Putative C2H2 Transcription Factor AflZKS3 Regulates Aflatoxin and Pathogenicity in Aspergillus flavus
Abstract
1. Introduction
2. Results
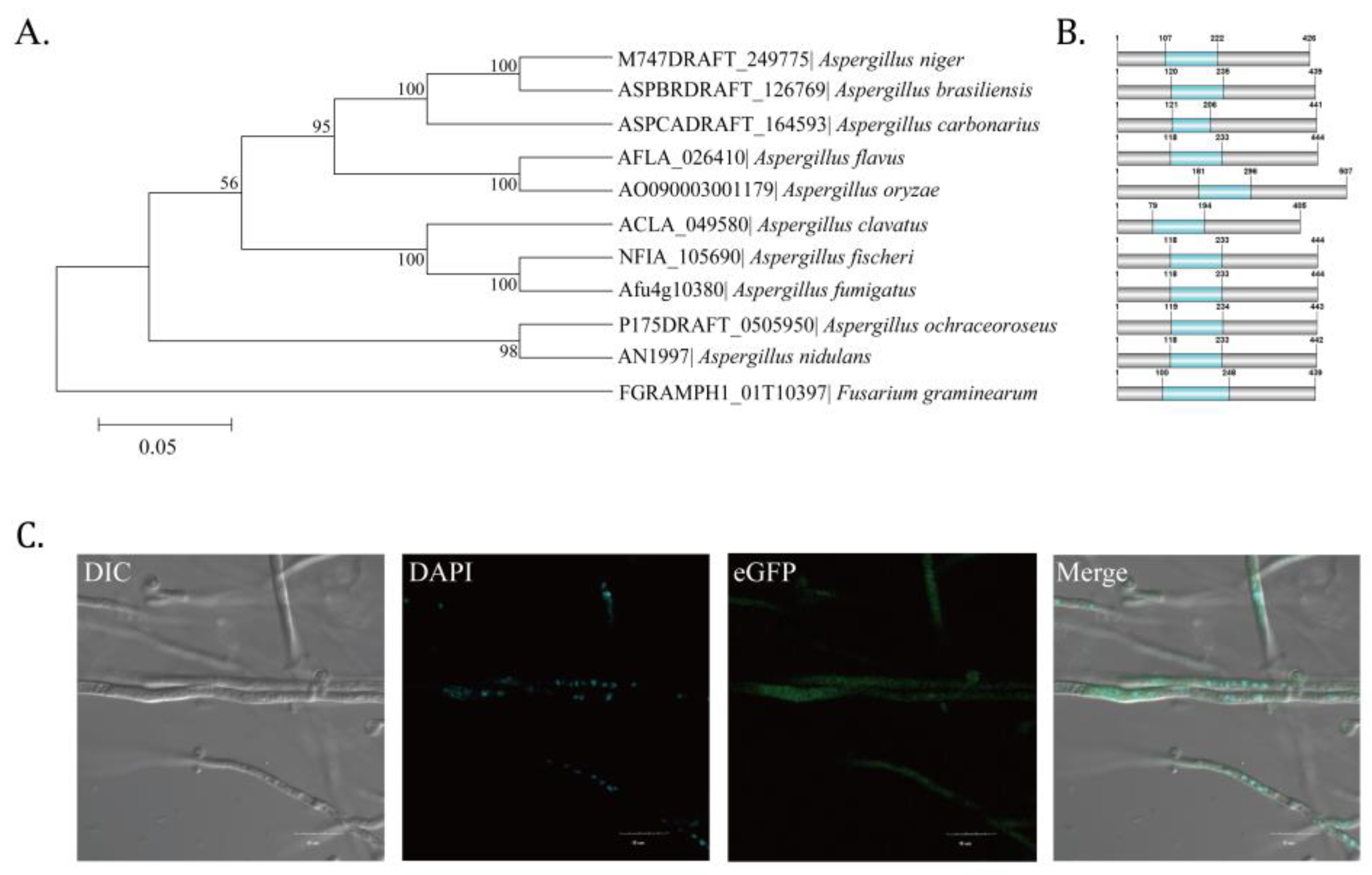
2.1. Identification of Putative C2H2 Zinc Finger Transcription Factor AflZKS3 in A. flavus
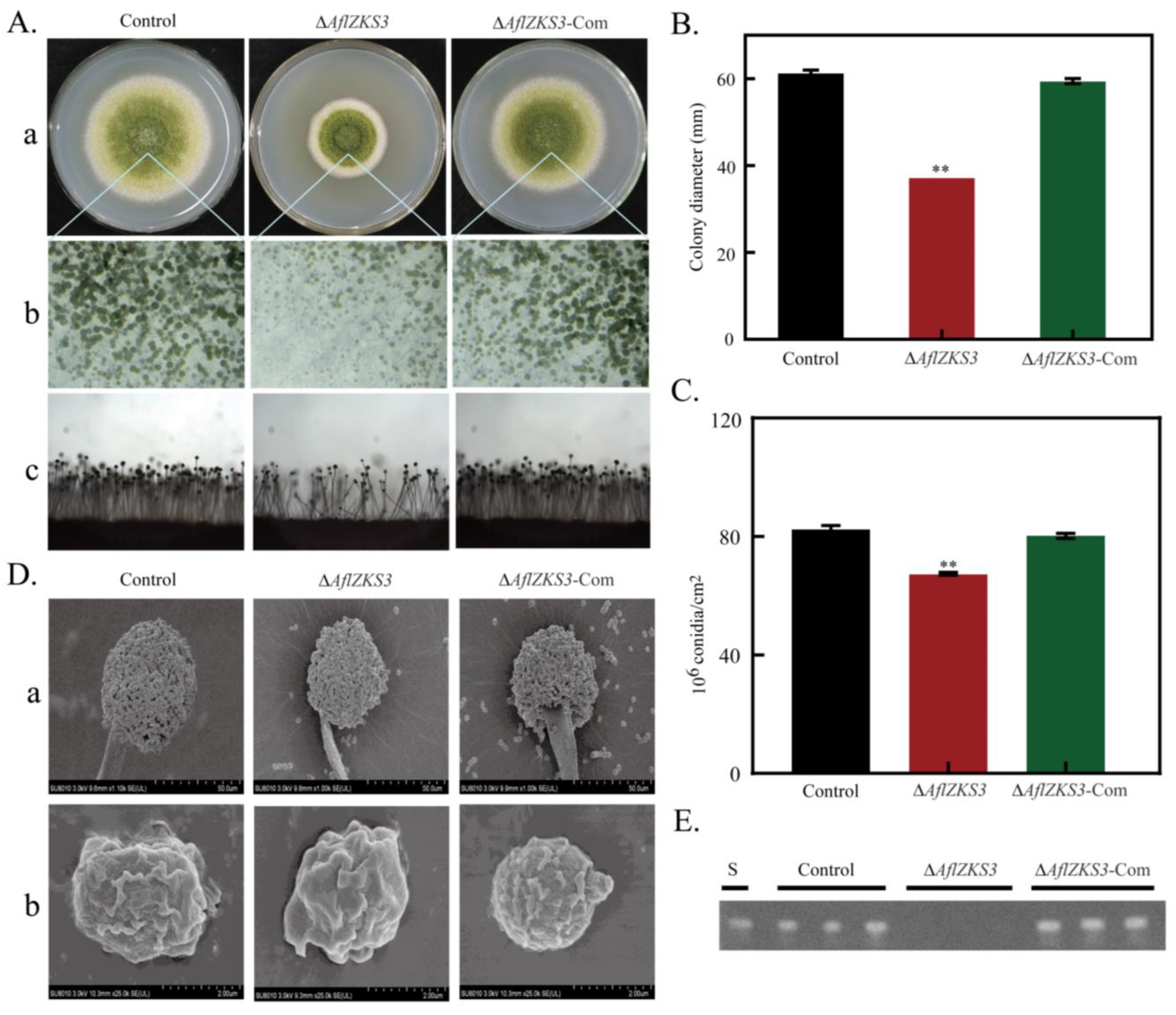
2.2. Deletion of AflZKS3 Affects Growth, Production of Conidia, and AF Biosynthesis
2.3. The ΔAflZKS3 Deletion Mutant Is Highly Sensitive to CFW and NaCl
2.4. Effects of AflZKS3 Deletion on the Pathogenicity of A. flavus on Grain Seeds
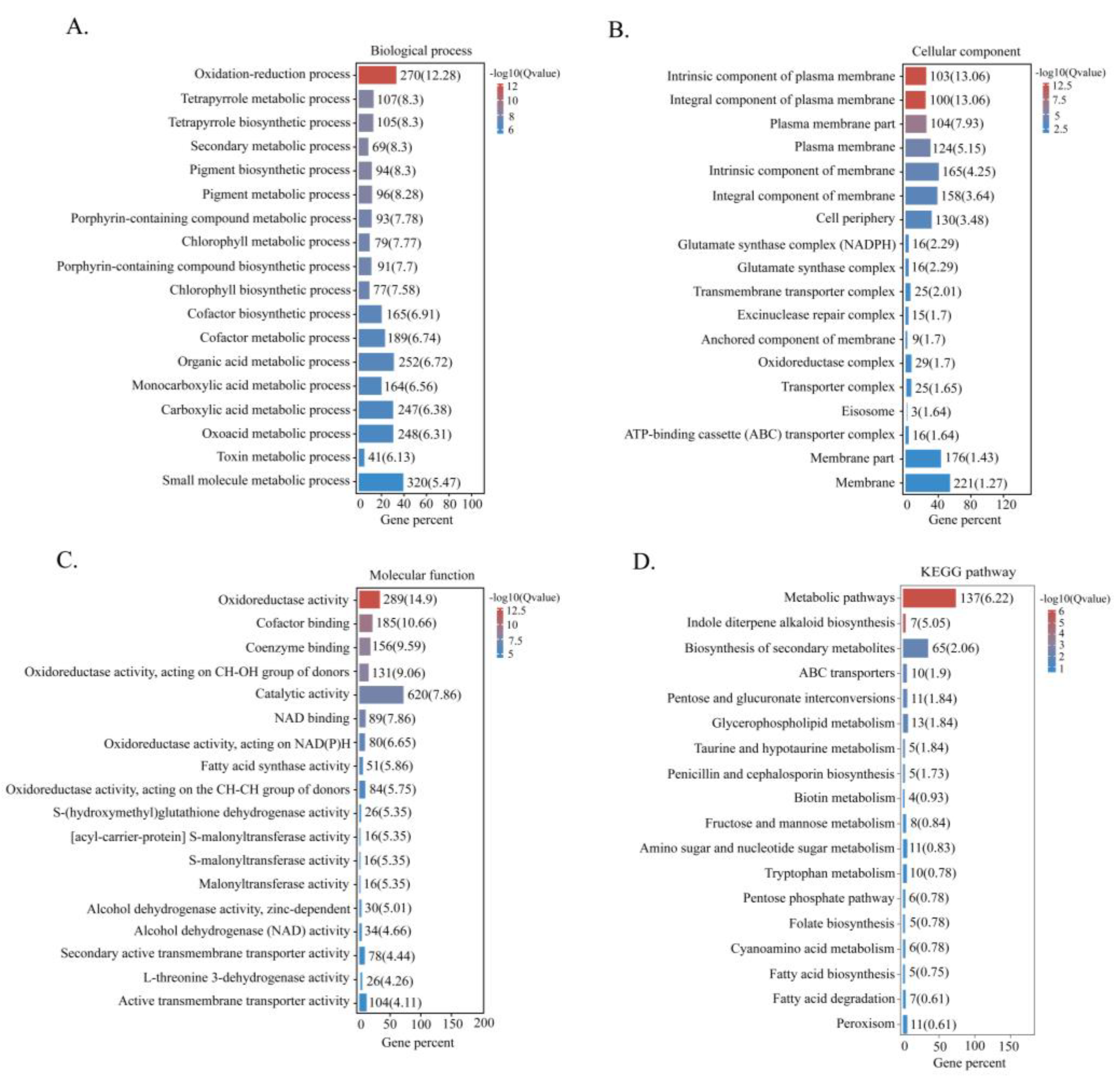
2.5. Transcriptome Analysis
2.6. Categorisation of DEGs
2.7. Validation of RNA-Seq
3. Discussion
4. Materials and Methods
4.1. Strains, Media, and Culture Conditions
4.2. Sequence Homology Analysis
4.3. Construction of Deletion, Complementation, and Localization Strains
4.4. Localization Analysis of AflZKS3 in A. flavus
4.5. Morphological and Physiological Analysis
4.6. Extraction and Detection of AFs
4.7. Evaluation of the Effect of AflZKS3 Deletion on the Growth of A. flavus Infecting Peanut and Corn
4.8. Transcriptome Analysis
4.9. Quantitative Real-Time PCR (qRT-PCR) Verification
4.10. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhuang, Z.; Lohmar, J.M.; Satterlee, T.; Cary, J.W.; Calvo, A.M. The master transcription factor mtfA governs aflatoxin production, morphological development and pathogenicity in the fungus Aspergillus flavus. Toxins 2016, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Omara, T.; Nassazi, W.; Omute, T.; Awath, A.; Laker, F.; Kalukusu, R.; Musau, B.; Nakabuye, B.V.; Kagoya, S.; Otim, G.; et al. Aflatoxins in uganda: An encyclopedic review of the etiology, epidemiology, detection, quantification, exposure assessment, reduction, and control. Int. J. Microbiol. 2020, 2020, 4723612. [Google Scholar] [CrossRef]
- Moretti, A.; Logrieco, A.F.; Susca, A. Mycotoxins: An underhand food problem. Methods Mol. Biol. 2017, 1542, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C.; Montalbano, B.G.; Cary, J.W. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 1999, 230, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xu, J.; Chang, P.K.; Liu, Z.; Kong, Q. New insights of transcriptional regulator AflR in Aspergillus flavus physiology. Microbiol. Spectr. 2022, 10, e0079121. [Google Scholar] [CrossRef]
- Price, M.S.; Yu, J.; Nierman, W.C.; Kim, H.S.; Pritchard, B.; Jacobus, C.A.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A. The aflatoxin pathway regulator AflR induces gene transcription inside and outside of the aflatoxin biosynthetic cluster. Fems Microbiol. Lett. 2006, 255, 275–279. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Mack, B.M.; Wei, Q.; Li, P.; Roze, L.V.; Dazzo, F.; Cary, J.W.; Bhatnagar, D.; Linz, J.E. Association with AflR in endosomes reveals new functions for AflJ in aflatoxin biosynthesis. Toxins 2012, 4, 1582–1600. [Google Scholar] [CrossRef]
- Chen, J.F.; Tan, J.J.; Wang, J.Y.; Mao, A.J.; Xu, X.P.; Zhang, Y.; Zheng, X.L.; Liu, Y.; Jin, D.; Li, X.B.; et al. The zinc finger transcription factor BbCmr1 regulates conidium maturation in Beauveria bassiana. Microbiol. Spectr. 2022, 10, e0206621. [Google Scholar] [CrossRef]
- Jun, S.C.; Choi, Y.H.; Lee, M.W.; Yu, J.H.; Shin, K.S. The putative APSES transcription factor RgdA governs growth, development, toxigenesis, and virulence in Aspergillus fumigatus. Msphere 2020, 5, e00998-20. [Google Scholar] [CrossRef]
- Guan, X.; Zhao, Y.; Liu, X.; Shang, B.; Xing, F.; Zhou, L.; Wang, Y.; Zhang, C.; Bhatnagar, D.; Liu, Y. The bZIP transcription factor Afap1 mediates the oxidative stress response and aflatoxin biosynthesis in Aspergillus flavus. Rev. Argent. Microbio. 2019, 51, 292–301. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, G.; Zhang, D.; Liu, Y.; Li, Y.; Lin, G.; Guo, Z.; Wang, S.; Zhuang, Z. The PHD transcription factor Rum1 regulates morphogenesis and aflatoxin biosynthesis in Aspergillus flavus. Toxins 2018, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Shelest, E. Transcription factors in fungi. FEMS Microbiol. Lett. 2008, 286, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Cary, J.W.; Harris-Coward, P.Y.; Ehrlich, K.C.; Mack, B.M.; Kale, S.P.; Larey, C.; Calvo, A.M. NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot. Cell 2012, 11, 1104–1111. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Wiemann, P.; Garvey, G.S.; Lim, F.Y.; Haas, B.; Wortman, J.; Keller, N.P. Illumina identification of RsrA, a conserved C2H2 transcription factor coordinating the NapA mediated oxidative stress signaling pathway in Aspergillus. BMC Genom. 2014, 15, 1011. [Google Scholar] [CrossRef]
- Wang, P.; Li, B.; Pan, Y.T.; Zhang, Y.Z.; Li, D.W.; Huang, L. Calcineurin-responsive transcription factor CgCrzA is required for cell wall integrity and infection-related morphogenesis in Colletotrichum gloeosporioides. Plant Pathol. J. 2020, 36, 385–397. [Google Scholar] [CrossRef]
- Görner, W.; Durchschlag, E.; Martinez-Pastor, M.T.; Estruch, F.; Ammerer, G.; Hamilton, B.; Ruis, H.; Schüller, C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998, 12, 586–597. [Google Scholar] [CrossRef]
- Kwon, N.J.; Garzia, A.; Espeso, E.A.; Ugalde, U.; Yu, J.H. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 2010, 77, 1203–1219. [Google Scholar] [CrossRef]
- Trofimov, K.; Ivanov, R.; Eutebach, M.; Acaroglu, B.; Mohr, I.; Bauer, P.; Brumbarova, T. Mobility and localization of the iron deficiency-induced transcription factor bHLH039 change in the presence of FIT. Plant Direct. 2019, 3, e00190. [Google Scholar] [CrossRef]
- Xiong, D.; Wang, Y.; Deng, C.; Hu, R.; Tian, C. Phylogenic analysis revealed an expanded C2H2-homeobox subfamily and expression profiles of C2H2 zinc finger gene family in Verticillium dahliae. Gene 2015, 562, 169–179. [Google Scholar] [CrossRef]
- Cao, Y.; He, Q.; Qi, Z.; Zhang, Y.; Lu, L.; Xue, J.; Li, J.; Li, R. Dynamics and endocytosis of Flot1 in Arabidopsis require CPI1 function. Int. J. Mol. Sci. 2020, 21, 1552. [Google Scholar] [CrossRef] [PubMed]
- Rehman, L.; Su, X.; Li, X.; Qi, X.; Guo, H.; Cheng, H. FreB is involved in the ferric metabolism and multiple pathogenicity-related traits of Verticillium dahliae. Curr. Genet. 2018, 64, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Muñiz, J.M.; Renshaw, H.; Richards, A.D.; Lamoth, F.; Soderblom, E.J.; Moseley, M.A.; Juvvadi, P.R.; Steinbach, W.J. The Aspergillus fumigatus septins play pleiotropic roles in septation, conidiation, and cell wall stress, but are dispensable for virulence. Fungal Genet. Biol. 2015, 81, 41–51. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Takagi, K.; Kikkawa, A.; Iwama, R.; Fukuda, R.; Horiuchi, H. Type II phosphatidylserine decarboxylase is crucial for the growth and morphogenesis of the filamentous fungus Aspergillus nidulans. J. Biosci. Bioeng. 2021, 131, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, X.; Ma, X.; Yu, Q.; Yu, X.; Liu, Y.; Nie, C.; Zhang, Y.; Xing, F. The regulatory mechanism of water activities on aflatoxins biosynthesis and conidia development, and transcription Factor AtfB is involved in this regulation. Toxins 2021, 13, 431. [Google Scholar] [CrossRef]
- Zhang, G.; Zheng, Y.; Ma, Y.; Yang, L.; Xie, M.; Zhou, D.; Niu, X.; Zhang, K.Q.; Yang, J. The velvet proteins VosA and VelB play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 2019, 10, 1917. [Google Scholar] [CrossRef]
- Friesen, H.; Hepworth, S.R.; Segall, J. An Ssn6-Tup1-dependent negative regulatory element controls sporulation-specific expression of DIT1 and DIT2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997, 17, 123–134. [Google Scholar] [CrossRef][Green Version]
- Peng, Y.J.; Zhang, H.; Feng, M.G.; Ying, S.H. Sterylacetyl hydrolase 1 (BbSay1) links lipid homeostasis to conidiogenesis and virulence in the entomopathogenic fungus Beauveria bassiana. J. Fungi 2022, 8, 292. [Google Scholar] [CrossRef]
- Ram, A.F.; Klis, F.M. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 2006, 1, 2253–2256. [Google Scholar] [CrossRef]
- Kiełkowska, A. Allium cepa root meristem cells under osmotic (sorbitol) and salt (NaCl) stress in vitro. Acta Bot. Croat. 2017, 76, 146–153. [Google Scholar] [CrossRef][Green Version]
- Wankhade, S.D.; Bahaji, A.; Mateu-Andrés, I.; Cornejo, M.D. Phenotypic indicators of NaCl tolerance levels in rice seedlings: Variations in development and leaf anatomy. Acta Physiol. Plant. 2010, 32, 1161–1169. [Google Scholar] [CrossRef][Green Version]
- Yamazaki, H.; Tanaka, A.; Kaneko, J.; Ohta, A.; Horiuchi, H. Aspergillus nidulans ChiA is a glycosylphosphatidylinositol (GPI)-anchored chitinase specifically localized at polarized growth sites. Fungal Genet. Biol. 2008, 45, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Takaya, N.; Yamazaki, D.; Horiuchi, H.; Ohta, A.; Takagi, M. Cloning and characterization of a chitinase-encoding gene (chiA) from Aspergillus nidulans, disruption of which decreases germination frequency and hyphal growth. Biosci. Biotechno. Biochem. 1998, 62, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Villalobos-Duno, H.; San-Blas, G.; Paulinkevicius, M.; Sánchez-Martín, Y.; Nino-Vega, G. Biochemical characterization of Paracoccidioides brasiliensis α-1,3-glucanase Agn1p, and its functionality by heterologous expression in Schizosaccharomyces pombe. PLoS ONE 2013, 8, e66853. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Lü, Y.; Ouyang, H.; Zhou, H.; Yan, J.; Du, T.; Jin, C. N-Glycosylation of Gel1 or Gel2 is vital for cell wall β-glucan synthesis in Aspergillus fumigatus. Glycobiology 2013, 23, 955–968. [Google Scholar] [CrossRef] [PubMed]
- Cabello, L.; Gómez-Herreros, E.; Fernández-Pereira, J.; Maicas, S.; Martínez-Esparza, M.C.; de Groot, P.; Valentín, E. Deletion of GLX3 in Candida albicans affects temperature tolerance, biofilm formation and virulence. Fems Yeast Res. 2019, 19, 124. [Google Scholar] [CrossRef]
- Bayram, O.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. Fems Yeast Res. 2012, 36, 1–24. [Google Scholar] [CrossRef]
- Yu, J.; Chang, P.K.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Linz, J.E.; Woloshuk, C.P.; Bennett, J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microb. 2004, 70, 1253–1262. [Google Scholar] [CrossRef]
- Reding, C.L.; Harrison, M.A. Possible relationship of succinate dehydrogenase and fatty acid synthetase activities to Aspergillus parasiticus (NRRL 5139) growth and aflatoxin production. Mycopathologia 1994, 127, 175–181. [Google Scholar] [CrossRef]
- Payne, G.A.; Brown, M.P. Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 1998, 36, 329–362. [Google Scholar] [CrossRef]
- Ren, X.; Branà, M.T.; Haidukowski, M.; Gallo, A.; Zhang, Q.; Logrieco, A.F.; Li, P.; Zhao, S.; Altomare, C. Potential of Trichoderma spp. for biocontrol of aflatoxin-producing Aspergillus flavus. Toxins 1998, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Jamali, M.; Karimipour, M.; Shams-Ghahfarokhi, M.; Amani, A.; Razzaghi-Abyaneh, M. Expression of aflatoxin genes aflO (omtB) and aflQ (ordA) differentiates levels of aflatoxin production by Aspergillus flavus strains from soils of pistachio orchards. Res. Microbiol. 2013, 164, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism-from biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Sugui, J.A. What do we know about the role of gliotoxin in the pathobiology of Aspergillus fumigatus? Med. Mycol. 2009, 47 (Suppl. S1), 97–103. [Google Scholar] [CrossRef]
- Wang, D.N.; Toyotome, T.; Muraosa, Y.; Watanabe, A.; Wuren, T.; Bunsupa, S.; Aoyagi, K.; Yamazaki, M.; Takino, M.; Kamei, K. GliA in Aspergillus fumigatus is required for its tolerance to gliotoxin and affects the amount of extracellular and intracellular gliotoxin. Med. Mycol. 2014, 52, 506–518. [Google Scholar] [CrossRef]
- Gallo, A.; Ferrara, M.; Perrone, G. Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins 2013, 5, 717–742. [Google Scholar] [CrossRef]
- Huang, Z.; Su, B.; Xu, Y.; Li, L.; Li, Y. Determination of two potential toxicity metabolites derived from the disruption of the pksCT gene in Monascus aurantiacus Li As3.4384. J. Sci. Food Agr. 2017, 97, 4190–4197. [Google Scholar] [CrossRef]
- Zhu, Z.; Yang, M.; Bai, Y.; Ge, F.; Wang, S. Antioxidant-related catalase CTA1 regulates development, aflatoxin biosynthesis, and virulence in pathogenic fungus Aspergillus flavus. Environ. Microbiol. 2020, 22, 2792–2810. [Google Scholar] [CrossRef]
- Domènech, A.; Ayté, J.; Antunes, F.; Hidalgo, E. Using in vivo oxidation status of one- and two-component redox relays to determine H2O2 levels linked to signaling and toxicity. BMC Biol. 2018, 16, 61. [Google Scholar] [CrossRef]
- Su, L.D.; Zhang, Q.L.; Lu, Z. Oxidation resistance 1 (OXR1) participates in silkworm defense against bacterial infection through the JNK pathway. Insect Sci. 2017, 24, 17–26. [Google Scholar] [CrossRef]
- Park, C.; Shin, B.; Park, W. Protective role of bacterial alkanesulfonate monooxygenase under oxidative stress. Appl. Environ. Microb. 2020, 86, e00692-20. [Google Scholar] [CrossRef]
- Dos Santos, T.B.; Baba, V.Y.; Vieira, L.; Pereira, L.; Domingues, D.S. The urea transporter DUR3 is differentially regulated by abiotic and biotic stresses in coffee plants. Physiol. Mol. Biol. Plants 2021, 27, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, H.; Tripathi, K.; Rai, A.K.; Cha-Um, S.; Waditee-Sirisattha, R.; Takabe, T. An alkaline phosphatase/phosphodiesterase, PhoD, induced by salt stress and secreted out of the cells of Aphanothece halophytica, a halotolerant cyanobacterium. Appl. Environ. Microb. 2011, 77, 5178–5183. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Yang, H.; Wang, J.; Wei, S.; Zhai, H.; Zhang, S.; Hu, Y. Afper1 contributes to cell development and aflatoxin biosynthesis in Aspergillus flavus. Int. J. Food Microbiol. 2022, 377, 109828. [Google Scholar] [CrossRef] [PubMed]
- Zhi, Q.Q.; He, L.; Li, J.Y.; Li, J.; Wang, Z.L.; He, G.Y.; He, Z.M. The kinetochore protein spc105, a novel interaction partner of LaeA, regulates development and secondary metabolism in Aspergillus flavus. Front. Microbiol. 2019, 10, 1881. [Google Scholar] [CrossRef]
- Mengjuan, Z.; Guanglan, L.; Xiaohua, P.; Weitao, S.; Can, T.; Xuan, C.; Yanling, Y.; Zhenhong, Z. The PHD transcription factor Cti6 is involved in the fungal colonization and aflatoxin B1 biological synthesis of Aspergillus flavus. IMA Fungus 2021, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lv, Y.; Lv, A.; Wei, S.; Zhang, S.; Li, C.; Hu, Y. Sub3 inhibits Aspergillus flavus growth by disrupting mitochondrial energy metabolism, and has potential biocontrol during peanut storage. J. Sci. Food Agric. 2021, 101, 486–496. [Google Scholar] [CrossRef]



| Gene Category | Log2(fc) | Name | Description |
|---|---|---|---|
| Growth | |||
| AFLA_046830 | −2.37 | FLOT1 | flotillin domain protein |
| AFLA_089670 | −1.38 | freB | ferric reductase transmembrane component 4 precursor |
| AFLA_061400 | −1.25 | aspC | aminotransferase |
| AFLA_014230 | −1.49 | psd2 | phosphatidylserine decarboxylase |
| AFLA_074470 | −2.25 | vosA | nuclear division Rft1 protein |
| AFLA_044800 | −1.70 | con-6 | conidiation protein Con-6 |
| AFLA_085140 | −1.18 | cetA | extracellular thaumatin domain protein |
| AFLA_058960 | −1.03 | DIT2 | hypothetical protein AFLA_058960 |
| AFLA_041620 | −6.80 | AQY1 | aquaporin |
| AFLA_016100 | −5.30 | betA | glucose-methanol-choline (gmc) oxidoreductase |
| AFLA_122440 | −1.40 | SAY1 | lipase/thioesterase family protein |
| Cell wall | |||
| AFLA_006590 | −1.19 | chiA | class III chitinase ChiA1 |
| AFLA_077910 | −2.41 | agn1 | alpha-1,3-glucanase |
| AFLA_108860 | −1.03 | gel2 | 1,3-beta-glucanosyltransferase Gel2 |
| AFLA_064920 | 1.96 | gel4 | 1,3-beta-glucanosyltransferase gel4 precursor |
| AFLA_124160 | −1.39 | glx3 | intracellular protease/amidase |
| AFLA_018750 | −1.09 | gpi13 | phosphatidylinositol glycan |
| Secondary metabolism | |||
| AFLA_038640 | −1.12 | fasA | fatty acid synthase alpha subunit |
| AFLA_139370 | −1.31 | aflB | aflB/fas-1/fatty acid synthase beta subunit |
| AFLA_093600 | −2.77 | aflF | oxidoreductase |
| AFLA_002920 | −1.94 | aflQ | flavonoid 3-hydroxylase |
| AFLA_064290 | −4.26 | imqG | O-methyltransferase |
| AFLA_059990 | −3.93 | aclH | O-methyltransferase |
| AFLA_101720 | −2.06 | lnaC | cytochrome P450 |
| AFLA_097510 | −8.69 | BOT4 | cytochrome P450 monooxygenase |
| AFLA_118990 | −2.08 | gliA | efflux pump antibiotic resistance protein |
| AFLA_006170 | −1.43 | albA | polyketide synthetase PksP |
| AFLA_060010 | −4.70 | nscA | PKS-like enzyme |
| AFLA_127090 | −1.43 | pksCT | polyketide synthase |
| AFLA_101700 | −2.34 | lnaA | NRPS-like enzyme |
| Oxidative stress | |||
| AFLA_033420 | −2.26 | sodB | Mn superoxide dismutase MnSOD |
| AFLA_034380 | −2.25 | cat1 | catalase |
| AFLA_124620 | −9.14 | oxr1 | disulfide oxidoreductase |
| AFLA_117020 | −3.24 | ssuD | alkanesulfonate monooxygenase |
| AFLA_089810 | −2.34 | dur3 | sodium/solute symporter |
| AFLA_075170 | −2.26 | phoD | alkaline phosphatase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, L.; Yang, H.; Wei, S.; Zhang, S.; Chen, L.; Hu, Y.; Lv, Y. Putative C2H2 Transcription Factor AflZKS3 Regulates Aflatoxin and Pathogenicity in Aspergillus flavus. Toxins 2022, 14, 883. https://doi.org/10.3390/toxins14120883
Liang L, Yang H, Wei S, Zhang S, Chen L, Hu Y, Lv Y. Putative C2H2 Transcription Factor AflZKS3 Regulates Aflatoxin and Pathogenicity in Aspergillus flavus. Toxins. 2022; 14(12):883. https://doi.org/10.3390/toxins14120883
Chicago/Turabian StyleLiang, Liuke, Haojie Yang, Shan Wei, Shuaibing Zhang, Liang Chen, Yuansen Hu, and Yangyong Lv. 2022. "Putative C2H2 Transcription Factor AflZKS3 Regulates Aflatoxin and Pathogenicity in Aspergillus flavus" Toxins 14, no. 12: 883. https://doi.org/10.3390/toxins14120883
APA StyleLiang, L., Yang, H., Wei, S., Zhang, S., Chen, L., Hu, Y., & Lv, Y. (2022). Putative C2H2 Transcription Factor AflZKS3 Regulates Aflatoxin and Pathogenicity in Aspergillus flavus. Toxins, 14(12), 883. https://doi.org/10.3390/toxins14120883





