Tissular Genomic Responses to Oral FB1 Exposure in Pigs
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Analysis of FB1 Effects on Gene Expression in Different Tissues
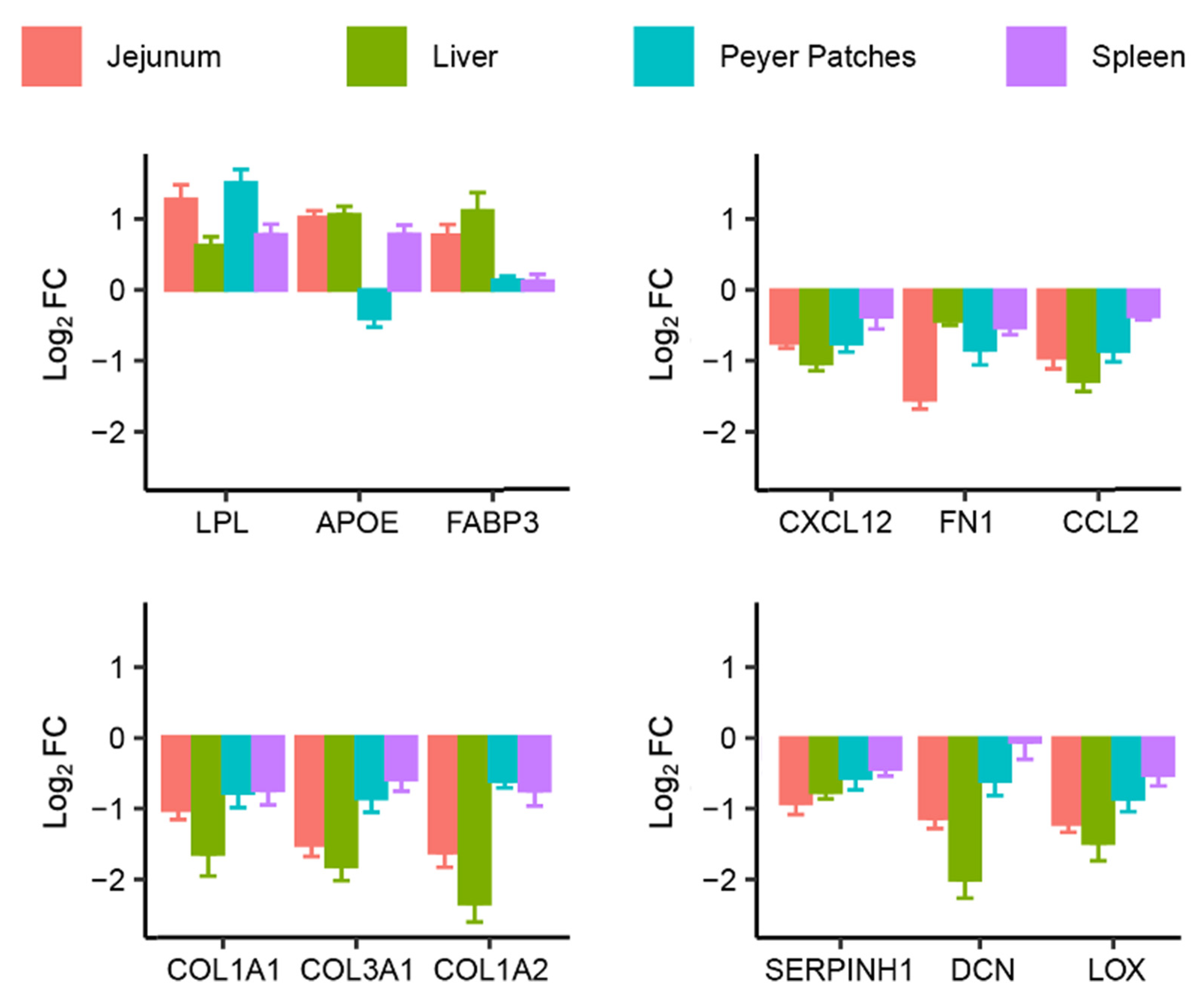
2.2. Identification of Common Fumonisin B1-Sensitive Biological Processes across Different Tissues
2.3. Identification of Tissue-Specific Biological Processes Sensitive to FB1 Exposure
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Animals
5.2. Feeding Trial and Sampling
5.3. Gene Expression Analysis by Microarray
5.4. Bioinformatic Data Analysis
5.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cano, P.M.; Puel, O.; Oswald, I.P. Mycotoxins: Fungal Secondary Metabolites with Toxic Properties, in: Fungi: Applications and Management Strategies; CRC Press: Boca Raton, FL, USA, 2016; pp. 318–371. [Google Scholar]
- Nnadi, N.E.; Carter, D.A. Climate change and the emergence of fungal pathogens. PLoS Pathog. 2021, 17, e1009503. [Google Scholar] [CrossRef] [PubMed]
- Goertz, A.; Zuehlke, S.; Spiteller, M.; Steiner, U.; Dehne, H.W.; Waalwijk, C.; de Vries, I.; Oerke, E.C. Fusarium species and mycotoxin profiles on commercial maize hybrids in Germany. Eur. J. Plant Pathol. 2010, 128, 101–111. [Google Scholar] [CrossRef]
- Knutsen, H.-K.; Alexander, J.; Barregård, L.; Bignami, M.; Brüschweiler, B.; Ceccatelli, S.; Cottrill, B.; Dinovi, M.; Edler, L.; Gra-lKraupp, B.; et al. Risks for animal health related to the presence of fumonisins, their modified forms and hidden forms in feed. EFSA J. 2018, 16, e05242. [Google Scholar] [CrossRef]
- European Commission. Commission Recommendation of 17 August 2006 on the presence of deoxynivalenol, zearalenone, ochratoxin A, T-2 and HT-2 and fumonisins in products intended for animal feeding. Off. J. Eur. Union 2006, 2006, 576. [Google Scholar]
- European Commission. Commission Regulation (EC) No 1126/2007 of 28 September 2007 setting maximum levels for certain contaminants in foodstuffs as regards Fusarium toxins in maize and maize products. Off. J. Eur. Union 2007, 2007, 1126. [Google Scholar]
- Terciolo, C.; Bracarense, A.P.; Souto, P.C.; Cossalter, A.-M.; Dopavogui, L.; Loiseau, N.; Oliveira, C.A.F.; Pinton, P.; Oswald, I.P. Fumonisins at Doses below EU Regulatory Limits Induce Histological Alterations in Piglets. Toxins 2019, 11, 548. [Google Scholar] [CrossRef] [Green Version]
- Devriendt, B.; Gallois, M.; Verdonck, F.; Wache, Y.; Bimczok, D.; Oswald, I.P.; Goddeeris, B.; Cox, E. The food contaminant fumonisin B1reduces the maturation of porcine CD11R1+intestinal antigen presenting cells and antigen-specific immune responses, leading to a prolonged intestinal ETEC infection. Veter-Res. 2009, 40, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Halloy, D.J.; Gustin, P.G.; Bouhet, S.; Oswald, I.P. Oral exposure to culture material extract containing fumonisins predisposes swine to the development of pneumonitis caused by Pasteurellamultocida. Toxicology 2005, 213, 34–44. [Google Scholar] [CrossRef]
- Loiseau, N.; Debrauwer, L.; Sambou, T.; Bouhet, S.; Miller, J.D.; Martin, P.G.; Viadère, J.-L.; Pinton, P.; Puel, O.; Pineau, T.; et al. Fumonisin B1 exposure and its selective effect on porcine jejunal segment: Sphingolipids, glycolipids and trans-epithelial passage disturbance. Biochem. Pharmacol. 2007, 74, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Norred, W.P.; Bacon, C.W.; Riley, R.T.; Merrill, A.H., Jr. Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 1991, 266, 14486–14490. [Google Scholar] [CrossRef]
- Grenier, B.; Bracarense, A.-P.F.; Schwartz-Zimmermann, H.; Trumel, C.; Cossalter, A.-M.; Schatzmayr, G.; Kolf-Clauw, M.; Moll, W.-D.; Oswald, I.P. The low intestinal and hepatic toxicity of hydrolyzed fumonisin B1 correlates with its inability to alter the metabolism of sphingolipids. Biochem. Pharmacol. 2012, 83, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; An, N.; Showker, J.; Yoo, H.; Norred, W.; Chamberlain, W.; Wang, E.; Merrill, A.; Motelin, G.; Beasley, V.; et al. Alteration of Tissue and Serum Sphinganine to Sphingosine Ratio: An Early Biomarker of Exposure to Fumonisin-Containing Feeds in Pigs. Toxicol. Appl. Pharmacol. 1993, 118, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Bouhet, S.; Le Dorze, E.; Peres, S.; Fairbrother, J.M.; Oswald, I.P. Mycotoxin fumonisin B1 selectively down-regulates the basal IL-8 expression in pig intestine: In vivo and in vitro studies. Food Chem. Toxicol. 2006, 44, 1768–1773. [Google Scholar] [CrossRef] [PubMed]
- Grenier, B.; Bracarense, A.P.L.; Schwartz-Zimmermann, H.; Lucioli, J.; Cossalter, A.-M.; Moll, W.-D.; Schatzmayr, G.; Oswald, I.P. Biotransformation Approaches To Alleviate the Effects Induced by Fusarium Mycotoxins in Swine. J. Agric. Food Chem. 2013, 61, 6711–6719. [Google Scholar] [CrossRef]
- Régnier, M.; Gourbeyre, P.; Pinton, P.; Napper, S.; Laffite, J.; Cossalter, A.; Bailly, J.; Lippi, Y.; Bertrand-Michel, J.; Bracarense, A.P.F.; et al. Identification of Signaling Pathways Targeted by the Food Contaminant FB1: Transcriptome and Kinome Analysis of Samples from Pig Liver and Intestine. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Régnier, M.; Polizzi, A.; Lukowicz, C.; Smati, S.; Lasserre, F.; Lippi, Y.; Naylies, C.; Laffitte, J.; Bétoulières, C.; Montagner, A.; et al. The protective role of liver X receptor (LXR) during fumonisin B1-induced hepatotoxicity. Arch. Toxicol. 2019, 93, 505–517. [Google Scholar] [CrossRef]
- Lallès, J.-P.; Lessard, M.; Oswald, I.P.; David, J.-C. Consumption of fumonisin B1 for 9 days induces stress proteins along the gastrointestinal tract of pigs. Toxicon 2010, 55, 244–249. [Google Scholar] [CrossRef]
- Yu, S.; Jia, B.; Liu, N.; Yu, D.; Zhang, S.; Wu, A. Fumonisin B1 triggers carcinogenesis via HDAC/PI3K/Akt signalling pathway in human esophageal epithelial cells. Sci. Total Environ. 2021, 787, 147405. [Google Scholar] [CrossRef]
- Lessard, M.; Boudry, G.; Sève, B.; Oswald, I.P.; Lallès, J.-P. Intestinal Physiology and Peptidase Activity in Male Pigs Are Modulated by Consumption of Corn Culture Extracts Containing Fumonisins. J. Nutr. 2009, 139, 1303–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burel, C.; Tanguy, M.; Guerre, P.; Boilletot, E.; Cariolet, R.; Queguiner, M.; Postollec, G.; Pinton, P.; Salvat, G.; Oswald, I.P.; et al. Effect of Low Dose of Fumonisins on Pig Health: Immune Status, Intestinal Microbiota and Sensitivity to Salmonella. Toxins 2013, 5, 841–864. [Google Scholar] [CrossRef] [Green Version]
- Taranu, I.; Marin, D.E.; Bouhet, S.; Pascale, F.; Bailly, J.-D.; Miller, J.D.; Pinton, P.; Oswald, I.P. Mycotoxin Fumonisin B1 Alters the Cytokine Profile and Decreases the Vaccinal Antibody Titer in Pigs. Toxicol. Sci. 2005, 84, 301–307. [Google Scholar] [CrossRef] [Green Version]
- Marin, D.E.; Taranu, I.; Pascale, F.; Lionide, A.; Burlacu, R.; Bailly, J.-D.; Oswald, I.P. Sex-related differences in the immune response of weanling piglets exposed to low doses of fumonisin extract. Br. J. Nutr. 2006, 95, 1185–1192. [Google Scholar] [CrossRef]
- Voss, K.A.; Riley, R.T. Fumonisin Toxicity and Mechanism of Action: Overview and Current Perspectives. Food Saf. 2013, 1, 2013006. [Google Scholar] [CrossRef] [Green Version]
- Humphreys, S.H.; Carrington, C.; Bolger, M. A quantitative risk assessment for fumonisins B1and B2in US corn. Food Addit. Contam. 2001, 18, 211–220. [Google Scholar] [CrossRef]
- Bouhet, S.; Oswald, I.P. The intestine as a possible target for fumonisin toxicity. Mol. Nutr. Food Res. 2007, 51, 925–931. [Google Scholar] [CrossRef]
- Mateos, I.; Combes, S.; Pascal, G.; Cauquil, L.; Barilly, C.; Cossalter, A.-M.; Laffitte, J.; Botti, S.; Pinton, P.; Oswald, I.P. Fumonisin-Exposure Impairs Age-Related Ecological Succession of Bacterial Species in Weaned Pig Gut Microbiota. Toxins 2018, 10, 230. [Google Scholar] [CrossRef] [Green Version]
- Knutsen, H.; Barregård, L.; Bignami, M.; Brüschweiler, B.; Ceccatelli, S.; Cottrill, B.; Dinovi, M.; Edler, L.; Grasl-Kraupp, B.; Hogstrand, C.; et al. Appropriateness to set a group health-based guidance value for fumonisins and their modified forms. EFSA J. 2018, 16, e05172. [Google Scholar] [CrossRef]
- Haschek, W.M.; Motelin, G.; Ness, D.K.; Harlin, K.S.; Hall, W.F.; Vesonder, R.F.; Peterson, R.E.; Beasley, V.R. Characterization of fumonisin toxicity in orally and intravenously dosed swine. Mycopathologia 1992, 117, 83–96. [Google Scholar] [CrossRef]
- Loiseau, N.; Polizzi, A.; Dupuy, A.; Therville, N.; Rakotonirainy, M.; Loy, J.; Viadere, J.-L.; Cossalter, A.-M.; Bailly, J.-D.; Puel, O.; et al. New insights into the organ-specific adverse effects of fumonisin B1: Comparison between lung and liver. Arch. Toxicol. 2015, 89, 1619–1629. [Google Scholar] [CrossRef]
- Szabó, A.; Szabó-Fodor, J.; Fébel, H.; Mézes, M.; Bajzik, G.; Kovács, M. Oral administration of fumonisin B1and T-2 individually and in combination affects hepatic total and mitochondrial membrane lipid profile of rabbits. Physiol. Int. 2016, 103, 321–333. [Google Scholar] [CrossRef] [Green Version]
- Riedel, S.; Abel, S.; Burger, H.-M.; van der Westhuizen, L.; Swanevelder, S.; Gelderblom, W. Differential modulation of the lipid metabolism as a model for cellular resistance to fumonisin B1–induced cytotoxic effects in vitro. Prostaglandins Leukot. Essent. Fat. Acids 2016, 109, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Bracarense, A.-P.F.L.; Lucioli, J.; Grenier, B.; Pacheco, G.D.; Moll, W.-D.; Schatzmayr, G.; Oswald, I.P. Chronic ingestion of deoxynivalenol and fumonisin, alone or in interaction, induces morphological and immunological changes in the intestine of piglets. Br. J. Nutr. 2012, 107, 1776–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fodor, J.; Balogh, K.; Weber, M.; Miklós, M.; Kametler, L.; Pósa, R.; Mamet, R.; Bauer, J.; Horn, P.; Kovács, F.; et al. Absorption, distribution and elimination of fumonisin B(1) metabolites in weaned piglets. Food Addit. Contam. Part Chem. Anal. Control Expo. Risk Assess. 2008, 25, 88–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piva, A.; Casadei, G.; Pagliuca, G.; Cabassi, E.; Galvano, F.; Solfrizzo, M.; Riley, R.T.; Diaz, D.E. Activated carbon does not prevent the toxicity of culture material containing fumonisin B1 when fed to weanling piglets1. J. Anim. Sci. 2005, 83, 1939–1947. [Google Scholar] [CrossRef] [Green Version]
- Meissonnier, G.; Laffitte, J.; Raymond, I.; Benoit, E.; Cossalter, A.-M.; Pinton, P.; Bertin, G.; Oswald, I.; Galtier, P. Subclinical doses of T-2 toxin impair acquired immune response and liver cytochrome P450 in pigs. Toxicology 2008, 247, 46–54. [Google Scholar] [CrossRef]
- Pierron, A.; Mimoun, S.; Murate, L.S.; Loiseau, N.; Lippi, Y.; Bracarense, A.-P.F.L.; Schatzmayr, G.; He, J.W.; Zhou, T.; Moll, W.-D.; et al. Microbial biotransformation of DON: Molecular basis for reduced toxicity. Sci. Rep. 2016, 6, 29105. [Google Scholar] [CrossRef] [Green Version]
- Pierron, A.; Neves, M.; Puel, S.; Lippi, Y.; Soler, L.; Miller, J.D.; Oswald, I.P. Intestinal toxicity of the new type A trichothecenes, NX and 3ANX. Chemosphere 2021, 288, 132415. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
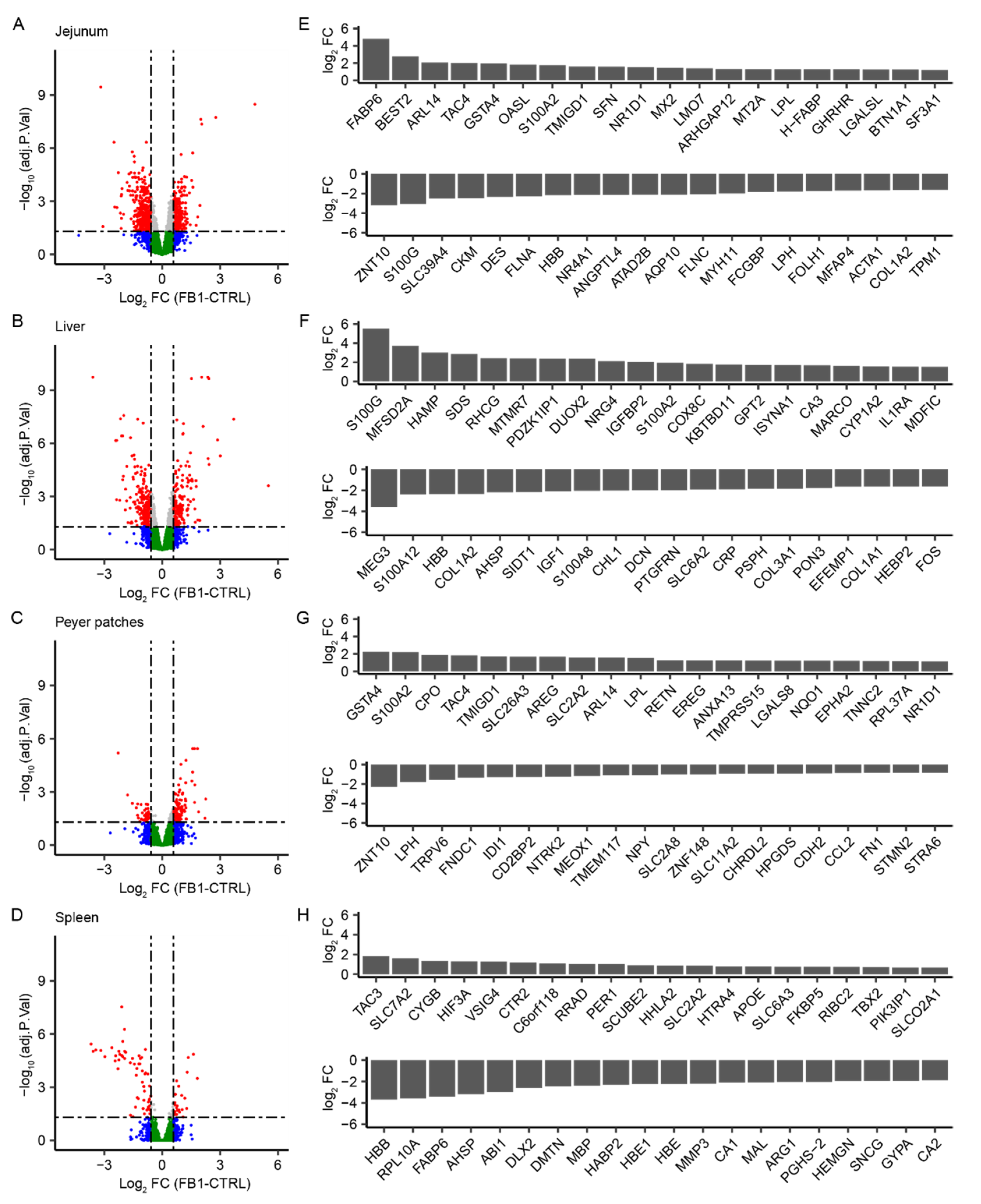
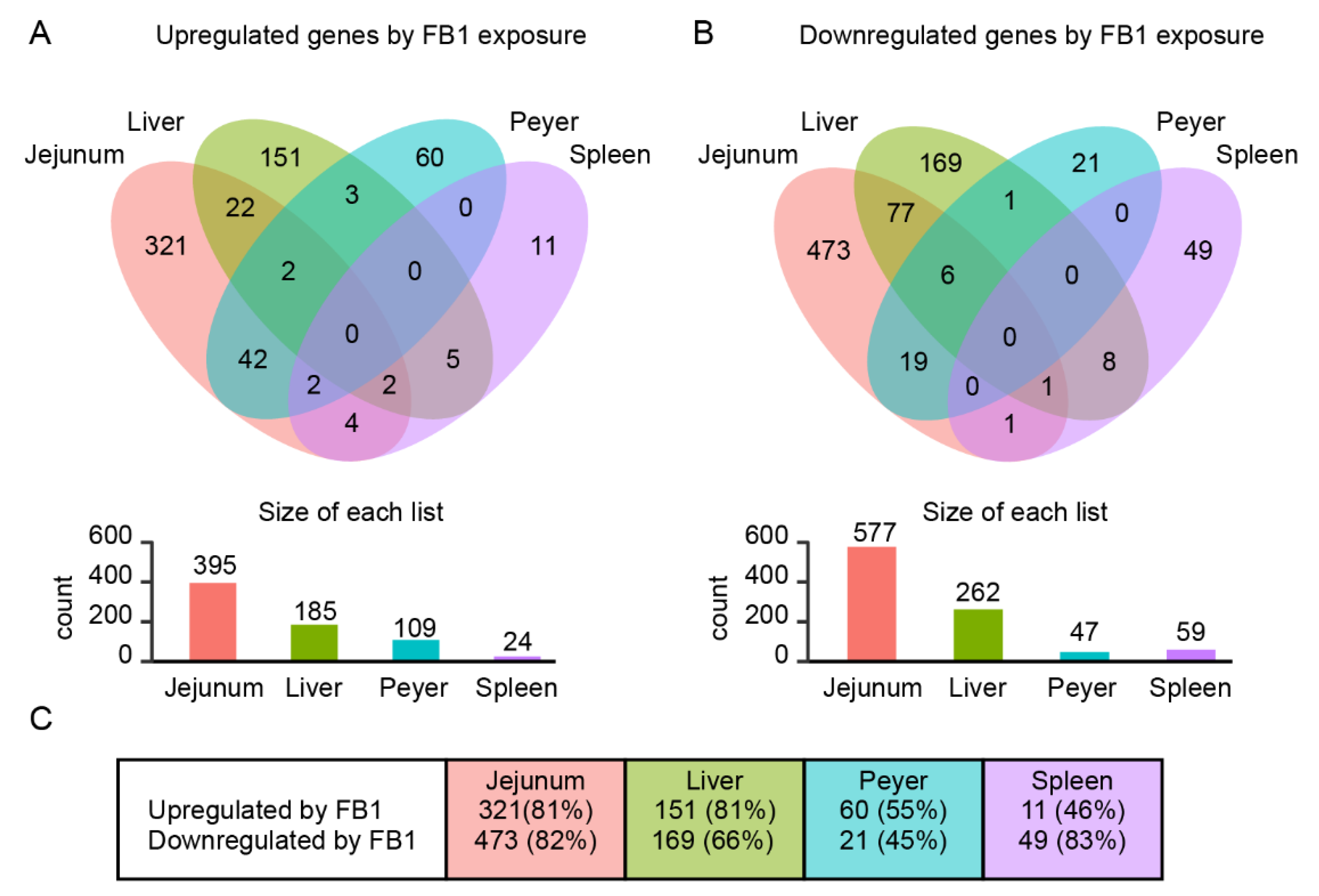

| Comparison | P < 0.05 | FC > 1.0 | FC > 1.2 | FC > 1.5 | FC > 2 | FC > 4 | FC > 6 | FC > 10 |
|---|---|---|---|---|---|---|---|---|
| Jejunum FB1- Jejunum Ctrl | Up | 1536 | 1521 | 538 | 71 | 7 | 4 | 1 |
| Down | 1172 | 1168 | 715 | 193 | 17 | 3 | 0 | |
| Total | 2708 | 2689 | 1253 | 264 | 24 | 7 | 1 | |
| Liver FB1- Liver Ctrl | Up | 477 | 477 | 241 | 81 | 10 | 4 | 2 |
| Down | 454 | 454 | 194 | 102 | 12 | 2 | 2 | |
| Total | 931 | 931 | 435 | 183 | 22 | 6 | 4 | |
| Peyer FB1- Peyer Ctrl | Up | 154 | 154 | 123 | 38 | 3 | 0 | 0 |
| Down | 73 | 73 | 62 | 17 | 2 | 0 | 0 | |
| Total | 227 | 227 | 185 | 55 | 5 | 0 | 0 | |
| Spleen FB1- Spleen Ctrl | Up | 53 | 53 | 33 | 10 | 1 | 0 | 0 |
| Down | 81 | 81 | 73 | 44 | 19 | 7 | 3 | |
| Total | 134 | 134 | 106 | 54 | 20 | 7 | 3 |
| Term | Overlap | Adjusted p Value | Genes | Ref |
|---|---|---|---|---|
| Common upregulated genes (a) | ||||
| triglyceride homeostasis (GO:0070328) | 4/31 | 0.003 | HNF4A; NR1H4; LPL; APOE | [16,17] |
| cholesterol homeostasis (GO:0042632) | 5/71 | 0.003 | FABP3; HNF4A; LPL; APOE; NR1D1 | [16,17] |
| sterol homeostasis (GO:0055092) | 5/72 | 0.003 | FABP3; HNF4A; LPL; APOE; NR1D1 | [16,17] |
| sulfur compound biosynthetic process (GO:0044272) | 5/113 | 0.019 | GSTA4; CTH; MGST1; GCLM; PAPSS2 | [18] |
| acylglycerol homeostasis (GO:0055090) | 3/25 | 0.023 | HNF4A; LPL; APOE | [16,17] |
| regulation of primary metabolic process (GO:0080090) | 5/130 | 0.027 | NQO1; HNF4A; FASN; APOE; NR1D1 | [16,17,18] |
| apoptotic mitochondrial changes (GO:0008637) | 3/33 | 0.036 | BAD; ATP2A1; SFN | [19] |
| positive regulation of phospholipid biosynthetic process (GO:0071073) | 2/7 | 0.036 | FABP3; NR1H4 | [16,17] |
| negative regulation of defense response (GO:0031348) | 5/85 | 0.036 | NR1H4; APOE; NR1D1; KLF4 | [16,19] |
| sodium ion transmembrane transport (GO:0035725) | 5/87 | 0.036 | SCN8A; SCNN1A; ATP1B1; SLC6A3 | [20] |
| Common downregulated genes (b) | ||||
| extracellular matrix organization (GO:0030198) | 27/300 | 5.46 × 10−22 | COL15A1; COL16A1; SPARC; ECM2; COL14A1; COL12A1; HTRA1; DPT; LAMC1; LOXL2; FLRT2; SERPINH1; ITGAV; POSTN; FN1; DCN; COL1A1; GREM1; COL3A1; BMP1; COL1A2; LOX; MMP16; COL4A1; COL5A2; COL6A3; FBN1 | [16] |
| collagen fibril organization (GO:0030199) | 16/89 | 2.63 × 10−17 | COL15A1; COL16A1; COL14A1; COL12A1; DPT; LOXL2; GREM1; COL1A1; COL3A1; BMP1; COL1A2; LOX; COL4A1; COL5A2; SERPINH1; COL6A3 | [16] |
| supramolecular fiber organization (GO:0097435) | 22/351 | 9.89 × 10−15 | COL15A1; COL16A1; TNXB; COL14A1; COL12A1; TPM1; STMN2; DPT; LOXL2; COL1A1; GREM1; EFEMP2; COL3A1; BMP1; COL1A2; ACTC1; LOX; COL4A1; COL5A2; SERPINH1; COL6A3; EMILIN1 | [16] |
| cellular protein metabolic process (GO:0044267) | 12/147 | 7.14 × 10−4 | IL6; RCN1;L GALS1; LOX; SDC2; IGFBP2; FN1; IGFBP7; LAMC1; FSTL1; LOXL2; FBN1 | [12,16,21] |
| integrin activation (GO:0033622) | 5/8 | 0.0012 | COL16A1; CXCL12; FN1 | [16] |
| positive regulation of cellular process (GO:0048522) | 13/625 | 0.0047 | SLC30A2; TPM1; FN1; HBB; GREM1; IL6; CXCL12; CXCR2; ITGAV; S100A9; SOX4; S100A8; TLR2 | [12,16,21] |
| granulocyte chemotaxis (GO:0071621) | 5/73 | 0.0047 | CXCR2; CCL2; S100A12; S100A9; S100A8 | [19,22,23] |
| positive regulation of macromolecule metabolic process (GO:0010604) | 10/384 | 0.0047 | GREM1; ACTA2; GJA1; IL6; ACTC1; FN1; EMILIN1; ANK3; ROBO1; TLR2 | [12,16,21] |
| endothelial cell migration (GO:0043542) | 4/39 | 0.0047 | PLXND1; FAP; FSTL1; LOXL2 | [19] |
| neutrophil migration (GO:1990266) | 5/77 | 0.0049 | CXCR2; CCL2; S100A12; S100A9; S100A8 | [19,22,23] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dopavogui, L.; Polizzi, A.; Fougerat, A.; Gourbeyre, P.; Terciolo, C.; Klement, W.; Pinton, P.; Laffite, J.; Cossalter, A.-M.; Bailly, J.-D.; et al. Tissular Genomic Responses to Oral FB1 Exposure in Pigs. Toxins 2022, 14, 83. https://doi.org/10.3390/toxins14020083
Dopavogui L, Polizzi A, Fougerat A, Gourbeyre P, Terciolo C, Klement W, Pinton P, Laffite J, Cossalter A-M, Bailly J-D, et al. Tissular Genomic Responses to Oral FB1 Exposure in Pigs. Toxins. 2022; 14(2):83. https://doi.org/10.3390/toxins14020083
Chicago/Turabian StyleDopavogui, Léonie, Arnaud Polizzi, Anne Fougerat, Pascal Gourbeyre, Chloé Terciolo, Wendy Klement, Philippe Pinton, Joëlle Laffite, Anne-Marie Cossalter, Jean-Denis Bailly, and et al. 2022. "Tissular Genomic Responses to Oral FB1 Exposure in Pigs" Toxins 14, no. 2: 83. https://doi.org/10.3390/toxins14020083
APA StyleDopavogui, L., Polizzi, A., Fougerat, A., Gourbeyre, P., Terciolo, C., Klement, W., Pinton, P., Laffite, J., Cossalter, A.-M., Bailly, J.-D., Puel, O., Lippi, Y., Naylies, C., Guillou, H., Oswald, I. P., & Loiseau, N. (2022). Tissular Genomic Responses to Oral FB1 Exposure in Pigs. Toxins, 14(2), 83. https://doi.org/10.3390/toxins14020083








