Abstract
Mycotoxins are secondary metabolites produced by fungi. Food/feed contamination by mycotoxins is a great threat to food safety. The contamination can occur along the food chain and can cause many diseases in humans and animals, and it also can cause economic losses. Many detoxification methods, including physical, chemical, and biological techniques, have been established to eliminate mycotoxins in food/feed. The biological method, with mycotoxin detoxification by microorganisms, is reliable, efficient, less costly, and easy to use compared with physical and chemical ones. However, it is important to discover the metabolite’s toxicity resulting from mycotoxin biodegradation. These compounds can be less or more toxic than the parent. On the other hand, mechanisms involved in a mycotoxin’s biological control remain still unclear. Mostly, there is little information about the method used by microorganisms to control mycotoxins. Therefore, this article presents an overview of the most toxic mycotoxins and the different microorganisms that have a mycotoxin detoxification ability. At the same time, different screening methods for degradation compound elucidation are given. In addition, the review summarizes mechanisms of mycotoxin biodegradation and gives some applications.
Key Contribution:
This review highlights the current research in mycotoxin biodegradation and bioadsorption. As such, an emphasis is placed on microorganism species; mechanisms; resulting compounds after biodegradation; and main important applications.
1. Introduction
Mycotoxins are secondary metabolites with low molecular weight produced by filamentous fungal species [1,2,3]. Their chemical structures are very different [4], and they cause various degrees of toxicity in humans and animals. Mycotoxins are often genotypically specific but can be produced by one or more fungal species; one species can produce more than one kind of mycotoxin. In the environment, there are more than 200 kinds of mycotoxins [5]. Some of them can exhibit carcinogenic, teratogenic, mutagenic, and neurotoxic properties, and others can show antitumor capacity and cytotoxic and antimicrobial properties [4,6].
Mycotoxin contamination can occur throughout the whole food process, from pre-harvest to food storage [5,7,8,9]. It is estimated that 25% of the world’s agricultural products may be contaminated by mycotoxins each year [10], which leads to economic losses and causes a variety of toxic effects in humans and animals. According to the RASFF (Rapid Alert System for Food and Feed), for the 10-year period from 2010–2019, almost 98.9% of U.S. food notifications on mycotoxins were due to aflatoxin contamination in almonds, peanuts, and pistachio nuts [11]. A multi-mycotoxin analysis of sorghum and finger millet in 2014 showed that these two types of cereals were contaminated with major mycotoxins, with a prevalence of 6 to 52% for finger millet and less than 15% for sorghum [12]. A similar study about the occurrence of mycotoxins in peanuts and peanut products showed that the level of aflatoxins was higher than the maximum limit in 90% of the samples [13]. A study by Monyo et al. on the occurrence of aflatoxin contamination in groundnut demonstrated that the amount of aflatoxin was higher than the maximal limit in 11 to 28% of the samples and below the limit in 2 to 10% of the samples [14]. A study on the occurrence of ochratoxin A (OTA) in food products available in Silesia markets showed that around 22% of the samples were contaminated [15]. Up to 30 or 31% of total wheat-based product samples collected from some districts of Punjab were found to be contaminated with aflatoxins and zearalenone (ZEN) [16]. A three-year survey about Fusarium mycotoxin contamination in wheat samples showed the presence of deoxynivalenol (DON) and nivalenol (NIV) in about 540 and 337 μg/kg, respectively [17]. To deal with this worldwide problem, many detoxification methods have been found against mycotoxins: physical methods, chemical methods, and biological methods [18,19].
Physical control refers to all methods that use the physical properties of a detoxication agent. This can include adsorption, extrusion, cooking, ozonation, the mechanical separation of the clean product from contaminated one, heating at high temperatures, use of radiation and light, grinding, and washing [20,21]. At present, the utilization of mycotoxin-binding adsorbents is the most frequently applied method to protect animals from contaminated feed [22]. Agro-product processing can also reduce mycotoxin contamination. Fermentation has been useful for some Fusarium mycotoxins [23]. It is considered an excellent technique for mycotoxin control in African countries [24].
Chemical control refers to methods that require the use of chemical compounds. This includes techniques such as ammonization [25], the influence of acids and bases, and the influence of oxidizing agents or various inorganic and organic chemicals [20]. However, these methods have some limitations because of the possible deterioration of animal health caused by excessive residual chemical substances in the feed and even some environmentally negative impacts [22].
Nowadays, the biological control of mycotoxins has gained great interest because most chemical and physical detoxification pathways have limitations such as high cost, residual compounds in food and feed, and loss of nutrients. Biological methods include the action of yeasts, bacteria, and enzymes against mycotoxins [26,27]. This detoxification pathway offers an excellent alternative to eliminate toxins and safeguards the nutritional value of food and feed. Nonetheless, biodegradation can result in more toxic compounds. Therefore, there is a need to study the toxicity of the resulting compounds [28].
This paper first describes the most common mycotoxins, then it provides a summary of different mycotoxin detoxification methods by microorganisms and detoxification mechanisms already found. Finally, some important microorganism applications are provided.
2. Major Mycotoxin Overview
Along the food chain, aflatoxins, ochratoxin A, zearalenone, deoxynivalenol, nivalenol, fumonisin B1 and B2, and patulin are the most common mycotoxins that can contaminate food and feed [29].
Aflatoxins are secondary fungi metabolites mostly produced by Aspergillus flavus, Aspergillus parasiticus, Aspergillus nominus, and Aspergillus niger [5,30,31]. Approximately 18 aflatoxins have been identified [32], but the most common are aflatoxin B1 (AFB1), aflatoxin B2 (AFB2), aflatoxin G1 (AFG1), aflatoxin G2 (AFG2), aflatoxin M1 (AFM1), and aflatoxin M2 (AFM2). Due to their capacity to bind with the DNA of cells, aflatoxins affect protein synthesis [33]. Group B has blue fluorescence and group G has green fluorescence under ultraviolet light [33]. Aflatoxin contamination occurs mainly in hot and humid regions [34]. AFB1 is the most toxic and is cancerogenic, teratogenic, and mutagenic [35]. It is included in category 1A of active carcinogenic compounds (IARC, 1993). The liver is its number one target [36]. On the other hand, AFM1 is a metabolite of AFB1 mainly present in dairy products [37] and is included in group 2B by International Agency for Research on Cancer (IARC) (1993) with a maximum of 0.5 μg/kg in milk [38]. Aflatoxin B1 is bio-transformed into AFB1-8,9-epoxide via cytochrome p450 enzymes, which can induce DNA damage [39].
Patulin (PAT) is a mycotoxin produced mostly by penicillium [40], Byssochlamys, and Aspergillus species [41]. Patulin contamination can cause a lot of damage to animals, such as cancer, by affecting different organs, including the kidney, liver, and intestine [42]. It can contaminate foodstuffs such as fruits and vegetables, especially apples and apple by-products [43,44,45].
Ochratoxin A (OTA) is the most common toxin in grapes and grape-derived products [46], but it can also contaminate food such as coffee, spices, beer [47], and some meat products [48]. OTA is mainly produced by Aspergillus ochraceus and Penicillium verrucosum [49]. Aspergillus carbonarius, and Aspergillus niger can also produce OTA, especially in grapes and wines [50]. OTA is very stable at high temperatures [51]. It has neurotoxicological effects [52,53] nephrotoxic effects and can affect mammary functions [54]. OTA production in grapes and grape-derived products is a severe problem in the wine production field, especially in European countries where the climate conditions favor the growth of ochratoxigenic Aspergillus species. Thus, since March 2002, maximum OTA levels in cereals and dried vine fruits are regulated by the EU [55,56].
Fumonisin B1 (FB1) is the most abundant and toxic of the more than 15 types of fumonisins that have been identified [57]. FB1 can cause many diverse toxic effects in animals, including neurotoxicity, hepatotoxicity, and nephrotoxicity [58]. FB1 is a mycotoxin produced by Fusarium species such as Fusarium verticilloides and Fusarium proliferatum [59]. It is found in various crops, but mostly in corn and corn-based food or feed products. It is classified by the IARC 2002 as a carcinogen to humans (group 2B) [60].
Trichothecene mycotoxins are a group of sesquiterpenoid metabolites produced by Fusarium species. They usually contaminate cereals and threaten human and animal health [61]. Around 200 tetracyclic sesquiterpenoids have been identified as part of the trichothecene group [62]. Deoxynivalenol (DON) and nivalenol (NIV), and T-2 Toxin (T-2) are the more significant trichothecenes [63]. Type-B trichothecenes include deoxynivalenol (DON), nivalenol (NIV), and their acetylated derivatives, whereas Type-A includes T-2 and HT-2 toxins [10]. They are distinguished by the presence or absence of a carbonyl group in the C8 position.
Deoxynivalenol (DON) has been found to contaminate cereal crops such as barley [64], wheat [65], and maize, as well as their by-products [66]. It is mainly produced by Fusarium species [67]. DON may cause toxic and immune-toxic effects in animal species [6]. It is a potent inhibitor of protein synthesis. Fusarium mycotoxins such as DON and ZEN have been shown to affect liver morphology [68] and to have an immunosuppressive effect [69].
Zearalenone is a β-resorcylic acid lactone [70] that is produced by several species of Fusarium, including Fusarium graminearum, Fusarium culmorum, Fusarium cerealis, Fusarium equiseti, and Fusarium semitectum [71]. This mycotoxin infects cereals such as maize and wheat and can cause many hazards to humans and animals, such as cytogenetic toxicity, decrease fertility, embryotoxicity, and immunotoxicity [72,73,74]. ZEN has the ability to bind to the estrogen receptors of a cell, making it hazardous to humans and animals [75]. ZEN is mostly bio-transforming in α-ZEN and β-ZEN [76].
Due to their toxicity and effects on human health, many countries and international organizations, such as the World Health Organization (WHO), the Food and Agriculture Organization (FAO), and the European Union (EU) through the European Food Safety Authority (EFSA) [77], have set up strict controls of maximum residue levels in foodstuffs. Figure 1 provides some examples of mycotoxin structures.
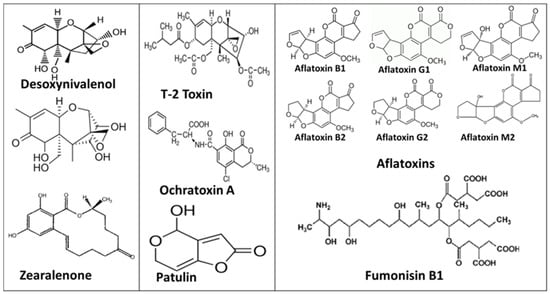
Figure 1.
Most common mycotoxin structures.
Table 1 provides an overview of the characteristics of some mycotoxins, including their effects and corresponding recommendations from the World Health Organization [38].

Table 1.
Some mycotoxin characterizations.
3. Microorganism Degradation
3.1. Toxin Detoxification by Bacteria
Many species of bacteria have the ability to degrade mycotoxins, including lactic acid bacteria [97] and other species [98]. Tetragenococcus halophilus [99], Rhodococcus erythropolis, and Mycobacterium fluoranthenivorans [100] were proven to degrade AFB1; Pediococcus parvulus [101] and Lactobacillus acidophilus [102,103] are effective for OTA, AFB1, and AFM1 biocontrol; Bifidobacterium animalis [104] is useful for patulin control; Pseudomonas otitidis [105] and Bacillus velezensis Strain ANSB01E [106] are able to detoxify ZEN. The degradation process depends on many factors, such as the incubation time, the medium, the microorganism species, the concentration of the bacteria cells, and the pH.
The degradation time changes according to the bacteria strain; the microbiota from the thermophilic compost of agricultural waste have degraded AFB1 in 5 days, with a degradation yield of more than 95% after cultivation in a PCS medium at 55 °C [107], and Rhodococcus pyridinivorans K408 took 12 days to detoxify AFB1 in bioethanol [26]; the Lacticaseibacillus rhamnosus (previously Lactobacillus rhamnosus) strains LBGG and LC705, however, removed AFB1 very rapidly [108].
The detoxification rate can depend on the medium; Bacillus subtilis UTBSP1 is able to detoxify AFB1 in a higher yield in pistachio nuts than in a medium culture [109], and Pseudomonas fluorescens strain 3JW1 can degrade AFB1 in potato dextrose broth and peanut medium by 97.8% and 99.4%, respectively [18].
Many bacteria have been reported to be able to degrade more than one mycotoxin [110]. AFB1 and ZEN have been degraded simultaneously by a microbial consortium, TADC7 [111]; Rhodococcus pyridinivorans strains (K408 and AK37) are able to degrade AFB1, T-2, and ZEN simultaneously [22], but also, some lactic acid bacteria strains can degrade multi-mycotoxins [112,113]. On the other hand, Pseudomonas fluorescens strain 3JW1 is able not only to degrade AFB1 but also to inhibit the AFB1 production of Aspergillus flavus. It reduces the amount of AFB1 produced by Aspergillus by 97.8%, 99.4%, and 55.8%, respectively, in the medium culture, peanut medium, and peanut kernels [18].
pH also plays an important role in mycotoxin biodegradation. An Alcaligenes faecalis strain called ANSA176 is able to detoxify OTA at a rate of 97.43% per 1 mg/mL OTA into OTα within 12 h at 37 °C. The optimal pH is between 6.0–9.0. The bacterial species subjected to the tested pH, ranging from 2.5 to 5.0, were unable to grow [114].
Therefore, mycotoxin biodegradation is an effective method, but it depends on multiple factors. Strict studies are needed for each biocontrol strain to determine the optimal conditions for its use. Table 2 provides an overview of AFB1 detoxification by bacteria with regard to the medium culture used and the main effect on the mycotoxin.

Table 2.
Aflatoxin B1 detoxification by bacteria.
Table 3 provides a global vision of mycotoxin detoxification by bacteria. The main effects on each mycotoxin are provided, as well as the medium culture used.

Table 3.
Other mycotoxins detoxification by bacteria.
3.2. Mycotoxin Detoxification by Yeast
Yeasts are able to detoxify mycotoxins in different ways: biodegradation, bioadsorption, or the inhibition of mycotoxin production [126].
The biodegradation method can happen with an enzyme isolated from the yeast or the use of the yeast itself. Hong Cao et al. [127] demonstrated the aflatoxin B1 degradation activity of an oxidase enzyme from the fungus Armillariella tabescens. The degradation ability of aflatoxin oxidase has been shown by high-performance thin-layer chromatography (HPTLC). The main mechanism was thought to be the cleavage of the bis-furan ring of the aflatoxin molecule. Meyerozyma guilliermondii has been shown to be able to control patulin in pear. The patulin degradation ability of Meyerozyma guilliermondii in pear wounds increases with a higher concentration of yeast cells. The optimal temperatures are 20 °C and 4 °C in wounds, as well as in whole fruits [128].
On the other hand, yeast biocontrol can involve bioadsorption mechanisms. Some Saccharomyces strains are able to remove OTA contamination via adsorption; the mechanism of removal can be enhanced from 45% to 90% by heat treatment of the microorganism and with a lower pH in the medium [129]. In another case, during OTA reduction caused by Saccharomyces cerevisiae, the addition of sugar at a temperature of 30 °C enhanced the OTA reduction rate in a semi-synthetic medium [130]. The binding capacity of AFB1, ZEN, OTA, and DON with respect to the Saccharomyces cerevisiae contained in beer fermentation residue was studied by Campagnollo et al. [131]. The results showed that beer fermentation residue has a higher binding capacity for ZEN at levels of 75.1% and 77.5% at pH 3.0 and 6.5, respectively. The volatiles of non-fermenting yeasts have shown significant binding activity against mycotoxins. The highest mycotoxin binding activities of these strains were noted against ochratoxin A (92%), AFB2 (66%), AFG2 (59%), and AFB1 (31%) [132]. One issue concerning mycotoxin biocontrol by yeast is that it can sometimes be a reversible mechanism, as has been noted with S. cerevisiae CECT 1891 and L. acidophilus 24, which were able to remove FB1 from a liquid medium. The removal was a fast and reversible process [133]. Yeasts’ complicated interactions with mycotoxins indicate that cell wall structural integrity, physical structure and morphology, and chemical components all play important roles in the adsorption process. On this basis, future approaches may rely on combinations of different microorganisms to provide complementary advantages in mycotoxin adsorption by yeast [134].
Finally, mycotoxin biocontrol by yeast can concern the inhibition of mycotoxin production. Ponsone et al. studied the activity of some yeast strains isolated from Argentinean vineyards against the growth of the ochratoxigenic Aspergillus strain Nigri and also evaluated their effects on OTA. This study demonstrated the natural occurrence of biocontrol agents in the environment to reduce fungi and mycotoxin problems. The results showed that these yeast strains have the ability, under different water activity (aw) and temperature conditions, to control Aspergillus carbonarius and A. niger aggregate growth and OTA accumulation with a reduction of at least 50% [135]. The same results were obtained when non-fermenting and low-fermenting yeasts were used by Fiori et al. to reduce OTA contamination in grape juice [136]. Nonetheless, some yeast strains are just able to inhibit growth parameters but not mycotoxin production.
Table 4 provides a summary of mycotoxin detoxification by yeast. Emphasis is given to related medium culture and its main effects on mycotoxins.

Table 4.
Toxin detoxification by yeasts.
3.3. Toxin Detoxification by Enzymes
Some enzymes isolated from microorganisms or mushrooms are able to degrade one or multiple mycotoxins. This is the case for the Ery4 laccase from Pleurotus eryngii, which can degrade AFB1, FB1, OTA, ZEN, and T-2 at the same time [142]. Other enzymes can detoxify only one mycotoxin; this is the case for Armillariella tabescens, which has been demonstrated to have an AFB1 degradation ability [127]. The degradation mechanism depends on the enzyme type and the type of mycotoxins. Enzymes can transform the parent into a new compound [91,127,143] or digest it completely [122]. Zeinvand-Lorestani et al. studied the action of a laccase enzyme against AFB1. Under optimal conditions, 67% of the total amount of AFB1 was degraded by the laccase after two days. The degraded product’s prooxidative properties and mutagenicity were lower than the AFB1 one [144]. Bacillus amyloliquefaciens ASAG1 can detoxify OTA by 98.5% after 24 h of incubation and 100% after 72 h. On the other hand, the carboxypeptidase cloned from the bacterium is also able to degrade OTA at a level of 41% and 72%, respectively, when cultivated with the supernatant and the purified protein of the carboxypeptidase [145]. Another study showed the effect of carboxypeptidases against OTA. Commercial protease A, commercial pancreatin, and an enzyme extract isolated from Aspergillus niger MUM have been proven to degrade OTA to Otα, respectively, by 87.3%, 43.4%, and 99.8% under the optimal conditions of pH 7.5 and temperature 37 °C after 25 h [146]. Porcine pancreatic lipase degraded PAT in pear juice [147].
Table 5 provides an overview of mycotoxin degradation by enzymes with emphasis given to the medium culture and its main effects.

Table 5.
Toxin detoxification by enzymes.
4. Detoxification Mechanism
4.1. Biodegradation Mechanism
The toxin biodegradation mechanism depends on the microorganism and toxin nature. In their study of AFB1 biodegradation, J. Li et al. demonstrated that aflatoxin B1 degradation by Tetragenococcus halophilus is first caused by adsorption and then by the enzymatical pathway. The amount of AFB1 binding caused by adsorption was smaller than the one degraded by the enzymatical pathway. Two mechanisms have been offered as possible pathways for enzymatical action, and six degradation products have been identified: C14H10O4, C18H16O8, C14H12O3, C16H20O4, C14H16O2, and C14H20O2. The first pathway involves the lactone ring, and the second one involves the double bond of the furan ring. Both mechanisms result in the same compound: C14H20O2 [99]. The same results were obtained with another salt-tolerant Candida versatilis, CGMCC 3790 [139]. In that case, four resulting compounds were identified by LC/TOF-MS: C14H10O4, C14H12O3, C13H12O2, and C11H10O4. Elsewhere, Hong Cao et al. suggested that the aflatoxin oxidase (AFO) extracted from Armillariella tabescens detoxifies the AFB1 by cleaving the bis-furan ring [127]. Adebo et al. found that the pathway of AFB1 degradation by the culture and lysate of a Pontibacter species is enzymatical and suggested that when the AFB1 is hydrolyzed, it has probably been transformed into new compounds, which were not identified in that paper [118]. AFB1 has been partially bio-transformed into aflatoxin D1 (AFD1) by deleting a mutant of the bacC gene in Baccilus subtilis UTB1. The mechanism was a reduction in the double bond of the lactone ring in the coumarin moiety, followed by the hydrolysis of the ester bond and, finally, the des-carboxylation of the yield to aflatoxin D1 (AFD1); all the processes were catalyzed by the BacC [150]. AFD1, AFD2, and AFD3 have been shown to be degradation compounds of AFB1 detoxification by Pseudomonas putida. The mechanism might be lactone [151]. Phanerochaetesordida YK-624 is able to transform AFB1 into AFB1-8,9-epoxide by, firstly, the oxidation of the manganese protease; thereafter, hydrolysis obtains the final product, AFB1-8,9-dihydrodiol [143]
A yeast enzyme, orotate phosphoribosyltransferase, from Rhodotorula mucilaginosa was tested against patulin in apple juice samples and under optimum degradation conditions, which are 0.15 g/L of orotate phosphoribosyltransferase for every 1 mg/L patulin at 25 °C for 18 h; the degradation rate of patulin reached over 80% [148]. During a study of patulin degradation by the yeast Rhodosporidium paludigenum, the authors of [138] made the statement that the enzyme(s) responsible for patulin degradation synthesis was enhanced by the presence of patulin. In fact, an assay with protein extracted from cells contaminated by patulin was more active than those with proteins from cells grown without patulin. This difference was attributed to the synthesis of the enzyme. Patulin degradation screening of Saccharomyces cerevisiae, tested by M. Li et al., showed that the mechanism was enzymatical and that the PAT-metabolizing enzyme production by the yeast cells is not induced by PAT preincubation [27]. These results were not in accordance with those of Ianiri et al., who concluded in their study that the patulin degradation mechanism by the yeast Sporobolomyces sp. IAM 13481 can be induced via pretreatment with the mycotoxin; the pre-incubation with patulin can induce the earlier activation of the gene-encoding proteins of the antioxidant system and the proteins involved in the patulin efflux and patulin degradation [152].
Young et al., in their study, showed that microbial isolate microbiota and pure cultures from chicken intestines have the ability to degrade twelve trichothecenes. The degradation compound identification by MS has suggested that the mechanism includes de-epoxidation and or a diacylation, with the route depending on the presence and position of acyl functionalities [153]. In addition, Gao et al. isolated a bacterium, Eggerthella sp. DII-9, which has the ability to degrade some types of trichothecenes, including DON, HT-2, T-2 triol, and T-2 tetraol, into other compounds. T-2 triol was degraded into de-epoxy T-2triol (88.0%), de-epoxy HT-2 (8.6%), and de-epoxy T-2tetraol (2.3%). T-2 tetraol was converted into de-epoxy T-2 tetraol (85.9%), and about 2.3% de-epoxy T-2 triol. HT-2 was transformed into de-epoxy HT-2 (81.4%) and 4.7% de-epoxy T-2 triol. To identify the molecular mechanism, the complete genome of DII-9 was sequenced, but the location of the responsible genes was not found. After the enzymatical study, de-epoxidation was found to be a complex phenomenon [62].
The zearalenone degradation of Bacillus pumilus ES-21 was studied by G. Wang et al. The degradation rate was more than 95.7%, and the degradation compound was identified as 1-(3,5-dihydroxyphenyl)-60-hydroxy-l0-undecen-l00-one. Nonetheless, the compound was not very stable and degraded very rapidly. The mechanism was found to be enzymatical and was thought to be due to esterase activity [91]. on the other hand, during the process of ZEN degradation by Bacillus amyloliquefaciens [122], no resulting compounds were detected. It was concluded that during the biodegradation of Zen by the bacteria’s extracellular enzyme, no ZEN derivatives were produced; in fact, a study of ZEN derivative biodegradation by Bacillus amyloliquefaciens, including α-zearalenol, β-zearalenol, α-zearalanol, and β-Zearalanol, resulted in no metabolites. Koch et al. (2014) studied the ZEN detoxification ability of nine different fungal strains of the genera Rhizopus and Aspergillus, which are known to produce and transform steroids. The results showed that all the strains were able to detoxify ZEN. Biodegradation and adsorption happen simultaneously. Five resulting compounds were identified: ZEN-14-sulfate, ZEN-O-14, ZEN- O-16-glucoside, α-zearalenol, and α- zearalenol-sulfate. The nine biocontrol agents were divided into three groups: (1) Rhizhopus oryzae DSM 907 and Rhizhopus stolonifera DSM 855, which can catalyze ZEN glycosylation; (2) Rhizhopus oryzae DSM 906 and Rhizhopus oligosporus DSM 1964 and Aspergillus oryzae DSM 1864 and Aspergillus oryzae NBRC 100959, which are involved in the formation of sulfated ZEN metabolites; (3) Rhizopus DSM 908, DSM 1834, and Rhizopus oligosporus LMH 1133 T, which have shown the ability to produce the metabolite of both patterns [154]. The bacterial gut flora of pigs are able to transform ZEN into α-zearalenol and an unidentified compound via hydrolysis and DON into de-epoxy-DON via a de-epoxydation reaction [155].
OTA biodegradation by Pediococcus parvulus UTAD depended on the inoculum size and the incubation temperature coupled with a latency phase before biodegradation initiation. This later effect is due to the biodegradation enzyme synthesis of the bacteria [101]. OTA has been biodegraded into Otα by OTA amide group hydrolysis. On the other hand, OTA reduction by Debaryomyces hansenii involves neither absorption nor detoxification. It is a repression of the expression of the non-ribosomal peptide synthetase (otanpsPN) gene linked to the OTA biosynthetic pathway, which was observed in [48].
Generally, mechanisms of mycotoxin degradation by microorganisms include different types of enzymes (protease, esterase, intracellular enzymes, etc.). The degradation process can include one or two types of reactions. The mechanisms elucidated by now include oxidation, hydrolysis, the cleavage of the lactone ring, des-carboxylation, de-epoxidation, glycosylation, and sulfate-conjugation reactions. Figure 2 provides a general scheme of different enzymes that participate in mycotoxin degradation caused by microorganisms and the involved reactions.
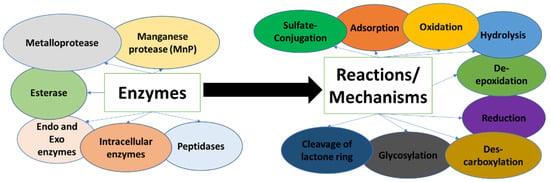
Figure 2.
Mycotoxin biodegradation: Enzymes and reactions/mechanisms.
Many studies have focused on the mycotoxin detoxification abilities of microorganisms, but a better understanding of responsible enzymes and the mechanisms involved is still needed. In some specific cases, no resulting metabolites were detected after mycotoxin biodegradation caused by microorganisms, but mostly, one or multiple compounds are usually detected. Table 6 provides an overview of mycotoxin degradation caused by microorganisms with a focus on the involved enzymes, degradation reactions, and resulting metabolites.

Table 6.
Toxin degradation by microorganism mechanisms summary.
4.2. Decontamination by Removal Mechanism
The use of microorganisms as agents for toxin sequestration in order to remove them from food and feed is an approach that has shown many good results.
Taheur et al. showed that strains isolated from a kefir culture are efficient in binding mycotoxins. The binding ability was dependent on the strain and the mycotoxin type [158]. From the same perspective, Saccharomyces cerevisiae CECT 1891 and Lactobacillus acidophilus 24 FB1 were shown to have a binding ability by Pizzolitto et al. The binding process needed a little time (1 min), and the mechanism involved was demonstrated to be a toxin molecule via the physical adsorption of the microorganism’s cell wall components. Cell viability was not necessary for FB1 binding, but the microorganism’s cell wall structural integrity was required, and the process did not involve FB1 chemical modification [133]. From the same perspective, two strains of Enterococcus faecium, which are present in dairy products, particularly in cheese, are efficient in AFB1 and PAT removal [110]. The same results were obtained by Elsanhoty et al. when they studied the AFM1 removal ability of some strains of Lactobacillus in milk samples [159].
OTA removal by Saccharomyces strains was demonstrated by Bejaoui et al. to be an adsorption mechanism. This mechanism was dependent on the OTA molecule’s ionic properties, the yeast membrane state, and the biomass concentration [129].
Lactococcus lactis and Bifidobacterium sp. Isolated from milk are able to neutralize ZEN contents via absorption. The Lactococcus lactis absorption is not homogeneous, and the process happens in two different steps. The first one includes a ZEN absorption of 88%, and the second one consists of ZEN diffusion into bacterial cells. This was contrary to that of Bifidobacterium sp., where the adsorption mechanism only had a single homogeneous step. The deprotonated carboxyl groups of the bacterial proteins and peptidoglycan play a significant role in the absorption process [71].
AFB1 binding via the Saccharomyces cerevisiae mannoprotein is possible because of AFB1 absorption onto mannose sites, where the new structure is maintained. Indeed, the new structure nature does not match that of a natural AFB1 molecule, so AFB1 can be removed from the media [160].
4.3. Degradation Compound Toxicity
Knowing the degraded compound’s toxicity is very important because it can be more or less toxic than the parent. Therefore, many cytotoxicity studies have been conducted.
Adebo et al. studied the toxicity of the compounds resulting from AFB1 degradation caused by Staphylococcus warneri, Sporosarcina sp., and Lysinibacillus fusiformis. The experiment was conducted by monitoring the mortality of lymphocyte cells (from human blood) after the cells were exposed to degraded compounds. A lower mortality rate was recorded compared with aflatoxin B1. The authors concluded that there was lower toxicity [117]. On the other hand, Escherichia coli DH5a, Arabidopsis thaliana, and human hepatocyte LO2 were used by [138] to determine the degradation toxicity of the compound identified as desoxypatulinic acid (DPA) due to patulin detoxification caused by Rhodosporidium paludigenum. The lower toxicity of DPA compared with PAT was demonstrated.
Elsewhere, no toxicity reduction has been found after ZEN and FB1 biocontrol using lactic acid bacteria. One toxicity study was conducted using human esophageal carcinoma cell lines [124]. Some ZEN degradation products are known to be more toxic than ZEN. In the case of α-ZOL, it shows higher estrogenicity than ZEN [71]. The compounds derived from ZEN biocontrol toxicity can be ranked as follows: α-zearalenol > α-zearalanol > zearalenone > β-zearalenol [161].
Figure 3 provides some mycotoxin degradation pathways.
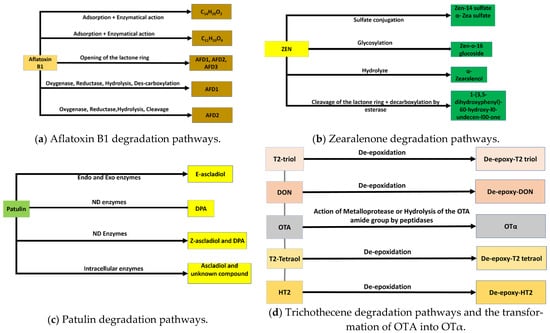
Figure 3.
Some mycotoxin degradation pathways.
5. Functional Enzymes Extraction from Bacteria
Nowadays, enzymes, as shown in Section 3.3, play a key role in mycotoxin biodegradation. Therefore, it is important to have a general method of enzyme extraction from microorganisms.
The process of enzyme extraction from microorganisms can be divided into three parts: extraction, purification, and characterization.
The extraction step’s main idea is to extract the enzyme outside the host. Some procedures are performed by harvesting the mycelia pellet via centrifugation and then washing it with phosphate buffer, followed by a second centrifugation to remove cell debris [127]. More recently, the homogenization of cells with protein extraction buffer followed by ultrasonication and centrifugation has been performed [148].
The purification step’s aim is, after the extraction step, to obtain an enzyme that is as pure as possible. Ammonium sulfate is the most used compound to precipitate enzymes [162]. This step is generally followed by centrifugation. In some cases, the precipitation step can be performed by using both organic solvents, such as methanol, ethanol, or acetone, and ammonium sulfate separately [163]. After enzyme activity determination, some purification techniques are used. Chromatography purification can be performed by using hydrophobic interaction chromatography (HIC) followed by immobilized metal ion affinity chromatography (IMAC) [127] or ion-exchange chromatography on a DEAE-Sepharose GE column, followed by dialysis and lyophilization [163]; dialysis can also be performed with a DEAE-Sepharose column [164]. Further purification can be performed using a Superdex 75 column followed by dialysis and lyophilization [163].
The last step is purified enzyme characterization. This step permits us to find the characteristics of the enzyme. It can be feasible to use SDS polyacrylamide gel electrophoresis (SDS-PAGE) to determine the molecular weight [127,163], HPTLC analysis to determine the enzymatic activity, and ESI-MS/MS to identify the enzyme [127]. Finally, the determination of the optimum pH, the optimum temperature, the ion metal effect on the enzyme activity [163], and the protein concentration (which can be determined using the method of Bradford) can be performed. Then, the enzyme can be stored at −85 °C until used.
Figure 4 provides a brief scheme of all the steps of enzyme extraction from microorganisms.
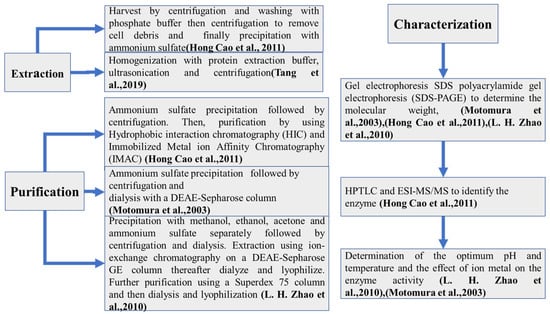
Figure 4.
Functional enzyme extraction from bacteria. [127,163,164,165].
6. Application and Perspectives
Microorganisms that can detoxify hazardous mycotoxins into low-toxicity compounds are of great importance. Being able to utilize them in the field and industries would be of great interest to food/feed safety. Therefore, it is advantageous to use microorganisms for mycotoxin detoxification on a large scale. Nevertheless, any applications to be set up must take into account both biocontrol agents and the life cycle of mycotoxigenic species, as well as the environmental conditions and plant agronomy [166]. Microorganisms that show activity against mycotoxins provide important properties because of the future possibility of exchanging the chemical and physical methods of preservation with a biological method based on those microorganisms and enzymes. Metabolism products of biocontrol agents are propitious for the bioconservation of food due to their ability to reduce the proliferation of mycotoxigenic fungi and mycotoxin production [167].
Microorganisms are used in many different ways. They are already used as probiotics to enhance the health of the host upon adequate administration. Lactobacillus species are most often used as probiotics [168], mainly via encapsulation [169], [170]. Encapsulation is one of the most effective methods of saving the viability and stability of microorganisms [113,171]. Therefore, it is a good alternative for microorganism applications in food and feed. Recently, the yeast Sporidiobolus pararoseus, which has a mycotoxin binding ability, was successfully produced with this approach on an industrial production scale with possible applications in feed additives [172].
Microorganisms can also be used as biopesticides [173]. The use of biofungicides is an approach that involves the application of different microorganisms that can suppress toxic fungi [174]. Recently, novel biofungicide formulations based on Bacillus subtilis 5, Bacillus cereus 3S5, and Pseudomonas fluorecens 10S2 were produced [175]. The same formulation has been created using other microorganisms [176,177,178].
Finally, the mycotoxin degradation enzyme can be especially valuable in the feed, food, and fermentation industry [109,120]. The α-amylase enzymes from some bacterial or fungal strains are widely used [179]. A carboxypeptidase that can degrade OTA has been cloned and used to detoxify the OTA mycotoxin [145].
7. Conclusions
This paper reviews mycotoxin degradation caused by microorganisms. Mycotoxins are secondary compounds produced by fungi with various chemical structures. Some of them are very hazardous for humans and animals, and strict regulations have been made for their content in food and feed. Physical, chemical, and biological methods can be used to control mycotoxin food/feed contamination. Biological control, which includes bacteria, yeast, and enzymatic activities against mycotoxins, is considered a very friendly control method compared with physical and chemical methods. However, more studies are needed to elucidate mycotoxin detoxification mechanisms. Unfortunately, most investigations do not address the real process involved with this biodegradation. In some cases, the degradation compound structures are elucidated, which helps to provide the hypothetically involved mechanisms. The detoxification of mycotoxins using bacterial strains augurs a new path for food/feed safety. From this perspective, more emphasis can be given to the toxicity of the resulting degradation compounds and the involved mechanisms of elucidation.
Author Contributions
S.N. wrote the article, conceptualized the idea, and designed the outline; Q.Z. conceptualized the idea, designed the outline, and revised and validated the manuscript; M.Z. helped to develop the figures; N.M.A. and M.F. revised the manuscript; P.L. supervised the work. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key Project of the National Natural Science Foundation of China (3203008), and the Major Project of the Hubei Hongshan Laboratory (2021hszd015).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sanzani, S.M.; Reverberi, M.; Geisen, R. Mycotoxins in harvested fruits and vegetables: Insights in producing fungi, biological role, conducive conditions, and tools to manage postharvest contamination. Postharvest Biol. Technol. 2016, 122, 95–105. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, X.; Li, J. Updating techniques on controlling mycotoxins—A review. Food Control 2018, 89, 123–132. [Google Scholar] [CrossRef]
- Ünüsan, N. Systematic review of mycotoxins in food and feeds in Turkey. Food Control 2019, 97, 1–14. [Google Scholar] [CrossRef]
- Hoyos Ossa, D.E.; Hincapié, D.A.; Peñuela, G.A. Determination of aflatoxin M1 in ice cream samples using immunoaffinity columns and ultra-high performance liquid chromatography coupled to tandem mass spectrometry. Food Control 2015, 56, 34–40. [Google Scholar] [CrossRef]
- Majer-Baranyi, K.; Zalán, Z.; Mörtl, M.; Juracsek, J.; Szendrő, I.; Székács, A.; Adányi, N. Optical waveguide lightmode spectroscopy technique-based immunosensor development for aflatoxin B1 determination in spice paprika samples. Food Chem. 2016, 211, 972–977. [Google Scholar] [CrossRef]
- Gerez, J.R.; Pinton, P.; Callu, P.; Oswald, I.P.; Bracarense, A.P.F.L. Deoxynivalenol alone or in combination with nivalenol and zearalenone induce systemic histological changes in pigs. Exp. Toxicol. Pathol. 2015, 67, 89–98. [Google Scholar] [CrossRef]
- Verheecke, C.; Liboz, T.; Mathieu, F. Microbial degradation of aflatoxin B1: Current status and future advances. Int. J. Food Microbiol. 2016, 237, 1–9. [Google Scholar] [CrossRef]
- Zhang, Z.; Hu, X.; Zhang, Q.; Li, P. Determination for multiple mycotoxins in agricultural products using HPLC-MS/MS via a multiple antibody immunoaffinity column. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1021, 145–152. [Google Scholar] [CrossRef]
- Mwakinyali, S.E.; Ding, X.; Ming, Z.; Tong, W.; Zhang, Q.; Li, P. Recent development of aflatoxin contamination biocontrol in agricultural products. Biol. Control 2019, 128, 31–39. [Google Scholar] [CrossRef]
- Ibáñez-Vea, M.; Lizarraga, E.; González-peñas, E.; López, A.; Cerain, D. Co-occurrence of type-A and type-B trichothecenes in barley from a northern region of Spain. Food Control 2012, 25, 81–88. [Google Scholar] [CrossRef]
- Alshannaq, A.; Yu, J. Analysis of E.U. Rapid Alert System (RASFF) Notifications for Aflatoxins in Exported U.S. Food and Feed Products for 2010–2019. Toxins 2021, 13, 90. [Google Scholar] [CrossRef]
- Chala, A.; Taye, W.; Ayalew, A.; Krska, R.; Sulyok, M.; Logrieco, A. Multimycotoxin analysis of sorghum (Sorghum bicolor L. Moench) and finger millet (Eleusine coracana L. Garten) from Ethiopia. Food Control 2014, 45, 29–35. [Google Scholar] [CrossRef]
- Ezekiel, C.N.; Sulyok, M.; Warth, B.; Odebode, A.C.; Krska, R. Natural occurrence of mycotoxins in peanut cake from Nigeria. Food Control 2012, 27, 338–342. [Google Scholar] [CrossRef]
- Monyo, E.; Njoroge, S.; Coe, R.; Osiru, M.; Madinda, F.; Waliyar, F.; Thakur, R.; Chilunjika, T.; Anitha, S. Occurrence and distribution of aflatoxin contamination in groundnuts (Arachis hypogaea L.) and population density of Aflatoxigenic Aspergilli in Malawi. Crop Prot. 2012, 42, 149–155. [Google Scholar] [CrossRef]
- Hajok, I.; Kowalska, A.; Piekut, A.; Ćwieląg-Drabek, M. A risk assessment of dietary exposure to ochratoxin A for the Polish population. Food Chem. 2019, 284, 264–269. [Google Scholar] [CrossRef]
- Zafar, S.; Ra, M.; Jinap, S.; Rashid, U. Detection of a fl atoxins and zearalenone contamination in wheat derived products. Food Control 2014, 35, 223–226. [Google Scholar]
- Del, E.M.; Garda-buffon, J.; Badiale-furlong, E. Deoxynivalenol and nivalenol in commercial wheat grain related to Fusarium head blight epidemics in southern Brazil. Food Chem. 2012, 132, 1087–1091. [Google Scholar]
- Yang, X.; Zhang, Q.; Chen, Z.Y.; Liu, H.; Li, P. Investigation of Pseudomonas fluorescens strain 3JW1 on preventing and reducing aflatoxin contaminations in peanuts. PLoS ONE 2017, 12, e0178810. [Google Scholar] [CrossRef]
- Vila-Donat, P.; Marín, S.; Sanchis, V.; Ramos, A.J. A review of the mycotoxin adsorbing agents, with an emphasis on their multi-binding capacity, for animal feed decontamination. Food Chem. Toxicol. 2018, 114, 246–259. [Google Scholar] [CrossRef]
- Agriopoulou, S.; Koliadima, A.; Karaiskakis, G.; Kapolos, J. Kinetic study of aflatoxins’ degradation in the presence of ozone. Food Control 2016, 61, 221–226. [Google Scholar] [CrossRef]
- Marković, M.; Daković, A.; Rottinghaus, G.E.; Kragović, M.; Petković, A.; Krajišnik, D.; Milić, J.; Mercurio, M.; de Gennaro, B. Adsorption of the mycotoxin zearalenone by clinoptilolite and phillipsite zeolites treated with cetylpyridinium surfactant. Colloids Surf. B Biointerfaces 2017, 151, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Cserháti, M.; Kriszt, B.; Krifaton, C.; Szoboszlay, S.; Háhn, J.; Tóth, S.; Nagy, I.; Kukolya, J. Mycotoxin-degradation profile of Rhodococcus strains. Int. J. Food Microbiol. 2013, 166, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Chilaka, C.A.; De Boevre, M.; Atanda, O.O.; De Saeger, S. Fate of Fusarium mycotoxins during processing of Nigerian traditional infant foods (ogi and soybean powder). Food Res. Int. 2019, 116, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Adebiyi, J.A.; Kayitesi, E.; Adebo, O.A.; Changwa, R.; Njobeh, P.B. Food fermentation and mycotoxin detoxification: An African perspective. Food Control 2019, 106, 106731. [Google Scholar] [CrossRef]
- Peng, W.-X.; Marchal, J.L.M.; van der Poel, A.F.B. Strategies to prevent and reduce mycotoxins for compound feed manufacturing. Anim. Feed Sci. Technol. 2018, 237, 129–153. [Google Scholar] [CrossRef]
- Prettl, Z.; Dési, E.; Lepossa, A.; Kriszt, B.; Kukolya, J.; Nagy, E. Biological degradation of aflatoxin B1by a Rhodococcus pyridinivorans strain in by-product of bioethanol. Anim. Feed Sci. Technol. 2017, 224, 104–114. [Google Scholar] [CrossRef]
- Li, M.; Chen, W.; Zhang, Z.; Zhang, Z.; Peng, B. Fermentative degradation of Patulin by Saccharomyces cerevisiae in aqueous solution. LWT—Food Sci. Technol. 2018, 97, 427–433. [Google Scholar] [CrossRef]
- Harkai, P.; Szabó, I.; Cserháti, M.; Krifaton, C.; Risa, A.; Radó, J.; Balázs, A.; Berta, K.; Kriszt, B. Biodegradation of aflatoxin-B1 and zearalenone by Streptomyces sp. collection. Int. Biodeterior. Biodegrad. 2016, 108, 48–56. [Google Scholar] [CrossRef]
- Siri-Anusornsak, W.; Kolawole, O.; Mahakarnchanakul, W.; Greer, B.; Petchkongkaew, A.; Meneely, J.; Elliott, C.; Vangnai, K. The Occurrence and Co-Occurrence of Regulated, Emerging, and Masked Mycotoxins in Rice Bran and Maize from Southeast Asia. Toxins 2022, 14, 567. [Google Scholar] [CrossRef]
- Xie, J.; Sun, Y.; Zheng, Y.; Wang, C.; Sun, S.; Li, J.; Ding, S.; Xia, X.; Jiang, H. Preparation and application of immunoaffinity column coupled with dcELISA detection for aflatoxins in eight grain foods. Food Control 2017, 73, 445–451. [Google Scholar] [CrossRef]
- Zhao, F.; Tian, Y.; Shen, Q.; Liu, R.; Shi, R.; Wang, H.; Yang, Z. A novel nanobody and mimotope based immunoassay for rapid analysis of aflatoxin B1. Talanta 2019, 195, 55–61. [Google Scholar] [CrossRef]
- Wang, X.; Niessner, R.; Tang, D.; Knopp, D. Nanoparticle-based immunosensors and immunoassays for aflatoxins. Anal. Chim. Acta 2016, 912, 10–23. [Google Scholar] [CrossRef]
- Edite Bezerra da Rocha, M.; Freire, F.C.O.; Erlan Feitosa Maia, F.; Izabel Florindo Guedes, M.; Rondina, D. Mycotoxins and their effects on human and animal health. Food Control 2014, 36, 159–165. [Google Scholar] [CrossRef]
- Xie, J.; Peng, T.; He, J.L.; Shao, Y. Preparation and characterization of an immunoaffinity column for the selective extraction of aflatoxin B1 in 13 kinds of foodstuffs. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015, 998–999, 50–56. [Google Scholar] [CrossRef]
- Peng, Z.; Chen, L.; Zhu, Y.; Huang, Y.; Hu, X.; Wu, Q.; Nüssler, A.K.; Liu, L.; Yang, W. Current major degradation methods for aflatoxins: A review. Trends Food Sci. Technol. 2018, 80, 155–166. [Google Scholar] [CrossRef]
- Ji, C.; Fan, Y.; Zhao, L. Review on biological degradation of mycotoxins. Anim. Nutr. 2016, 2, 127–133. [Google Scholar] [CrossRef]
- Mirzaei, A.; Hajimohammadi, A.; Badiei, K.; Pourjafar, M.; Naserian, A.A.; Razavi, S.A. Effect of dietary supplementation of bentonite and yeast cell wall on serum endotoxin, inflammatory parameters, serum and milk aflatoxin in high-producing dairy cows during the transition period. Comp. Clin. Path. 2020, 29, 433–440. [Google Scholar] [CrossRef]
- WHO/FAO. General Standard for Contaminants and Toxins in Food and Feed (Codex Stan 193-1995); World Health Organization: Geneva, Switzerland, 2015; Volume 51, pp. 39–54. [Google Scholar]
- Engin, A.B.; Engin, A. DNA damage checkpoint response to aflatoxin B1. Environ. Toxicol. Pharmacol. 2019, 65, 90–96. [Google Scholar] [CrossRef]
- Rodríguez-Bencomo, J.J.; Sanchis, V.; Viñas, I.; Martín-Belloso, O.; Soliva-Fortuny, R. Formation of patulin-glutathione conjugates induced by pulsed light: A tentative strategy for patulin degradation in apple juices. Food Chem. 2020, 315, 126283. [Google Scholar] [CrossRef]
- Li, B.; Chen, Y.; Zong, Y.; Shang, Y. Dissection of patulin biosynthesis, spatial control and regulation mechanism in Penicillium expansum. Environ. Microbiol. 2019, 21, 1124–1139. [Google Scholar] [CrossRef]
- Liu, M.; Wang, J.; Yang, Q.; Hu, N. Patulin removal from apple juice using a novel cysteine-functionalized metal-organic framework adsorbent. Food Chem. Toxicol. 2019, 270, 1–9. [Google Scholar] [CrossRef]
- Kapetanakou, A.E.; Nestora, S.; Evageliou, V.; Skandamis, P.N. Sodium alginate–cinnamon essential oil coated apples and pears: Variability of Aspergillus carbonarius growth and ochratoxin A production. Food Res. Int. 2019, 119, 876–885. [Google Scholar] [CrossRef]
- Sohrabi, H.; Arbabzadeh, O.; Khaaki, P.; Khataee, A.; Majidi, M.R.; Orooji, Y. Patulin and Trichothecene: Characteristics, occurrence, toxic effects and detection capabilities via clinical, analytical and nanostructured electrochemical sensing/biosensing assays in foodstuffs. Crit. Rev. Food Sci. Nutr. 2022, 62, 5540–5568. [Google Scholar] [CrossRef]
- Guo, Y.; Zhou, Z.; Yuan, Y.; Yue, T. Survey of patulin in apple juice concentrates in Shaanxi (China) and its dietary intake. Food Control 2013, 34, 570–573. [Google Scholar] [CrossRef]
- Pantelides, I.S.; Christou, O.; Tsolakidou, M.D.; Tsaltas, D.; Ioannou, N. Isolation, identification and in vitro screening of grapevine yeasts for the control of black aspergilli on grapes. Biol. Control 2015, 88, 46–53. [Google Scholar] [CrossRef]
- Covarelli, L.; Beccari, G.; Marini, A.; Tosi, L. A review on the occurrence and control of ochratoxigenic fungal species and ochratoxin A in dehydrated grapes, non-forti fi ed dessert wines and dried vine fruit in the Mediterranean area. Food Control 2012, 26, 347–356. [Google Scholar] [CrossRef]
- Peromingo, B.; Núñez, F.; Rodríguez, A.; Alía, A.; Andrade, M.J. Potential of yeasts isolated from dry-cured ham to control ochratoxin A production in meat models. Int. J. Food Microbiol. 2018, 268, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Abarca, M.L.; Bragulat, M.R.; Castellá, G.; Cabañes, F.J. Microbiology Impact of some environmental factors on growth and ochratoxin A production by Aspergillus niger and Aspergillus welwitschiae. Int. J. Food Microbiol. 2019, 291, 10–16. [Google Scholar] [CrossRef]
- Ponsone, M.L.; Nally, M.C.; Chiotta, M.L.; Combina, M.; Köhl, J.; Chulze, S.N. Evaluation of the effectiveness of potential biocontrol yeasts against black sur rot and ochratoxin A occurring under greenhouse and field grape production conditions. Biol. Control 2016, 103, 78–85. [Google Scholar] [CrossRef]
- Zhang, H.; Apaliya, M.T.; Mahunu, G.K.; Chen, L.; Li, W. Trends in Food Science & Technology Control of ochratoxin A-producing fungi in grape berry by microbial antagonists: A review. Trends Food Sci. Technol. 2016, 51, 88–97. [Google Scholar]
- Park, S.; Lim, W.; You, S.; Song, G. Ochratoxin A exerts neurotoxicity in human astrocytes through mitochondria-dependent apoptosis and intracellular calcium overload. Toxicol. Lett. 2019, 313, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Khoi, C.; Chen, J.; Lin, T.; Chiang, C. Ochratoxin A-Induced Nephrotoxicity: Up-to-Date Evidence. Int. J. Mol. Sci. 2021, 22, 11237. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lim, W.; Ryu, S.; Kim, J.; Song, G. Ochratoxin A mediates cytotoxicity through the MAPK signaling pathway and alters intracellular homeostasis in bovine mammary epithelial cells. Environ. Pollut. 2019, 246, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Vlachou, M.; Pexara, A.; Solomakos, N.; Govaris, A. Ochratoxin A in Slaughtered Pigs and Pork Products. Toxins 2022, 14, 67. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Abbas, F.; Rhouati, A.; Sun, Y.; Chu, X.; Cui, S.; Sun, B.; Xue, C. Design of a Quencher-Free Fluorescent Aptasensor for Ochratoxin A Detection in Red Wine Based on theGuanine-Quenching Ability. Biosensors 2022, 12, 297. [Google Scholar]
- Chen, J.; Wen, J.; Tang, Y.; Shi, J.; Mu, G.; Yan, R.; Cai, J.; Long, M. Research progress on fumonisin b1 contamination and toxicity: A review. Molecules 2021, 26, 5238. [Google Scholar] [CrossRef]
- Liu, X.; Fan, L.; Yin, S.; Chen, H.; Hu, H. Molecular mechanisms of fumonisin B1-induced toxicities and its applications in the mechanism-based interventions. Toxicon 2019, 167, 1–5. [Google Scholar] [CrossRef]
- Arumugam, T.; Pillay, Y.; Ghazi, T.; Nagiah, S.; Abdul, N.S.; Chuturgoon, A.A. Fumonisin B1-induced oxidative stress triggers Nrf2-mediated antioxidant response in human hepatocellular carcinoma (HepG2) cells. Mycotoxin Res. 2019, 35, 99–109. [Google Scholar] [CrossRef]
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajšlová, J.; Mayar, S.; Krska, R. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited ‘FAO estimate’ of 25 %. Crit. Rev. Food Sci. Nutr. 2020, 60, 2773–2789. [Google Scholar] [CrossRef]
- Schöneberg, T.; Jenny, E.; Wettstein, F.E.; Bucheli, T.D.; Mascher, F.; Bertossa, M.; Musa, T.; Seifert, K.; Gräfenhan, T.; Keller, B.; et al. Occurrence of Fusarium species and mycotoxins in Swiss oats—Impact of cropping factors. Eur. J. Agron. 2018, 92, 123–132. [Google Scholar] [CrossRef]
- Gao, X.; Mu, P.; Wen, J.; Sun, Y.; Chen, Q.; Deng, Y. Detoxification of trichothecene mycotoxins by a novel bacterium, Eggerthella sp. DII-9. Food Chem. Toxicol. 2018, 112, 310–319. [Google Scholar] [CrossRef]
- Zhang, J.; You, L.; Wu, W.; Wang, X.; Chrienova, Z.; Nepovimova, E.; Wu, Q.; Kuca, K. The neurotoxicity of trichothecenes T-2 toxin and deoxynivalenol (DON): Current status and future perspectives. Food Chem. Toxicol. 2020, 145, 111676. [Google Scholar] [CrossRef]
- Piacentini, K.C.; Rocha, L.O.; Savi, G.D.; Carnielli-Queiroz, L.; Almeida, F.G.; Minella, E.; Corrêa, B. Occurrence of deoxynivalenol and zearalenone in brewing barley grains from Brazil. Mycotoxin Res. 2018, 34, 173–178. [Google Scholar] [CrossRef]
- dos Santos, J.S.; Souza, T.M.; Ono, E.Y.S.; Hashimoto, E.H.; Bassoi, M.C.; de Miranda, M.Z.; Itano, E.N.; Kawamura, O. Natural occurrence of deoxynivalenol in wheat from Paraná State, Brazil and estimated daily intake by wheat products. Food Chem. 2013, 138, 90–95. [Google Scholar] [CrossRef]
- Bracarense, A.-P.F.L.; Lucioli, J.; Grenier, B.; Pacheco, G.D.; Moll, W.-D.; Schatzmayr, G.; Oswald, I.P. Chronic ingestion of deoxynivalenol and fumonisin, alone or in interaction, induces morphological and immunological changes in the intestine of piglets. Br. J. Nutr. 2012, 107, 1776–1786. [Google Scholar] [CrossRef]
- Bonnighausen, J.; Schauer, N.; Schafer, W.; Bormann, J. Metabolic profiling of wheat rachis node infection by Fusarium graminearum—Decoding deoxynivalenol-dependent susceptibility. New Phytol. 2019, 221, 459–469. [Google Scholar] [CrossRef]
- Skiepko, N.; Przybylska-Gornowicz, B.; Gajecka, M.; Gajecki, M.; Lewczuk, B. Effects of Deoxynivalenol and Zearalenone on the Histology and Ultrastructure of Pig Liver. Toxins 2020, 12, 463. [Google Scholar] [CrossRef]
- Ji, F.; He, D.; Olaniran, A.O.; Mokoena, M.P.; Xu, J.; Shi, J. Occurrence, toxicity, production and detection of Fusarium mycotoxin: A review. Food Prod. Process. Nutr. 2019, 1, 6. [Google Scholar] [CrossRef]
- Frizzell, C.; Uhlig, S.; Miles, C.O.; Verhaegen, S.; Elliott, C.T.; Eriksen, G.S.; Sørlie, M.; Ropstad, E. Toxicology in Vitro Biotransformation of zearalenone and zearalenols to their major glucuronide metabolites reduces estrogenic activity. Toxicol. In Vitro 2015, 29, 575–581. [Google Scholar] [CrossRef]
- Król, A.; Pomastowski, P.; Rafi, K.; Walczak, J. Microbiology neutralization of zearalenone using Lactococcus lactis and Bifidobacterium sp. Anal. Bioanal. Chem. 2018, 410, 943–952. [Google Scholar] [CrossRef]
- Hueza, I.M.; Raspantini, P.C.F.; Raspantini, L.E.R.; Latorre, A.O.; Górniak, S.L. Zearalenone, an estrogenic mycotoxin, is an immunotoxic compound. Toxins 2014, 6, 1080–1095. [Google Scholar] [CrossRef] [PubMed]
- Molina-Molina, J.-M.; Real, M.; Jimenez-Diaz, I.; Belhassen, H.; Hedhili, A.; Torné, P.; Fernández, M.F.; Olea, N. Assessment of estrogenic and anti-androgenic activities of the mycotoxin zearalenone and its metabolites using in vitro receptor-specific bioassays. Food Chem. Toxicol. 2014, 74, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Zhi, Y.; Xu, H.; Fang, H.; Jia, X. Zearalenone causes embryotoxicity and induces oxidative stress and apoptosis in differentiated human embryonic stem cells. Toxicol. In Vitro 2019, 54, 243–250. [Google Scholar] [CrossRef]
- Rogowska, A.; Pomastowski, P.; Sagandykova, G.; Buszewski, B. Zearalenone and its metabolites: Effect on human health, metabolism and neutralisation methods. Toxicon 2019, 162, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Abrunhosa, L.; Keller, K.; Rosa, C.A.; Cavaglieri, L.; Venâncio, A. Zearalenone and Its Derivatives α-Zearalenol and β-Zearalenol Decontamination by Saccharomyces cerevisiae Strains Isolated from Bovine Forage. Toxins 2015, 7, 3297–3308. [Google Scholar] [CrossRef]
- Schrenk, D.; Bodin, L.; Chipman, J.K.; Del Mazo, J.; GraslKraupp, B.; Hogstrand, C.; Hoogenboom, L.; Leblanc, J.-C.; Nebbia, C.S. Risk assessment of ochratoxin A in food. EFSA J. 2020, 18, 150. [Google Scholar]
- Abbès, S.; Ben Salah-Abbès, J.; Jebali, R.; Younes, R.B.; Oueslati, R. Interaction of aflatoxin B1 and fumonisin B1 in mice causes immunotoxicity and oxidative stress: Possible protective role using lactic acid bacteria. J. Immunotoxicol. 2016, 13, 46–54. [Google Scholar] [CrossRef]
- Hamid, A.S.; Tesfamariam, I.G. Aflatoxin B1-induced hepatocellular carcinoma in developing countries: Geographical distribution, mechanism of action and prevention (Review). Oncol. Lett. 2013, 5, 1087–1092. [Google Scholar] [CrossRef]
- Xiulan, S.; Xiaolian, Z.; Jian, T.; Zhou, J.; Chu, F.S. Preparation of gold-labeled antibody probe and its use in immunochromatography assay for detection of aflatoxin B1. Int. J. Food Microbiol. 2005, 99, 185–194. [Google Scholar] [CrossRef]
- Morales, H.; Sanchis, V.; Usall, J.; Ramos, A.J.; Marín, S. Effect of biocontrol agents Candida sake and Pantoea agglomerans on Penicillium expansum growth and patulin accumulation in apples. Int. J. Food Microbiol. 2008, 122, 61–67. [Google Scholar] [CrossRef]
- Jayashree, G.V.; Krupashree, K.; Rachitha, P.; Khanum, F. Patulin Induced Oxidative Stress Mediated Apoptotic Damage in Mice, and its Modulation by Green Tea Leaves. J. Clin. Exp. Hepatol. 2017, 7, 127–134. [Google Scholar] [CrossRef]
- Terezinha, F.; Melo, D.; Marques, I.; Greggio, S.; Dacosta, J.C.; Guecheva, T.N.; Saffi, J.; Rosa, R.M. DNA damage in organs of mice treated acutely with patulin, a known mycotoxin. Food Chem. Toxicol. 2012, 50, 3548–3555. [Google Scholar]
- Saxena, N.; Ansari, K.M.; Kumar, R.; Chaudhari, B.P.; Dwivedi, P.D.; Das, M. Role of mitogen activated protein kinases in skin tumorigenicity of Patulin. Toxicol. Appl. Pharmacol. 2011, 257, 264–271. [Google Scholar] [CrossRef]
- Drusch, S.; Kopka, S.; Kaeding, J. Stability of patulin in a juice-like aqueous model system in the presence of ascorbic acid. Food Chem. 2007, 100, 192–197. [Google Scholar] [CrossRef]
- De Curtis, F.; de Felice, D.V.; Ianiri, G.; De Cicco, V.; Castoria, R. Environmental factors affect the activity of biocontrol agents against ochratoxigenic Aspergillus carbonarius on wine grape. Int. J. Food Microbiol. 2012, 159, 17–24. [Google Scholar] [CrossRef]
- Paradells, S.; Rocamonde, B.; Llinares, C.; Garcia-verdugo, J.M.; Zipancic, I.; Miguel, J. Neurotoxic effects of ochratoxin A on the subventricular zone of adult mouse brain. J. Apply Toxicol. 2015, 737–751. [Google Scholar] [CrossRef]
- Wang, L.; Liao, Y.; Peng, Z.; Chen, L.; Zhang, W.; Nüssler, A.K.; Shi, S.; Liu, L.; Yang, W. Food raw materials and food production occurrences of deoxynivalenol in different regions. Trends Food Sci. Technol. 2019, 83, 41–52. [Google Scholar] [CrossRef]
- Feizollahi, E.; Iqdiam, B.; Vasanthan, T.; Thilakarathna, M.S.; Roopesh, M.S. Effects of Atmospheric-Pressure Cold Plasma Treatment on Deoxynivalenol Degradation, Quality Parameters, and Germination of Barley Grains. Appl. Sci. 2020, 10, 3530. [Google Scholar] [CrossRef]
- Chen, D.; Chen, P.; Cheng, Y.; Peng, P.; Liu, J.; Ma, Y.; Liu, Y.; Ruan, R. Deoxynivalenol Decontamination in Raw and Germinating Barley Treated by Plasma-Activated Water and Intense Pulsed Light. Food Bioprocess Technol. 2019, 12, 246–254. [Google Scholar] [CrossRef]
- Wang, G.; Yu, M.; Dong, F.; Shi, J.; Xu, J. Esterase activity inspired selection and characterization of zearalenone degrading bacteria Bacillus pumilus ES-21. Food Control 2017, 77, 57–64. [Google Scholar] [CrossRef]
- Abbasian, N.; Momtaz, S.; Baeeri, M.; Navaei-nigjeh, M. Molecular and biochemical evidence on the role of zearalenone in rat polycystic ovary. Toxicon 2018, 154, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Muthulakshmi, S.; Maharajan, K.; Habibi, H.R.; Kadirvelu, K.; Venkataramana, M. Zearalenone induced embryo and neurotoxicity in zebra fish model (Danio rerio): Role of oxidative stress revealed by a multi biomarker study. Chemosphere 2018, 198, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Antonissen, G.; Devreese, M.; De Baere, S.; Martel, A.; Van Immerseel, F.; Croubels, S. Impact of Fusarium mycotoxins on hepatic and intestinal mRNA expression of cytochrome P450 enzymes and drug transporters, and on the pharmacokinetics of oral enrofloxacin in broiler chickens. Food Chem. Toxicol. 2017, 101, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Chuturgoon, A.; Phulukdaree, A.; Moodley, D. Fumonisin B1 induces global DNA hypomethylation in HepG2 cells—An alternative mechanism of action. Toxicology 2014, 315, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Demirel, G.; Alpertunga, B.; Ozden, S. Role of fumonisin B1 on DNA methylation changes in rat kidney and liver cells. Pharm. Biol. 2015, 53, 1302–1310. [Google Scholar] [CrossRef]
- Sadiq, F.A.; Yan, B.; Tian, F.; Zhao, J. Lactic Acid Bacteria as Antifungal and Anti-Mycotoxigenic Agents: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1403–1436. [Google Scholar] [CrossRef]
- Akbar, A.; Khan, G.I.; Shafee, M.; Samad, A.; Masood, H.; Khan, S.A. Biocontrol of Aflatoxin through Biodegradation by Using Environment Friendly Microbes. Pol. J. Environ. Stud. 2022, 31, 4985–4988. [Google Scholar] [CrossRef]
- Li, J.; Huang, J.; Jin, Y.; Wu, C.; Shen, D.; Zhang, S.; Zhou, R. Aflatoxin B1 degradation by salt tolerant Tetragenococcus halophilus CGMCC 3792. Food Chem. Toxicol. 2018, 121, 430–436. [Google Scholar] [CrossRef]
- Teniola, O.; Addo, P.; Brost, I.; Farber, P.; Jany, K.; Alberts, J.; Vanzyl, W.; Steyn, P.; Holzapfel, W. Degradation of aflatoxin B1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp. nov. DSM44556. Int. J. Food Microbiol. 2005, 105, 111–117. [Google Scholar] [CrossRef]
- Abrunhosa, L.; Inês, A.; Rodrigues, A.I.; Guimarães, A.; Pereira, V.L.; Parpot, P.; Mendes-Faia, A.; Venâncio, A. Biodegradation of ochratoxin A by Pediococcus parvulus isolated from Douro wines. Int. J. Food Microbiol. 2014, 188, 45–52. [Google Scholar] [CrossRef]
- Fuchs, S.; Sontag, G.; Stidl, R.; Ehrlich, V.; Kundi, M.; Knasmüller, S. Detoxification of patulin and ochratoxin A, two abundant mycotoxins, by lactic acid bacteria. Food Chem. Toxicol. 2008, 46, 1398–1407. [Google Scholar] [CrossRef]
- Wochner, K.F.; Moreira, M.C.C.; Kalschne, D.L.; Drunkler, D.A. Detoxification of Aflatoxin B1 and M1 by Lactobacillus acidophilus and prebiotics in whole cow’s milk. J. Food Saf. 2019, 39, e12670. [Google Scholar] [CrossRef]
- Marina, S.; Sajid, M.; Yahong, Y.; Tianli, Y. Mycotoxin patulin in food matrices: Occurrence and its biological degradation strategies. Drug Metab. Rev. 2019, 51, 105–120. [Google Scholar]
- Tan, H.; Zhang, Z.; Hu, Y.; Wu, L.; Liao, F.; He, J.; Luo, B.; He, Y.; Zuo, Z.; Ren, Z.; et al. Isolation and characterization of Pseudomonas otitidis TH-N1 capable of degrading zearalenone. Food Control 2015, 47, 285–290. [Google Scholar] [CrossRef]
- Guo, Y.; Zhou, J.; Tang, Y.; Ma, Q.; Zhang, J.; Ji, C.; Zhao, L. Characterization and Genome Analysis of a Zearalenone—Degrading Bacillus velezensis Strain ANSB01E. Curr. Microbiol. 2019, 77, 273–278. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, C.; Zhang, D.; Zhao, M.; Zheng, D.; Lyu, Y.; Cheng, W.; Guo, P.; Cui, Z. Effective degradation of aflatoxin B1 using a novel thermophilic microbial consortium TADC7. Bioresour. Technol. 2017, 224, 166–173. [Google Scholar] [CrossRef]
- El-Nezami, H.; Kankaanpaa, P.; Salminen, S.; Ahokas, J. Ability of dairy strains of lactic acid bacteria to bind a common food carcinogen, aflatoxin B1. Food Chem. Toxicol. 1998, 36, 321–326. [Google Scholar] [CrossRef]
- Farzaneh, M.; Shi, Z.-Q.; Ghassempour, A.; Sedaghat, N.; Ahmadzadeh, M.; Mirabolfathy, M.; Javan-Nikkhah, M. Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control 2012, 23, 100–106. [Google Scholar] [CrossRef]
- Topcu, A.; Bulat, T.; Wishah, R.; BoyacI, I.H. Detoxification of aflatoxin B1 and patulin by Enterococcus faecium strains. Int. J. Food Microbiol. 2010, 139, 202–205. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, C.; Zhang, D.; Zhao, M.; Zheng, D.; Peng, M.; Cheng, W.; Guo, P.; Cui, Z. Simultaneous degradation of aflatoxin B1 and zearalenone by a microbial consortium. Toxicon 2018, 146, 69–76. [Google Scholar] [CrossRef]
- Juodeikiene, G.; Bartkiene, E.; Cernauskas, D.; Cizeikiene, D.; Zadeike, D.; Lele, V.; Bartkevics, V. Antifungal activity of lactic acid bacteria and their application for Fusarium mycotoxin reduction in malting wheat grains. LWT—Food Sci. Technol. 2018, 89, 307–314. [Google Scholar] [CrossRef]
- Bartkiene, E.; Zavistanaviciute, P.; Lele, V.; Ruzauskas, M.; Bartkevics, V.; Bernatoniene, J.; Gallo, P.; Tenore, G.C.; Santini, A. Lactobacillus plantarum LUHS135 and paracasei LUHS244 as functional starter cultures for the food fermentation industry: Characterisation, mycotoxin-reducing properties, optimisation of biomass growth and sustainable encapsulation by using dairy by-produc. LWT 2018, 93, 649–658. [Google Scholar] [CrossRef]
- Zheng, R.; Qing, H.; Ma, Q.; Huo, X.; Huang, S.; Zhao, L.; Zhang, J.; Ji, C. A Newly Isolated Alcaligenes faecalis ANSA176 with the Capability of Alleviating Immune Injury and Inflammation through Efficiently Degrading Ochratoxin A. Toxins 2022, 14, 569. [Google Scholar] [CrossRef] [PubMed]
- Hormisch, D.; Brost, I.; Kohring, G.-W.; Giffhorn, F.; Kroppenstedt, R.; Stackebradt, E.; Färber, P.; Holzapfel, W. Mycobacterium fluoranthenivorans sp. nov., a Fluoranthene and Aflatoxin B1 Degrading Bacterium from Contaminated Soil of a Former Coal Gas Plant. Syst. Appl. Microbiol. 2004, 27, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Mwakinyali, S.E.; Ming, Z.; Xie, H.; Zhang, Q.; Li, P. Investigation and Characterization of Myroides odoratimimus Strain 3J2MO Aflatoxin B1 Degradation. J. Agric. Food Chem. 2019, 67, 4595–4602. [Google Scholar] [CrossRef]
- Adebo, O.A.; Njobeh, P.B.; Mavumengwana, V. Degradation and detoxification of AFB1 by Staphylocococcus warneri, Sporosarcina sp. and Lysinibacillus fusiformis. Food Control 2016, 68, 92–96. [Google Scholar] [CrossRef]
- Adebo, O.A.; Njobeh, P.B.; Sidu, S.; Adebiyi, J.A.; Mavumengwana, V. Aflatoxin B1 degradation by culture and lysate of a Pontibacter specie. Food Control 2017, 80, 99–103. [Google Scholar] [CrossRef]
- Elsanhoty, R.M.; Ramadan, M.F.; El-Gohery, S.S.; Abol-Ela, M.F.; Azeke, M.A. Ability of selected microorganisms for removing aflatoxins invitro and fate of aflatoxins in contaminated wheat during baladi bread baking. Food Control 2013, 33, 287–292. [Google Scholar] [CrossRef]
- Alberts, J.F.; Engelbrecht, Y.; Steyn, P.S.; Holzapfel, W.H.; van Zyl, W.H. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 2006, 109, 121–126. [Google Scholar] [CrossRef]
- Mosallaie, F.; Jooyandeh, H.; Hojjati, M.; Fazlara, A. Biological reduction of aflatoxin B1 in yogurt by probiotic strains of Lactobacillus acidophilus and Lactobacillus rhamnosus. Food Sci. Biotechnol. 2020, 29, 793–803. [Google Scholar] [CrossRef]
- Xu, J.; Wang, H.; Zhu, Z.; Ji, F.; Yin, X.; Hong, Q.; Shi, J. Isolation and characterization of Bacillus amyloliquefaciens ZDS-1: Exploring the degradation of Zearalenone by Bacillus spp. Food Control 2016, 68, 244–250. [Google Scholar] [CrossRef]
- He, W.-J.; Yuan, Q.-S.; Zhang, Y.-B.; Guo, M.-W.; Gong, A.-D.; Zhang, J.-B.; Wu, A.-B.; Huang, T.; Qu, B.; Li, H.-P.; et al. Aerobic De-Epoxydation of Trichothecene Mycotoxins by a Soil Bacterial Consortium Isolated Using In Situ. Toxins 2016, 8, 277. [Google Scholar] [CrossRef] [PubMed]
- Mokoena, M.P.; Chelule, P.K. Reduction of Fumonisin B1 and Zearalenone by Lactic Acid Bacteria in Fermented Maize Meal. J. Food Prot. 2005, 68, 2095–2099. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Malaphan, W.; Xing, F.; Yu, B. Biodetoxification of fungal mycotoxins zearalenone by engineered probiotic bacterium Lactobacillus reuteri with surface-displayed lactonohydrolase. Appl. Microbiol. Biotechnol. 2019, 103, 8813–8824. [Google Scholar] [CrossRef] [PubMed]
- Papp, L.A.; Horváth, E.; Peles, F.; Pócsi, I.; Miklós, I. Insight into Yeast—Mycotoxin Relations. Agriculture 2021, 11, 1291. [Google Scholar] [CrossRef]
- Cao, H.; Liu, D.; Mo, X.; Xie, C.; Yao, D. A fungal enzyme with the ability of aflatoxin B1 conversion: Purification and ESI-MS/MS identification. Microbiol. Res. 2011, 166, 475–483. [Google Scholar] [CrossRef]
- Yang, Q.; Ma, J.; Solairaj, D.; Fu, Y.; Zhang, H. Efficacy of Meyerozyma guilliermondii in controlling patulin production by Penicillium expansum in shuijing pears. Biol. Control 2022, 168, 104856. [Google Scholar] [CrossRef]
- Bejaoui, H.; Mathieu, F.; Taillandier, P.; Lebrihi, A. Ochratoxin A removal in synthetic and natural grape juices by selected oenological Saccharomyces strains. J. Appl. Microbiol. 2004, 97, 1038–1044. [Google Scholar] [CrossRef]
- Petruzzi, L.; Bevilacqua, A.; Baiano, A.; Beneduce, L.; Corbo, M.R.; Sinigaglia, M. Study of Saccharomyces cerevisiae W13 as a functional starter for the removal of ochratoxin A. Food Control 2014, 35, 373–377. [Google Scholar] [CrossRef]
- Campagnollo, F.B.; Franco, L.T.; Rottinghaus, G.E.; Kobashigawa, E.; Ledoux, D.R.; Daković, A.; Oliveira, C.A. In vitro evaluation of the ability of beer fermentation residue containing Saccharomyces cerevisiae to bind mycotoxins. Food Res. Int. 2015, 77, 643–648. [Google Scholar] [CrossRef]
- Ul Hassan, Z.; Al Thani, R.; Atia, F.A.; Alsafran, M.; Migheli, Q.; Jaoua, S. Application of yeasts and yeast derivatives for the biological control of toxigenic fungi and their toxic metabolites. Environ. Technol. Innov. 2021, 22, 101447. [Google Scholar] [CrossRef]
- Pizzolitto, R.P.; Salvano, M.A.; Dalcero, A.M. Analysis of fumonisin B1 removal by microorganisms in co-occurrence with aflatoxin B1 and the nature of the binding process. Int. J. Food Microbiol. 2012, 156, 214–221. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, X.; Yuan, L.; Li, J. Complicated interactions between bio-adsorbents and mycotoxins during mycotoxin adsorption: Current research and future prospects. Trends Food Sci. Technol. 2020, 96, 127–134. [Google Scholar] [CrossRef]
- Ponsone, M.L.; Chiotta, M.L.; Combina, M.; Dalcero, A.; Chulze, S. Biocontrol as a strategy to reduce the impact of ochratoxin A and Aspergillus section Nigri in grapes. Int. J. Food Microbiol. 2011, 151, 70–77. [Google Scholar] [CrossRef]
- Fiori, S.; Paolo, P.; Hammami, W.; Razzu, S.; Jaoua, S.; Migheli, Q. International Journal of Food Microbiology Biocontrol activity of four non- and low-fermenting yeast strains against Aspergillus carbonarius and their ability to remove ochratoxin A from grape juice. Int. J. Food Microbiol. 2014, 189, 45–50. [Google Scholar] [CrossRef]
- Meca, G.; Ritieni, A.; Zhou, T.; Li, X.Z.; Mañes, J. Degradation of the minor Fusarium mycotoxin beauvericin by intracellular enzymes of Saccharomyces cerevisiae. Food Control 2013, 33, 352–358. [Google Scholar] [CrossRef]
- Zhu, R.; Feussner, K.; Wu, T.; Yan, F.; Karlovsky, P.; Zheng, X. Detoxification of mycotoxin patulin by the yeast Rhodosporidium paludigenum. Food Chem. 2015, 179, 1–5. [Google Scholar] [CrossRef]
- Li, J.; Huang, J.; Jin, Y.; Wu, C.; Shen, D.; Zhang, S.; Zhou, R. Mechanism and kinetics of degrading aflatoxin B1 by salt tolerant Candida versatilis CGMCC 3790. J. Hazard. Mater. 2018, 359, 382–387. [Google Scholar] [CrossRef]
- Varga, J.; Péteri, Z.; Tábori, K.; Téren, J.; Vágvölgyi, C. Degradation of ochratoxin A and other mycotoxins by Rhizopus isolates. Int. J. Food Microbiol. 2005, 99, 321–328. [Google Scholar] [CrossRef]
- Bdswald, C.; Engelhardt, G.; Vogel, H.; Wallnofer, P.R. Metabolism of the Fusatium Mycotoxins Zearalenone and Deoxynivalenol by Yeast Strains of Technological Relevance. Nat. Toxins 1995, 3, 138–144. [Google Scholar] [CrossRef]
- Loi, M.; Fanelli, F.; Cimmarusti, M.T.; Mirabelli, V.; Haidukowski, M.; Logrieco, A.F.; Caliandro, R.; Mule, G. In vitro single and combined mycotoxins degradation by Ery4 laccase from Pleurotus eryngii and redox mediators. Food Control 2018, 90, 401–406. [Google Scholar] [CrossRef]
- Wang, J.; Ogata, M.; Hirai, H.; Kawagishi, H. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 2011, 314, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Zeinvand-Lorestani, H.; Sabzevari, O.; Setayesh, N.; Amini, M.; Nili-Ahmadabadi, A.; Faramarzi, M.A. Comparative study of in vitro prooxidative properties and genotoxicity induced by aflatoxin B1 and its laccase-mediated detoxification products. Chemosphere 2015, 135, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Wu, Z.; Wu, S.; Dai, Y.; Sun, C. Degradation of ochratoxin A by Bacillus amyloliquefaciens ASAG1. Food Addit. Contam. Part A 2015, 32, 564–571. [Google Scholar] [CrossRef]
- Abrunhosa, L.; Santos, L.; Venâncio, A. Degradation of ochratoxin A by proteases and by a crude enzyme of Aspergillus niger. Food Biotechnol. 2006, 20, 231–242. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, B.; Wang, Z.; Han, C.; Meng, X.; Zhang, F. Effective degradation of the mycotoxin patulin in pear juice by porcine pancreatic lipase. Food Chem. Toxicol. 2019, 133, 110769. [Google Scholar] [CrossRef]
- Tang, H.; Li, X.; Zhang, F.; Meng, X.; Liu, B. Biodegradation of the mycotoxin patulin in apple juice by Orotate phosphoribosyltransferase from Rhodotorula mucilaginosa. Food Control 2019, 100, 158–164. [Google Scholar] [CrossRef]
- Alberts, J.F.; Gelderblom, W.C.A.; Botha, A.; van Zyl, W.H. Degradation of aflatoxin B1 by fungal laccase enzymes. Int. J. Food Microbiol. 2009, 135, 47–52. [Google Scholar] [CrossRef]
- Afsharmanesh, H.; Perez-Garcia, A.; Zeriouh, H.; Ahmadzadeh, M.; Romero, D. Aflatoxin degradation by Bacillus subtilis UTB1 is based on production of an oxidoreductase involved in bacilysin biosynthesis. Food Control 2018, 94, 48–55. [Google Scholar] [CrossRef]
- Samuel, M.S.; Sivaramakrishna, A.; Mehta, A. Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeterior. Biodegrad. 2014, 86, 202–209. [Google Scholar] [CrossRef]
- Ianiri, G.; Pinedo, C.; Fratianni, A.; Panfili, G.; Castoria, R. Patulin degradation by the biocontrol yeast Sporobolomyces sp. Is an inducible process. Toxins 2017, 9, 61. [Google Scholar] [CrossRef]
- Young, J.C.; Zhou, T.; Yu, H.; Zhu, H.; Gong, J. Degradation of trichothecene mycotoxins by chicken intestinal microbes. Food Chem. Toxicol. 2007, 45, 136–143. [Google Scholar] [CrossRef]
- Koch, M.; Maul, R.; Brodehl, A.; Anne, M. Biotransformation of the mycotoxin zearalenone by fungi of the genera Rhizopus and Aspergillus. FEMS Microbiol. Lett. 2014, 359, 124–130. [Google Scholar]
- Kollarczik, B.; Gareis, M.; Hanelt, M. In Vitro Transformation of the Fusarium Mycotoxins Deoxynivalenol and ZearaLenone by the Normal Gut Microflora of Pigs. Nat. Toxins 1994, 2, 105–110. [Google Scholar] [CrossRef]
- Péteri, Z.; Téren, J.; Vágvölgyi, C.; Varga, J. Ochratoxin degradation and adsorption caused by astaxanthin-producing yeasts. Food Microbiol. 2007, 24, 205–210. [Google Scholar] [CrossRef]
- Zheng, X.; Li, Y.; Zhang, H.; Apaliya, M.T.; Zhang, X.; Zhao, L.; Jiang, Z.; Yang, Q.; Gu, X. Identification and toxicological analysis of products of patulin degradation by Pichia caribbica. Biol. Control 2018, 123, 127–136. [Google Scholar] [CrossRef]
- Taheur, F.B.; Fedhila, K.; Chaieb, K.; Kouidhi, B.; Bakhrouf, A.; Abrunhosa, L. Adsorption of aflatoxin B1, zearalenone and ochratoxin A by microorganisms isolated from Kefir grains. Int. J. Food Microbiol. 2017, 251, 1–7. [Google Scholar] [CrossRef]
- Elsanhoty, R.M.; Salam, S.A.; Ramadan, M.F.; Badr, F.H. Detoxification of aflatoxin M1 in yoghurt using probiotics and lactic acid bacteria. Food Control 2014, 43, 129–134. [Google Scholar] [CrossRef]
- Abdolshahi, A.; Tabatabaie Yazdi, F.; Shabani, A.A.; Mortazavi, S.A.; Marvdashti, L.M. Aflatoxin binding efficiency of Saccharomyces cerevisiae mannoprotein in contaminated pistachio nuts. Food Control 2018, 87, 17–21. [Google Scholar] [CrossRef]
- Alekinejad, H.M.; Franciscus, R.; Akker, M.A.A.S.; Remmels, J.F.I.N.K. Bioactivation of zearalenone by porcine hepatic biotransformation. Vet. Res. 2005, 36, 799–810. [Google Scholar] [CrossRef]
- Liang, Y.-F.; Zhou, X.-W.; Wang, F.; Shen, Y.-D.; Xiao, Z.-L.; Zhang, S.-W.; Li, Y.-J.; Wang, H. Development of a Monoclonal Antibody-Based ELISA for the Detection of Alternaria Mycotoxin Tenuazonic Acid in Food Samples. Food Anal. Methods 2020, 13, 1594–1602. [Google Scholar] [CrossRef]
- Zhao, L.; Guan, S.; Gao, X.; Ma, Q.; Lei, Y.; Bai, X.; Ji, C. Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 2010, 110, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Motomura, M.; Toyomasu, T.; Mizuno, K.; Shinozawa, T. Purification and characterization of an aflatoxin degradation enzyme from Pleurotus ostreatus. Microbiol. Res. 2003, 158, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Peng, X.; Li, X.; Meng, X.; Liu, B. Biodegradation of mycotoxin patulin in apple juice by calcium carbonate immobilized porcine pancreatic lipase. Food Control 2018, 88, 69–74. [Google Scholar] [CrossRef]
- Medina, A.; Mohale, S.; Samsudin, N.I.P.; Rodriguez-Sixtos, A.; Rodriguez, A.; Magan, N. Biocontrol of mycotoxins: Dynamics and mechanisms of action. Curr. Opin. Food Sci. 2017, 17, 41–48. [Google Scholar] [CrossRef]
- Illueca, F.; Vila-Donat, P.; Calpe, J.; Luz, C.; Meca, G.; Quiles, J.M. Antifungal activity of biocontrol agents in vitro and potential application to reduce mycotoxins (Aflatoxin B1 and ochratoxin A). Toxins 2021, 13, 752. [Google Scholar] [CrossRef]
- Zapasnik, A.; Sokołowska, B.; Bryła, M. Role of Lactic Acid Bacteria in Food Preservation and Safety. Foods 2022, 11, 1283. [Google Scholar] [CrossRef]
- Yao, M.; Wu, J.; Li, B.; Xiao, H.; McClements, D.J.; Li, L. Microencapsulation of Lactobacillus salivarious Li01 for enhanced storage viability and targeted delivery to gut microbiota. Food Hydrocoll. 2017, 72, 228–236. [Google Scholar] [CrossRef]
- Wang, M.; Yang, J.; Li, M.; Wang, Y.; Wu, H.; Xiong, L.; Sun, Q. Enhanced viability of layer-by-layer encapsulated Lactobacillus pentosus using chitosan and sodium phytate. Food Chem. 2019, 285, 260–265. [Google Scholar] [CrossRef]
- Alehosseini, A.; Sarabi-jamab, M.; Ghorani, B.; Kadkhodaee, R. Electro-encapsulation of Lactobacillus casei in high-resistant capsules of whey protein containing transglutaminase enzyme. LWT—Food Sci. Technol. 2019, 102, 150–158. [Google Scholar] [CrossRef]
- Tapingkae, W.; Srinual, O.; Lumsangkul, C.; Van Doan, H.; Chiang, H.-I.; Manowattana, A.; Boonchuay, P.; Chaiyaso, T. Industrial-Scale Production of Mycotoxin Binder from the Red Yeast Sporidiobolus pararoseus KM281507. J. Fungi 2022, 8, 353. [Google Scholar] [CrossRef]
- Hynes, R.K.; Boyetchko, S.M. Research initiatives in the art and science of biopesticide formulations. Soil Biol. Biochem. 2006, 38, 845–849. [Google Scholar] [CrossRef]
- Nešić, K.; Habschied, K.; Mastanjević, K. Possibilities for the Biological Control of Mycotoxins in Food and Feed. Toxins 2021, 13, 198. [Google Scholar] [CrossRef]
- Chen, W.C.; Chiou, T.Y.; Delgado, A.L.; Liao, C.S. The control of rice blast disease by the novel biofungicide formulations. Sustainability 2019, 11, 3449. [Google Scholar] [CrossRef]
- Zamoum, M.; Allali, K.; Benadjila, A.; Zitouni, A.; Goudjal, Y. Formulation of biofungicides based on Streptomyces lincolnensis strain SZ03 spores and efficacy against R. solani damping-off of tomato seedlings. ResearchSquare 2022, 1–22. [Google Scholar] [CrossRef]
- Mousumi Das, M.; Aguilar, C.N.; Haridas, M.; Sabu, A. Production of bio-fungicide, Trichoderma harzianum CH1 under solid-state fermentation using coffee husk. Bioresour. Technol. Rep. 2021, 15, 100708. [Google Scholar] [CrossRef]
- Sudantha, I.M.; Suwardji, S. Trichoderma biofungicides formulations on shallot growth, yield and fusarium wilt disease resistance. IOP Conf. Ser. Earth Environ. Sci. 2021, 824, 12032. [Google Scholar] [CrossRef]
- Souza, P.M.D.; Magalhães, P.D.O. Application of microbial α-amylase in industry—A review. Braz. J. Microbiol. 2010, 41, 850–861. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).