvaalicus) venom in relation to metabolic cost and toxicity. Toxicon 2012, 60, 315–323. [Google Scholar] [CrossRef]
- Vonk, F.J.; Bittenbinder, M.A.; Kerkkamp, H.M.I.; Grashof, D.G.B.; Archer, J.P.; Afonso, S.; Richardson, M.K.; Kool, J.; van der Meijden, A. A non-lethal method for studying scorpion venom gland transcriptomes, with a review of potentially suitable taxa to which it can be applied. PLoS ONE 2021, 16, e0258712. [Google Scholar] [CrossRef]
- Santibáñez-López, C.E.; Francke, O.F.; Ureta, C.; Possani, L.D. Scorpions from Mexico: From species diversity to venom complexity. Toxins 2015, 8, 2. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.; Scheer, M.; Grote, A.; Schomburg, I.; Schomburg, D. BRENDA, AMENDA and FRENDA the enzyme information system: New content and tools in 2009. Nucleic Acids Res. 2009, 37, D588–D592. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Sonderby, C.K.; Sonderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]






| Species (ID) | Year | Transcriptome | Proteome | Distribution | Enzymatic Activity | Reference |
|---|---|---|---|---|---|---|
| C. hentzi | 2018 | Hydrolases Oxidoreductases | Hydrolases Oxidoreductases | North America | ND | [22] |
| C. limpidus | 2019 | Hydrolases | Hydrolases Transferases | North America | Protease Hyaluronidase | [23] |
| C. hirsutipalpus | 2020 | Hydrolases Oxidoreductases | Hydrolases Oxidoreductases | North America | ND | [24] |
| C. vittatus | 2017 | Hydrolases | ND | North America | ND | [25] |
| M. martensii | 2021 | Hydrolases | ND | East Asia | Hyaluronidase | [26] |
| H. aztecus | 2019 | Hydrolases | ND | North America | ND | [20] |
| H. spadix | 2017 | Hydrolases Oxidoreductases Transferases | Hydrolases | North America | ND | [27] |
| H. concolorus | 2019 | Hydrolases | ND | North America | ND | [20] |
| M. gerstchi | 2017 | Hydrolases | Hydrolases | Mexico | Phospholipase | [28] |
| S. donensis | 2016 | Hydrolases | Hydrolases | North America | ND | [21] |
| P. schwenkmeyeri | 2018 | Hydrolases | Hydrolases | Mexico | Protease Phospholipase Hyaluronidase | [29] |
| S. gertschi | 2018 | Hydrolases | Hydrolases | North America | Hyaluronidase Phospholipase Gelatinolytic | [30] |
| T. atrox | 2017 | Hydrolases | Hydrolases | Mexico | ND | [31] |
| U. yaschenkoi | 2015 | Hydrolases Transferases | ND | Australia | ND | [32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delgado-Prudencio, G.; Cid-Uribe, J.I.; Morales, J.A.; Possani, L.D.; Ortiz, E.; Romero-Gutiérrez, T. The Enzymatic Core of Scorpion Venoms. Toxins 2022, 14, 248. https://doi.org/10.3390/toxins14040248
Delgado-Prudencio G, Cid-Uribe JI, Morales JA, Possani LD, Ortiz E, Romero-Gutiérrez T. The Enzymatic Core of Scorpion Venoms. Toxins. 2022; 14(4):248. https://doi.org/10.3390/toxins14040248
Chicago/Turabian StyleDelgado-Prudencio, Gustavo, Jimena I. Cid-Uribe, J. Alejandro Morales, Lourival D. Possani, Ernesto Ortiz, and Teresa Romero-Gutiérrez. 2022. "The Enzymatic Core of Scorpion Venoms" Toxins 14, no. 4: 248. https://doi.org/10.3390/toxins14040248
APA StyleDelgado-Prudencio, G., Cid-Uribe, J. I., Morales, J. A., Possani, L. D., Ortiz, E., & Romero-Gutiérrez, T. (2022). The Enzymatic Core of Scorpion Venoms. Toxins, 14(4), 248. https://doi.org/10.3390/toxins14040248







