Toxic Habits: An Analysis of General Trends and Biases in Snake Venom Research
Abstract
1. Introduction
2. Results
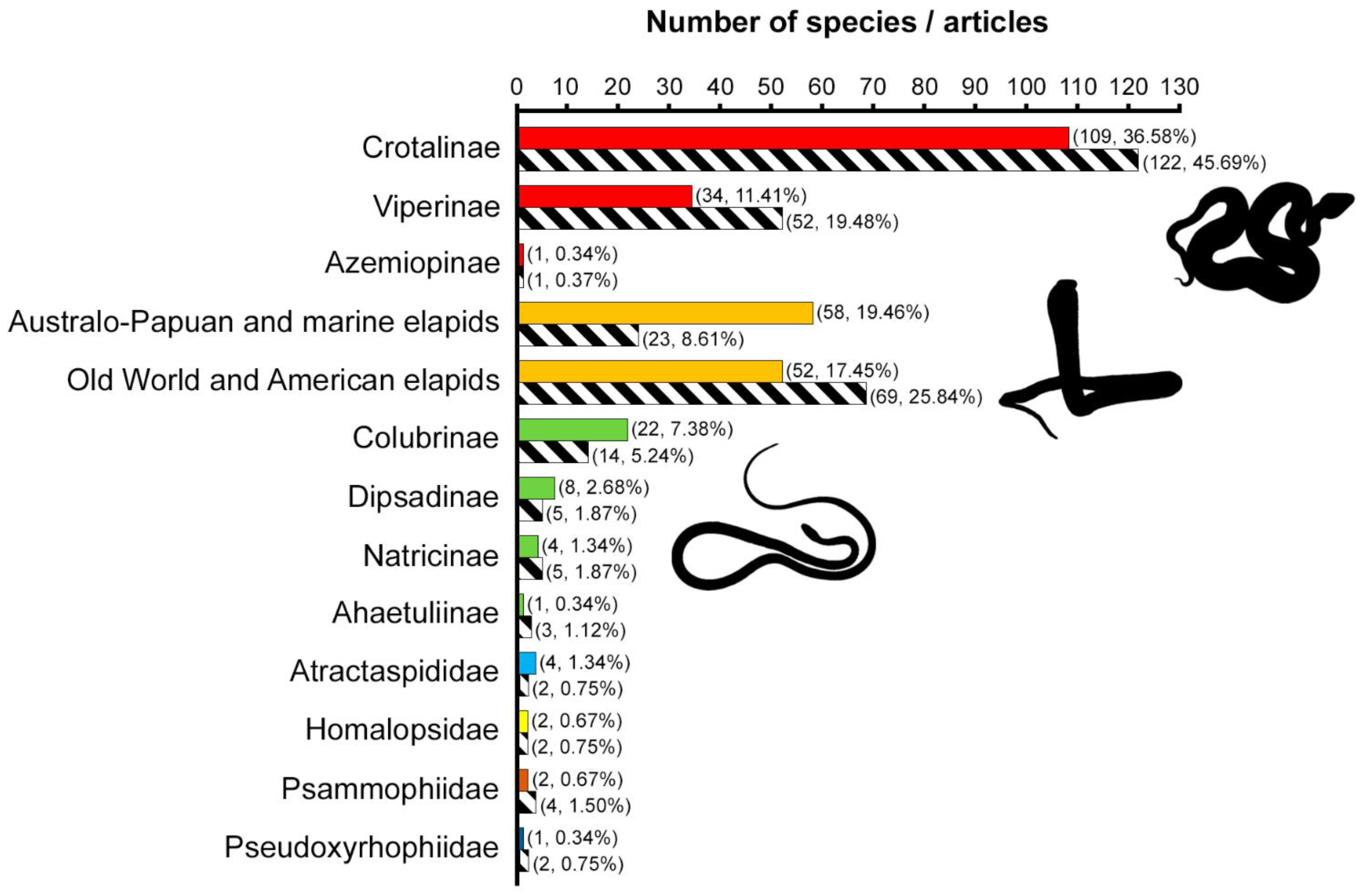
2.1. Taxonomic Information
2.2. Hazard Categories
2.3. Countries and Biogeographic Realms
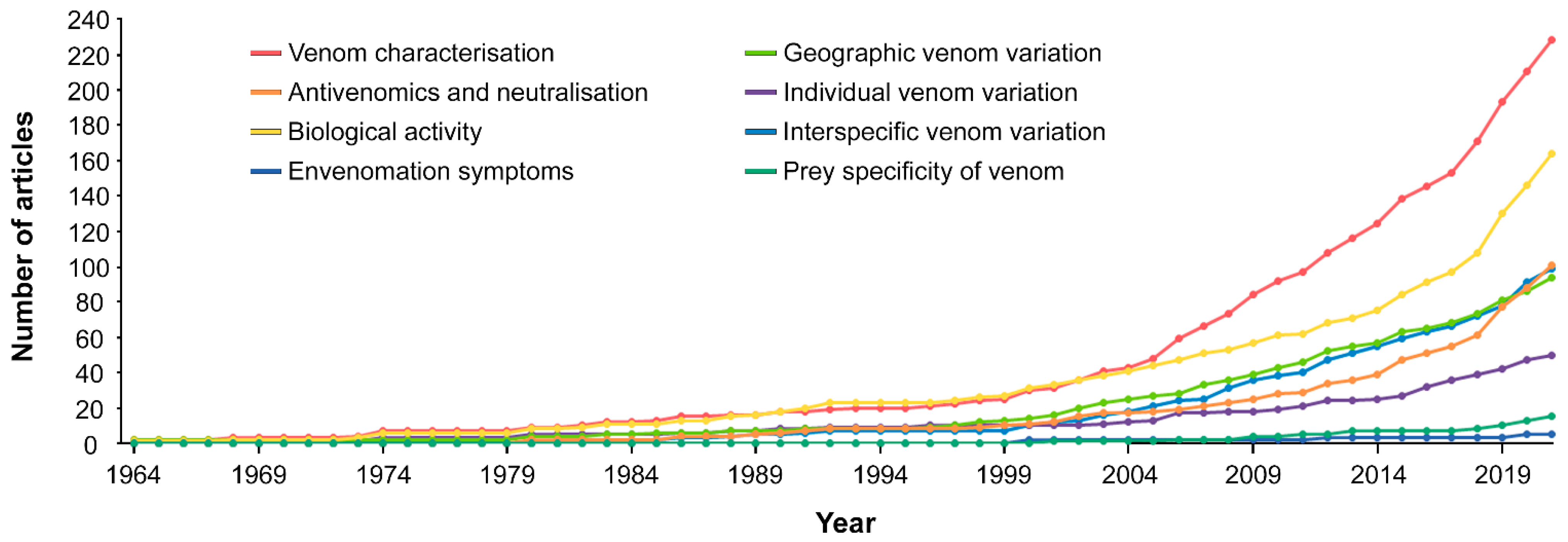
2.4. Topics Investigated
2.5. Chronological Trends
2.5.1. Taxonomic Information: Snake Families and Subfamilies
2.5.2. Topics Investigated
2.6. Factors Influencing the Differences in Number of Articles between Species
3. Discussion
3.1. Viperids Are the Most Studied Snakes
3.2. The Neotropics as a Gold Mine for Snake Venom Studies
3.3. The Neglect of the Ecological Context
4. Conclusions
5. Materials and Methods
5.1. Article Selection
5.2. Taxonomic Information
5.3. Hazard Categories
5.4. Origin of the Specimens
5.5. Topics Covered
5.6. Chronological Trends
5.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jenner, R.A.; Undheim, E. Venom: The Secrets of Nature’s Deadliest Weapon; Smithsonian Institution: Washington, DC, USA, 2017. [Google Scholar]
- Uetz, P.; Freed, P.; Aguilar, R.; Hošek, J. The Reptile Database. 2021. Available online: http://www.reptile-database.org (accessed on 3 October 2022).
- Longbottom, J.; Shearer, F.M.; Devine, M.; Alcoba, G.; Chappuis, F.; Weiss, D.J.; Ray, S.E.; Ray, N.; Warrell, D.A.; Ruiz de Castañeda, R.; et al. Vulnerability to snakebite envenoming: A global mapping of hotspots. Lancet 2018, 392, 673–684. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Snakebite Information and Data Platform. 2020. Available online: https://www.who.int/teams/control-of-neglected-tropical-diseases/snakebite-envenoming/snakebite-information-and-data-platform/overview#tab=tab_1,2020 (accessed on 1 March 2022).
- Calvete, J.J. Snake venomics: From the inventory of toxins to biology. Toxicon 2013, 75, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.S.; Cheung, R.C.F.; Xia, L.; Wong, J.H.; Ng, T.B.; Chan, W.Y. Snake venom toxins: Toxicity and medicinal applications. Appl. Microbiol. Biotechnol. 2016, 100, 6165–6181. [Google Scholar] [CrossRef]
- Cohen, L.; Karbat, I.; Gilles, N.; Ilan, N.; Benveniste, M.; Gordon, D.; Gurevitz, M. Common features in the functional surface of scorpion β-toxins and elements that confer specificity for insect and mammalian voltage-gated sodium channels. J. Biol. Chem. 2005, 280, 5045–5053. [Google Scholar] [CrossRef] [PubMed]
- Herzig, V.; Cristofori-Armstrong, B.; Israel, M.R.; Samantha, A.; Vetter, I.; King, G.F. Animal toxins—Nature’s evolutionary-refined toolkit for basic research and drug discovery. Biochem. Pharmacol. 2020, 181, 114096. [Google Scholar] [CrossRef] [PubMed]
- Lyons, K.; Dugon, M.M.; Healy, K. Diet Breadth Mediates the Prey Specificity of Venom Potency in Snakes. Toxins 2020, 12, 74. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Theakston, R.D.G.; Warrell, D.A. Confronting the neglected problem of snake bite envenoming: The need for a global partnership. PLoS Med. 2006, 3, e150. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Primers 2017, 3, 17063. [Google Scholar] [CrossRef]
- Kasturiratne, A.; Wickremasinghe, A.R.; De Silva, N.; Gunawardena, N.K.; Pathmeswaran, A.; Premaratna, R.; Savioli, L.; Lalloo, D.G.; De Silva, H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008, 5, e218. [Google Scholar] [CrossRef]
- Kasturi, S.; Gowda, T.V. Purification and characterization of a major phospholipase A2 from Russell’s viper (Vipera russelli) venom. Toxicon 1989, 27, 229–237. [Google Scholar] [CrossRef]
- Nishida, S.; Fujimura, Y.; Miura, S.; Yoshida, E.; Sugimoto, M.; Yoshioka, A.; Fukui, H.; Ozaki, Y.; Usami, Y. Purification and characterization of bothrombin, a fibrinogen-clotting serine protease from the venom of Bothrops jararaca. Biochemistry 1994, 33, 1843–1849. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.N.W.; Jouanne, H.; Vidal, N. Snake venom in context: Neglected clades and concepts. Front. Ecol. Evol. 2019, 7, 332. [Google Scholar] [CrossRef]
- Lomonte, B.; Calvete, J.J. Strategies in “snake venomics” aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P.; Saviola, A.J. Understanding biological roles of venoms among the Caenophidia: The importance of rear-fanged snakes. Integr. Comp. Biol. 2016, 56, 1004–1021. [Google Scholar] [CrossRef]
- Fuzita, F.J.; Pinkse, M.W.H.; Patane, J.S.L.; Verhaert, P.D.E.M.; Lopes, A.R. High throughput techniques to reveal the molecular physiology and evolution of digestion in spiders. BMC Genom. 2016, 17, 716. [Google Scholar] [CrossRef]
- Oldrati, V.; Arrell, M.; Violette, A.; Perret, F.; Sprüngli, X.; Wolfender, J.L.; Stöcklin, R. Advances in venomics. Mol. Biosyst. 2016, 12, 3530–3543. [Google Scholar] [CrossRef]
- Post, Y.; Puschhof, J.; Beumer, J.; Kerkkamp, H.M.; de Bakker, M.A.; Slagboom, J.; de Barbanson, B.; Wevers, N.R.; Spijkers, X.M.; Olivier, T.; et al. Snake venom gland organoids. Cell 2020, 180, 233–247. [Google Scholar] [CrossRef]
- Sajevic, T.; Leonardi, A.; Križaj, I. Haemostatically active proteins in snake venoms. Toxicon 2011, 57, 627–645. [Google Scholar] [CrossRef]
- Azim, S.; McDowell, D.; Cartagena, A.; Rodriguez, R.; Laughlin, T.F.; Ahmad, Z. Venom peptides cathelicidin and lycotoxin cause strong inhibition of Escherichia coli ATP synthase. Int. J. Biol. Macromol. 2016, 87, 246–251. [Google Scholar] [CrossRef]
- King, G.F. Venoms as a platform for human drugs: Translating toxins into therapeutics. Expert Opin. Biol. Ther. 2011, 11, 1469–1484. [Google Scholar] [CrossRef]
- Arbuckle, K. From molecules to macroevolution: Venom as a model system for evolutionary biology across levels of life. Toxicon X 2020, 6, 100034. [Google Scholar] [CrossRef] [PubMed]
- Stern, C.D. The ‘Omics Revolution: How an Obsession with Compiling Lists Is Threatening the Ancient Art of Experimental Design. Bioessays 2019, 41, e1900168. [Google Scholar] [CrossRef]
- Gutiérrez, J.M. Snakebite envenoming from an Ecohealth perspective. Toxicon X 2020, 7, 100043. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.A.; Martin, G.; Iwamura, T. Focus on snake ecology to fight snakebite. Lancet 2020, 395, e14. [Google Scholar] [CrossRef] [PubMed]
- Diz, A.P.; Calvete, J.J. Ecological proteomics: Is the field ripe for integrating proteomics into evolutionary ecology research? J. Proteom. 2016, 135, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, A. Bothrops atrox. In Reptiles of Ecuador: Life in the Middle of the World; Arteaga, A., Bustamante, L., Vieira, J., Guayasamin, J.M., Eds.; Universidad Tecnológica Indoamérica: Quito, Ecuador, 2020; Available online: www.reptilesofecuador.com (accessed on 12 February 2021).
- Málaque, C.M.S.; Gutiérrez, J.M. Snakebite envenomation in Central and South America. In Critical Care Toxicology: Diagnosis and Management of the Critically Poisoned Patient; Brent, J., Burkhart, K., Dargan, P., Hatten, B., Megarbane, B., Palmer, R., White, J., Eds.; Springer: Cham, Switzerland, 2017; pp. 1–22. [Google Scholar]
- Mamede, C.C.N.; de Sousa Simamoto, B.B.; da Cunha Pereira, D.F.; de Oliveira Costa, J.; Ribeiro, M.S.M.; de Oliveira, F. Edema, hyperalgesia and myonecrosis induced by Brazilian bothropic venoms: Overview of the last decade. Toxicon 2020, 187, 10–18. [Google Scholar] [CrossRef]
- Karunanayake, R.K.; Dissanayake, D.M.R.; Karunanayake, A.L. A study of snake bite among children presenting to a paediatric ward in the main teaching hospital of North Central Province of Sri Lanka. BMC Res. Notes 2014, 7, 482. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Guidelines for the Management of Snakebites, 2nd ed.; 2016. Available online: https://www.who.int/publications/i/item/9789290225300 (accessed on 15 September 2020).
- World Health Organization (WHO). World Health Organization Guidelines for the Prevention and Clinical Management of Snakebites in Africa. 2010. Available online: https://www.who.int/publications/i/item/9789290231684 (accessed on 15 September 2020).
- Otero-Patiño, R. Epidemiological, clinical and therapeutic aspects of Bothrops asper bites. Toxicon 2009, 54, 998–1011. [Google Scholar] [CrossRef]
- Warrell, D.A. Clinical toxicology of Snake bites in Asia. In Handbook of Clinical Toxicology of Animal Venoms and Poisons; White, M.A., Ed.; CRC Press: New York, NY, USA, 1995; pp. 493–588. [Google Scholar]
- Groneberg, D.A.; Geier, V.; Klingelhöfer, D.; Gerber, A.; Kuch, U.; Kloft, B. Snakebite Envenoming—A Combined Density Equalizing Mapping and Scientometric Analysis of the Publication History. PLoS Negl. Trop. Dis. 2016, 10, e0005046. [Google Scholar] [CrossRef]
- Sofyantoro, F.; Yudha, D.S.; Lischer, K.; Nuringtyas, T.R.; Putri, W.A.; Kusuma, W.A.; Purwestri, Y.A.; Swasono, R.T. Bibliometric Analysis of Literature in Snake Venom-Related Research Worldwide (1933–2022). Animals 2022, 12, 2058. [Google Scholar] [CrossRef]
- Tilbury, C.R.; Branch, W.R. Observations on the bite of the southern burrowing asp (Atractaspis bibronii) in Natal. S. Afr. Med. J. 1989, 75, 327–331. [Google Scholar] [PubMed]
- Weinstein, S.A.; Warrell, D.A.; White, J.; Keyler, D.E. Venomous Bites from Non-Venomous Snakes: A Critical Analysis of Risk and Management of Colubrid Snake Bites; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Guedes, T.B.; Sawaya, R.J.; Zizka, A.; Laffan, S.; Faurby, S.; Pyron, R.A.; Bérnils, R.S.; Jansen, M.; Passos, P.; Prudente, A.L.C.; et al. Patterns, biases and prospects in the distribution and diversity of Neotropical snakes. Glob. Ecol. Biogeogr. 2017, 27, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M. Reducing the impact of snakebite envenoming in Latin America and the Caribbean: Achievements and challenges ahead. Trans. R. Soc. Trop. Med. Hyg. 2014, 108, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.A.; Hargreaves, A.; Wagstaff, S.C.; Faragher, B.; Lalloo, D.G. Snake envenoming: A disease of poverty. PLoS Negl. Trop. Dis. 2009, 3, e569. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M. Exploring snake venom proteomes: Multifaceted analyses for complex toxin mixtures. Proteomics 2008, 8, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J. Venomics: Digging into the evolution of venomous systems and learning to twist nature to fight pathology. J. Proteom. 2009, 72, 121–126. [Google Scholar] [CrossRef]
- Calvete, J.J. Proteomic tools against the neglected pathology of snake bite envenoming. Expert Rev. Proteom. 2011, 8, 739–758. [Google Scholar] [CrossRef]
- Chippaux, J.P. Snake-bites: Appraisal of the global situation. Bull. World Health Organ. 1998, 76, 515–524. [Google Scholar]
- Debono, J.; Dobson, J.; Casewell, N.R.; Romilio, A.; Li, B.; Kurniawan, N.; Mardon, K.; Weisbecker, V.; Nouwens, A.; Kwok, H.F.; et al. Coagulating colubrids: Evolutionary, pathophysiological and biodiscovery implications of venom variations between boomslang (Dispholidus typus) and twig snake (Thelotornis mossambicanus). Toxins 2017, 9, 171. [Google Scholar] [CrossRef]
- Ainsworth, S.; Petras, D.; Engmark, M.; Süssmuth, R.D.; Whiteley, G.; Albulescu, L.O.; Kazandjian, T.D.; Wagstaff, S.C.; Rowley, P.; Wüster, W.; et al. The medical threat of mamba envenoming in sub-Saharan Africa revealed by genus-wide analysis of venom composition, toxicity and antivenomics profiling of available antivenoms. J. Proteom. 2018, 172, 173–189. [Google Scholar] [CrossRef]
- Habib, A.G.; Brown, N.I. The snakebite problem and antivenom crisis from a health-economic perspective. Toxicon 2018, 150, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.A.; Casewell, N.R.; Ainsworth, S.A.; Lalloo, D.G. The time is now: A call for action to translate recent momentum on tackling tropical snakebite into sustained benefit for victims. Trans. R. Soc. Trop. 2019, 113, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Schiermeier, Q. Snakebite crisis gets US$100-million boost for better antivenoms. Nature 2019, 28, 1–2. [Google Scholar] [CrossRef]
- Chippaux, J.P. Snakebite envenomation turns again into a neglected tropical disease! J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 1–2. [Google Scholar] [CrossRef]
- Williams, D.J.; Gutiérrez, J.M.; Calvete, J.J.; Wüster, W.; Ratanabanangkoon, K.; Paiva, O.; Brown, N.I.; Casewell, N.R.; Harrison, R.A.; Rowley, P.D.; et al. Ending the drought: New strategies for improving the flow of affordable, effective antivenoms in Asia and Africa. J. Proteom. 2011, 74, 1735–1767. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.Y.; Sanders, K.L.; King, B.; Palci, A. Diversification rates and phenotypic evolution in venomous snakes (Elapidae). R. Soc. Open Sci. 2016, 3, 150277. [Google Scholar] [CrossRef]
- Williams, D.J.; Wüster, W.; Fry, B.G. The good, the bad and the ugly: Australian snake taxonomists and a history of the taxonomy of Australia’s venomous snakes. Toxicon 2006, 48, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Zaher, H.; Murphy, R.W.; Arredondo, J.C.; Graboski, R.; Machado-Filho, P.R.; Mahlow, K.; Montingelli, G.G.; Quadros, A.B.; Orlov, N.L.; Wilkinson, M.; et al. Large-scale molecular phylogeny, morphology, divergence-time estimation, and the fossil record of advanced caenophidian snakes (Squamata: Serpentes). PLoS ONE 2019, 14, e0216148. [Google Scholar] [CrossRef]
- Scanlon, J.D.; Lee, M.S.Y. Phylogeny of Australasian venomous snakes (Colubroidea, Elapidae, Hydrophiinae) based on phenotypic and molecular evidence. Zool. Scr. 2004, 33, 335–366. [Google Scholar] [CrossRef]
- Slowinski, J.B.; Knight, A.; Rooney, A.P. Inferring Species Trees from Gene Trees: A Phylogenetic Analysis of the Elapidae (Serpentes) Based on the Amino Acid Sequences of Venom Proteins. Mol. Phylogenet. Evol. 1997, 8, 349–362. [Google Scholar] [CrossRef]
- Slowinski, J.B.; Keogh, J.S. Phylogenetic relationships of elapid snakes based on cytochrome b mtDNA sequences. Mol. Phylogenet. Evol. 2000, 15, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Dinerstein, E.; Olson, D.; Joshi, A.; Vynne, C.; Burgess, N.D.; Wikramanayake, E.; Hahn, N.; Palminteri, S.; Hedao, P.; Noss, R.; et al. An Ecoregion-Based Approach to Protecting Half the Terrestrial Realm. Bioscience 2017, 67, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach; Springer: New York, NY, USA, 2002. [Google Scholar]
- SPSS Inc. SPSS for Windows, Version 13.0.; SPSS Inc.: Chicago, IL, USA, 2003.
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021; Available online: https://www.R-project.org (accessed on 13 October 2021).
- Oksanen, J. Design Decisions and Implementation Details in Vegan. Vignette of the Package Vegan. 2016. Available online: https://cran.r-project.org/web/packages/vegan/vignettes/decision-vegan.pdf (accessed on 9 November 2021).
- Bartoń, K. Package “MuMIn”: Multi-Model Inference, R Package Version 1.43.17; 2020. Available online: https://cran.r-project.org/web/packages/MuMIn/MuMIn.pdf (accessed on 9 November 2021).




| Hazard Category | ||||||
|---|---|---|---|---|---|---|
| N Species | Cat. 1 | Cat. 2 | Cat. 3 | Cat. 4 | Unknown | |
| Viperidae | 144 | 73 (50.69%) | 65 (45.14%) | 6 (4.17%) | 0 | 0 |
| Azemiopinae | 1 | 0 | 1 (100%) | 0 | 0 | 0 |
| Crotalinae | 109 | 55 (50.46%) | 54 (49.54%) | 0 | 0 | 0 |
| Viperinae | 34 | 18 (52.94%) | 10 (29.41%) | 6 (17.65) | 0 | 0 |
| Elapidae | 110 | 55 (50%) | 33 (30%) | 15 (13.64%) | 6 (5.45%) | 1 (0.91%) |
| Australo-Papuan and marine elapids | 58 | 25 (43.1%) | 12 (20.69%) | 14 (24.14%) | 6 (10.35%) | 1 (1.72%) |
| Old World and American elapids | 52 | 30 (57.69%) | 21 (40.39%) | 1 (1.92%) | 0 | 0 |
| Colubridae | 35 | 2 (5.71%) | 2 (5.71%) | 12 (34.39%) | 19 (54.39%) | 0 |
| Ahaetuliinae | 1 | 0 | 0 | 0 | 1 (100%) | 0 |
| Colubrinae | 28 | 2 (9.1%) | 0 | 8 (36.4%) | 12 (54.5) | 0 |
| Dipsadinae | 4 | 0 | 0 | 4 (50.0%) | 4 (50.0%) | 0 |
| Natricinae | 2 | 0 | 2 (50.0%) | 0 | 2 (50.0%) | 0 |
| Atractaspididae | 4 | 0 | 4 (100%) | 0 | 0 | 0 |
| Homalopsidae | 2 | 0 | 0 | 0 | 2 (100%) | 0 |
| Psammophiidae | 2 | 0 | 0 | 1 (50.0%) | 1 (50.0%) | 0 |
| Pseudoxyrhophiidae | 1 | 0 | 0 | 0 | 1 (100%) | 0 |
| Total | 298 | 130 (43.62%) | 104 (34.9%) | 34 (11.41%) | 29 (9.73%) | 1 (0.34%) |
| Model | K | AICc | ΔAICc | wAICc |
|---|---|---|---|---|
| Biogeographic realm and Hazard category | 10 | 1058.710 | 0.000 | 0.991 |
| Hazard category | 4 | 1068.161 | 9.450 | 0.009 |
| Biogeographic realm | 7 | 1110.483 | 51.772 | 5.675 × 10−12 |
| Family | 8 | 1115.132 | 56.421 | 5.552 × 10−13 |
| Null | 1 | 1116.503 | 57.792 | 2.797 × 10−13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avella, I.; Wüster, W.; Luiselli, L.; Martínez-Freiría, F. Toxic Habits: An Analysis of General Trends and Biases in Snake Venom Research. Toxins 2022, 14, 884. https://doi.org/10.3390/toxins14120884
Avella I, Wüster W, Luiselli L, Martínez-Freiría F. Toxic Habits: An Analysis of General Trends and Biases in Snake Venom Research. Toxins. 2022; 14(12):884. https://doi.org/10.3390/toxins14120884
Chicago/Turabian StyleAvella, Ignazio, Wolfgang Wüster, Luca Luiselli, and Fernando Martínez-Freiría. 2022. "Toxic Habits: An Analysis of General Trends and Biases in Snake Venom Research" Toxins 14, no. 12: 884. https://doi.org/10.3390/toxins14120884
APA StyleAvella, I., Wüster, W., Luiselli, L., & Martínez-Freiría, F. (2022). Toxic Habits: An Analysis of General Trends and Biases in Snake Venom Research. Toxins, 14(12), 884. https://doi.org/10.3390/toxins14120884







