β-N-Methylamino-L-Alanine (BMAA) Causes Severe Stress in Nostoc sp. PCC 7120 Cells under Diazotrophic Conditions: A Proteomic Study
Abstract
1. Introduction
2. Results and Discussion
2.1. Proteins That Are Affected by BMAA under Diazotrophic Conditions
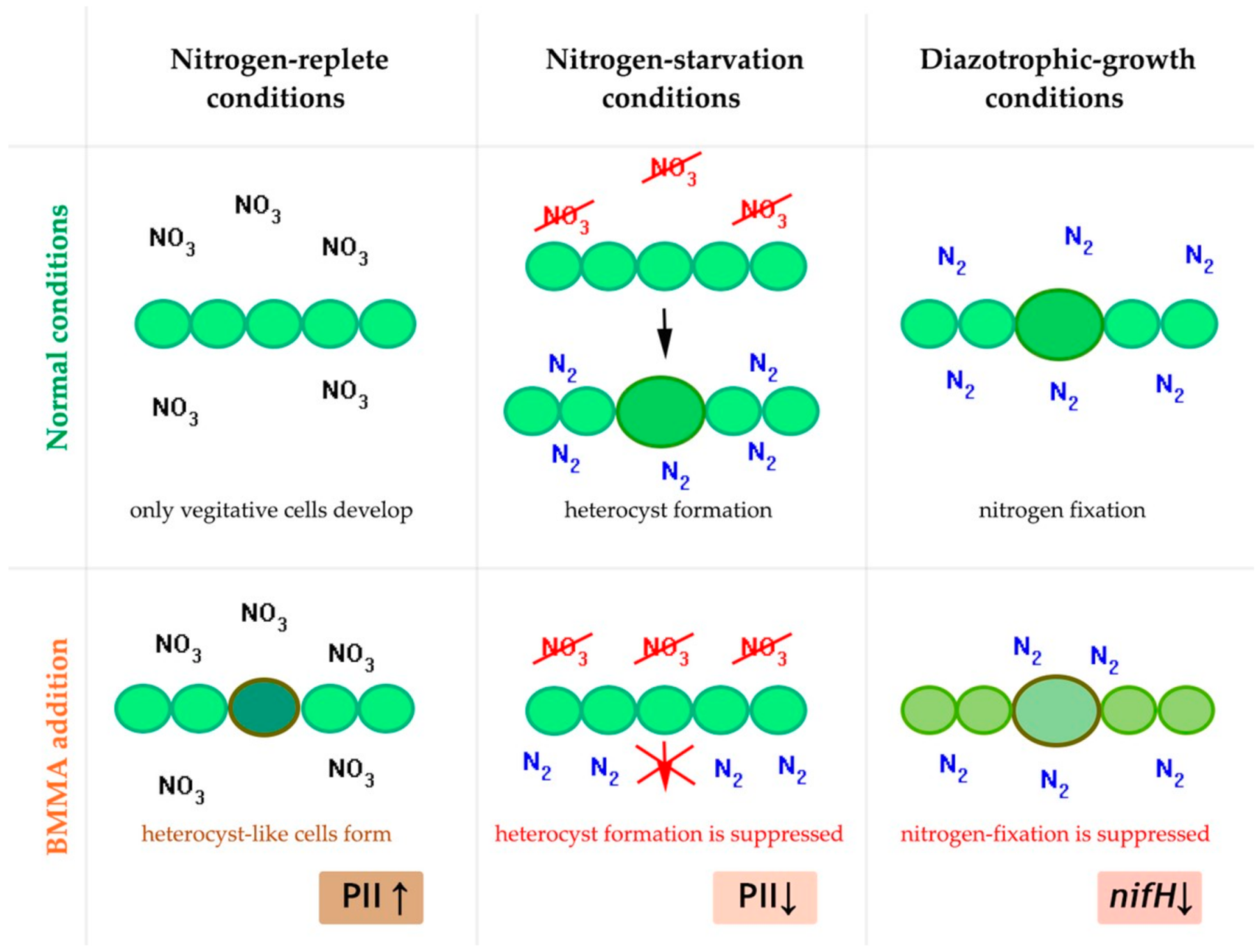
2.2. BMAA Downregulates Nitrogenase Proteins
2.3. BMAA Downregulates Photosynthesis and Oxidative Phosphorylation Proteins
2.4. BMAA Impact on the CO2 Concentrating Mechanism and Bicarbonate Transport
2.5. Changes in Carbohydrate Metabolism Proteins’ Regulation Caused by BMAA in Diazotrophic Conditions
2.6. Amino Acid Biosynthesis, Metabolism, and Transport
2.7. Transcription and Translation
2.8. Proteases, Stress Response Proteins, and DNA Repair Enzymes Are Significantly Upregulated at BMAA Presence
2.9. Hypothetical Proteins
3. Conclusions
4. Materials and Methods
4.1. Cyanobacterial Strain and Cultivation Conditions
4.2. Trypsin Digestion in Solution
4.3. LC-MS/MS Analysis
4.4. Protein Identification by Using LC-MS/MS Data Analysis
4.5. Pathway Analysis Based on LC-MS/MS Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pohnert, G. Influence of Algal Secondary Metabolites on Plankton Community Structure. In Algal Chemical Ecology; Amsler, C.D., Ed.; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Azam, F.; Worden, A.Z. Microbes, Molecules, and Marine Ecosystems. Science 2004, 303, 1622–1624. [Google Scholar] [CrossRef]
- Murch, S.J.; Cox, P.A.; Banack, S.A. A mechanism for slow release of biomagnified cyanobacterial neurotoxins and neurodegenerative disease in Guam. Proc. Natl. Acad. Sci. USA 2004, 101, 12228–12231. [Google Scholar] [CrossRef]
- Hirano, A.; Malamud, N.; Elizan, T.S.; Kurland, L.T. Amyotrophic lateral sclerosis and Parkinsonism-dementia complex on Guam. Further pathologic studies. Arch. Neurol. 1966, 15, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Murch, S.J.; Cox, P.A.; Banack, S.A.; Steele, J.C.; Sacks, O.W. Occurrence of beta-methylamino-l-alanine (BMAA) in ALS/PDC patients from Guam. Actaneurol. Scand. 2004, 110, 267–269. [Google Scholar] [CrossRef]
- Cox, P.A.; Banack, S.A.; Murch, S.J.; Rasmussen, U.; Tien, G.; Bidigare, R.R.; Metcalf, J.S.; Morrison, L.F.; Codd, G.A.; Bergman, B. Diverse taxa of cyanobacteria produce β-N-methylamino-L-alanine, a neurotoxic amino acid. Proc. Natl. Acad. Sci. USA 2005, 102, 5074–5078. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Eriksson, J.; Lage, S.; Jonasson, S.; Shams, S.; Mehine, M.; Ilag, L.L.; Rasmussen, U. Diatoms: A novel source for the neurotoxin BMAA in aquatic environments. PLoS ONE 2014, 9, e84578. [Google Scholar] [CrossRef] [PubMed]
- Réveillon, D.; Séchet, V.; Hess, P.; Amzil, Z. Production of BMAA and DAB by diatoms (Phaeodactylum tricornutum, Chaetoceros sp., Chaetoceros calcitrans and, Thalassiosira pseudonana) and bacteria isolated from a diatom culture. Harmful Algae 2016, 58, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Downing, T.G.; Phelan, R.R.; Downing, S. A potential physiological role for cyanotoxins in cyanobacteria of arid environments. J. Arid Environ. 2015, 112, 147–151. [Google Scholar] [CrossRef]
- Downing, S.; Downing, T.G. The metabolism of the non proteinogenic amino acid β-N-methylamino-L-alanine (BMAA) in the cyanobacterium Synechocystis PCC 6803. Toxicon 2016, 115, 41–48. [Google Scholar] [CrossRef]
- Popova, A.A.; Koksharova, O.A. Neurotoxic non-proteinogenic amino acid β-N-methylamino-L-alanine and its role in biological systems. Biochemistry 2016, 81, 794–805. [Google Scholar] [CrossRef]
- Nunn, P.B.; Codd, G.A. Metabolic solutions to the biosynthesis of some diaminomono carboxylic acids in nature: Formation in cyanobacteria of the neurotoxins 3-N-methyl-2,3 diaminopropanoic acid (BMAA) and 2,4-diaminobutanoic acid (2,4-DAB). Phytochemistry 2017, 144, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Downing, S.; Banack, S.A.; Metcalf, J.S.; Cox, P.A.; Downing, T.G. Nitrogen starvation of cyanobacteria results in the production of β-N-methylamino-L-alanine. Toxicon 2011, 58, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Downing, S.; Van de Venter, M.; Downing, T.G. The effect of exogenous β-N-methylamino-L-alanine on the growth of Synechocystis PCC 6803. Microb. Ecol. 2012, 63, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Berntzon, L.; Erasmie, S.; Celepli, N.; Eriksson, J.; Rasmussen, U.; Bergman, B. BMAA inhibits nitrogen fixation in the cyanobacterium Nostoc sp. PCC 7120. Mar. Drugs 2013, 11, 3091–3108. [Google Scholar] [CrossRef]
- Wang, Z.-Q.; Wang, S.; Zhang, J.-Y.; Lin, G.-M.; Gan, N.; Song, L.; Zhang, C.-C. The Proposed Neurotoxin β-N-Methylamino-l-Alanine (BMAA) Is Taken up through Amino-Acid Transport Systems in the Cyanobacterium Anabaena PCC 7120. Toxins 2020, 12, 518. [Google Scholar] [CrossRef]
- Popova, A.; Rasmussen, U.; Semashko, T.; Govorun, V.; Koksharova, O. Stress effects of cyanotoxin β-methylamino-L-alanine (BMAA) on cyanobacterial heterocyst formation and functionality. Environ. Microbiol. Rep. 2018, 10, 369–377. [Google Scholar] [CrossRef]
- Popova, A.; Semashko, T.; Kostina, N.; Rasmussen, U.; Govorun, V.; Koksharova, O. The cyanotoxin BMAA induces heterocyst specific gene expression in Anabaena sp. PCC 7120 under repressive conditions. Toxins 2018, 10, 478. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakamura, Y.; Wolk, C.P.; Kuritz, T.; Sasamoto, S.; Watanabe, A.; Iriguchi, M.; Ishikawa, A.; Kawashima, K.; Kimura, T.; et al. Complete genomic sequence of the ¢lamentous nitrogen-fixing Cyanobacterium Anabaena sp strain PCC 7120. DNA Res. 2001, 8, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Castenholz, R.W. Phylum BX. Cyanobacteria (Oxygenic Photosynthetic Bacteria). Bergey’s Manual® of Systematic Bacteriology: The Archaea and the Deeply Branching and Phototrophic Bacteria; Springer: New York, NY, USA, 2001; Volume 1, pp. 473–599. [Google Scholar]
- Herrero, A.; Stavans, J.; Flores, E. The multicellular nature of filamentous heterocyst-forming cyanobacteria. FEMS Microbiol. Rev. 2016, 40, 831–854. [Google Scholar] [CrossRef]
- Olli, K.; Klais, R.; Tamminen, T. Rehabilitating the cyanobacteria—Niche partitioning, resource use efficiency and phytoplankton community structure during diazotrophic cyanobacterial blooms. J. Ecol. 2015, 103, 1153–1164. [Google Scholar] [CrossRef]
- Koksharova, O.A.; Butenko, I.O.; Pobeguts, O.V.; Safronova, N.A.; Govorun, V.M. The first proteomic study of Nostoc sp. PCC 7120 exposed to cyanotoxin BMAA under nitrogen starvation. Toxins 2020, 12, 310. [Google Scholar] [CrossRef] [PubMed]
- Koksharova, O.A.; Butenko, I.O.; Pobeguts, O.V.; Safronova, N.A.; Govorun, V.M. Proteomic Insights into Starvation of Nitrogen-Replete Cells of Nostoc sp. PCC7120 under BMAA Treatment. Toxins 2020, 12, 372. [Google Scholar] [CrossRef] [PubMed]
- Picossi, S.; Flores, E.; Herrero, A. ChIP analysis unravels an exceptionally wide distribution of DNA binding sites for the NtcA transcription factor in a heterocyst-forming cyanobacterium. BMC Genom. 2014, 15, 22. [Google Scholar] [CrossRef]
- Mishra, A.K.; Tewari, D.N.; Rai, A.N. Cyanobacteria 1st Edition from Basic Science to Applications; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Igarashi, R.Y.; Seefeldt, L.C. Nitrogen fixation: The mechanism of the Mo-dependent nitrogenase. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 351–384. [Google Scholar] [CrossRef] [PubMed]
- Seefeldt, L.C.; Hoffman, B.M.; Dean, D.R. Mechanism of Mo-dependent nitrogenase. Annu. Rev. Biochem. 2009, 78, 701–722. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.G. Heterocyst formation in cyanobacteria. Curr. Opin. Microbiol. 2000, 3, 618–624. [Google Scholar] [CrossRef]
- Gallon, J.R. N2 fixation in phototrophs: Adaptation to a specialized way of life. Plant Soil. 2001, 230, 39–48. [Google Scholar] [CrossRef]
- Smith, R.L.; Van Baalen, C.; Tabita, F.R. Alteration of the Fe protein of nitrogenase by oxygen in the cyanobacterium Anabaena sp. strain CA. J. Bacteriol. 1987, 169, 2537–2542. [Google Scholar] [CrossRef]
- Singh, H.N.; Rai, U.N.; Rao, V.V.; Bagchi, S.N. Evidence for ammonia as an inhibitor of heterocyst and nitrogenase formation in the cyanobacterium Anabaena cycadeae. Biochem. Biophys. Res. Commun. 1983, 111, 180–187. [Google Scholar] [CrossRef]
- Martín-Nieto, J.; Herrero, A.; Flores, E. Control of Nitrogenase mRNA Levels by Products of Nitrate Assimilation in the Cyanobacterium Anabaena sp. Strain PCC 7120. Plant Physiol. 1991, 97, 825–828. [Google Scholar] [CrossRef]
- Rawson, D.M. The effects of exogenous amino acids on growth and nitrogenase activity in the cyanobacterium Anabaena cylindrica PCC 7122. Microbiology 1985, 131, 2549–2554. [Google Scholar] [CrossRef]
- Ramos, J.L.; Madueño, F.; Guerrero, M.G. Regulation of nitrogenase levels in Anabaena sp. ATCC 33047 and other filamentous cyanobacteria. Arch. Microbiol. 1985, 141, 105–111. [Google Scholar] [CrossRef]
- Nunn, P.B.; Ponnusamy, M. β-N-methyl-aminoalanine (BMAA): Metabolism and metabolic effects in model systems and in neural and other tissues of the rat in vitro. Toxicon 2009, 54, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Nunn, P.B. Three phases of research on β-N-methylamino_L_alanine (BMAA)—A neurotoxic amino acid. Amyotroph. Lateral Scler. 2009, 10, 26–33. [Google Scholar] [CrossRef]
- Nunn, P.B.; Nunn, P.B.; Bell, E.A.; Watson, A.A.; Nash, R.J. Toxicity of Non-protein Amino Acids to Humans and Domestic Animals. Nat. Prod. Commun. 2010, 5. [Google Scholar] [CrossRef]
- Liu, X.; Rush, T.; Zapata, J.; Lobner, D. β-N-methylamino-l-alanine induces oxidative stress and glutamate release through action on system Xc(−). Exp. Neurol. 2009, 217, 429–433. [Google Scholar] [CrossRef]
- Yan, B.; Liu, Z.; Huang, R.; Xu, Y.; Liu, D.; Wang, W.; Zhao, Z.; Cui, F.; Shi, W. Impact factors on the production of β-methylamino-L-alanine (BMAA) by cyanobacteria. Chemosphere 2020, 243, 125355. [Google Scholar] [CrossRef]
- Koksharova, O.A.; Wolk, C.P. Novel DNA-Binding Proteins in the Cyanobacterium Anabaena sp. Strain PCC 7120. J. Bacteriol. 2002, 184, 3931–3940. [Google Scholar] [CrossRef]
- Nelson, N.; Junge, W. Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem. 2015, 84, 659–683. [Google Scholar] [CrossRef]
- Wilde, A.; Hihar, Y. Transcriptional and posttranscriptional regulation of cyanobacterial photosynthesis. Biochim. Biophys. Acta 2016, 1857, 296–308. [Google Scholar] [CrossRef]
- Herrero, A.; Flores, E. Genetic responses to carbon and nitrogen availability in Anabaena. Environ. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.; Picossi, S.; Valladares, A.; Herrero, A. Transcriptional regulation of development in heterocyst forming cyanobacteria. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2019, 1862, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Sosa, F.M.; Gil-Martínez, J.; Molina-Heredia, F.P. Cytochrome c 6-like protein as a putative donor of electrons to photosystem I in the cyanobacterium Nostoc sp. PCC 7119. Photosynth. Res. 2011, 110, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Torrado, A.; Ramírez-Moncayo, C.; Navarro, J.A.; Mariscal, V.; Molina-Heredia, F.P. Cytochrome c6 is the main respiratory and photosynthetic soluble electron donor in heterocysts of the cyanobacterium Anabaena sp. PCC 7120. Biochim. Biophys. Acta BBA Bioenergy 2019, 1860, 60–68. [Google Scholar] [CrossRef]
- Kaplan, A.; Reinhold, L. The CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 539–570. [Google Scholar] [CrossRef]
- Ghoshal, D.; Goyal, A. Carbon concentration mechanisms in photosynthetic microorganisms. Indian J. Biochem. Biophys. 2000, 37, 383–394. [Google Scholar]
- Kupriyanova, E.V.; Sinetova, M.A.; Cho, S.M.; Park, Y.I.; Los, D.; Pronina, N.A. CO2-concentrating mechanism in cyanobacterial photosynthesis: Organization, physiological role, and evolutionary origin. Photosynth. Res. 2013, 117, 133–146. [Google Scholar] [CrossRef]
- Price, G.D. Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth. Res. 2011, 109, 47–57. [Google Scholar] [CrossRef]
- Wang, C.; Sun, B.; Zhang, X.; Huang, X.; Zhang, M.; Guo, H.; Chen, X.-Q.; Huang, F.; Chen, T.; Mi, H.L.; et al. Structural mechanism of the active bicarbonate transporter from cyanobacteria. Nat. Plants 2019, 5, 1184–1193. [Google Scholar] [CrossRef]
- Koropatkin, N.M.; Koppenaal, D.W.; Pakrasi, H.B.; Smith, T.J. The Structure of a Cyanobacterial Bicarbonate Transport Protein, CmpA. J. Biol. Chem. 2006, 282, 2606–2614. [Google Scholar] [CrossRef]
- Tamoi, M.; Takeda, T.; Shigeoka, S. Functional Analysis of Fructose-1,6-Bisphosphatase Isozymes (fbp-I and fbp-II Gene Products) in Cyanobacteria. Plant Cell Physiol. 1999, 40, 257–261. [Google Scholar] [CrossRef]
- Tamoi, M.; Murakami, A.; Takeda, T.; Shigeoka, S. Acquisition of a new type of fructose-1,6-bisphosphatase with resistance to hydrogen peroxide in cyanobacteria: Molecular characterization of the enzyme from Synechocystis PCC 6803. Biochim. Biophys. Acta 1998, 1383, 232–244. [Google Scholar] [CrossRef]
- Tamoi, M.; Nagaoka, M.; Miyagawa, Y.; Shigeoka, S. Contribution of fructose-1,6-bisphosphatase and sedoheptulose-1,7-bisphosphatase to the photosynthetic rate and carbon flow in the Calvin cycle in transgenic plants. Plant Cell Physiol. 2006, 47, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Cotton, C.A.R.; Kabasakal, B.V.; Miah, N.A.; Murray, J.W. Structure of the dual-function fructose-1,6/sedoheptulose-1,7-bisphosphatase from Thermosynechococcus elongates bound with sedoheptulose-7-phosphate. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 1341–1345. [Google Scholar] [CrossRef] [PubMed]
- Doello, S.; Klotz, A.; Makowka, A.; Gutekunst, K.; Forchhammer, K. A Specific Glycogen Mobilization Strategy Enables Rapid Awakening of Dormant Cyanobacteria from Chlorosis. Plant Physiol. 2018, 177, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Ankel, H.; Feingold, D.S. Biosynthesis of uridine diphosphate D-xylose. 1. Uridine diphosphate glucuronate carboxy-lyase of wheat germ. Biochemistry 1965, 4, 2468–2475. [Google Scholar] [CrossRef]
- Bakker, H. UDP-Glucuronate Decarboxylase 1 (UXS1). In Handbook of Glycosyltransferases and Related Genes; Taniguchi, N., Honke, K., Fukuda, M., Narimatsu, H., Yamaguchi, Y., Angata, T., Eds.; Springer: Tokyo, Japan, 2014. [Google Scholar] [CrossRef]
- Mackerras, A.H.; Smith, G.D. Urease Activity of the Cyanobacterium Anabaena cylindrica. Microbiology 1986, 132, 2749–2752. [Google Scholar] [CrossRef][Green Version]
- Veaudor, T.; Cassier-Chauvat, C.; Chauvat, F. Genomics of urea transport and catabolism in cyanobacteria: Biotechnological implications. Front. Microbiol. 2019, 10, 2052. [Google Scholar] [CrossRef]
- Rai, A.K.; Singh, S. Urease of blue-green algae (Cyanobacteria) Anabaena doliolum and Anacystis nidulans. Curr. Microbiol. 1987, 16, 113–117. [Google Scholar] [CrossRef]
- Singh, S. Regulation of urease activity in the cyanobacterium Anabaena doliolum. FEMS Microbiol. Lett. 1990, 67, 79–83. [Google Scholar] [CrossRef]
- Pernil, R.; Picossi, S.; Mariscal, V.; Herrero, A.; Flores, E. ABC-type amino acid uptake transporters Bgt and N-II of Anabaena sp. strain PCC 7120 share an ATPase subunit and are expressed in vegetative cells and heterocysts. Mol. Microbiol. 2008, 67, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Zhang, Y.X.; Zhou, Q.; Liu, Q.-E.; Chen, D.-B.; Wang, H.; Cheng, S.-H.; Cao, L.-Y.; Shen, X.-H. UMP Kinase Regulates Chloroplast Development and Cold Response in Rice. Int. J. Mol. Sci. 2019, 20, 2107. [Google Scholar] [CrossRef]
- Ha, K.S.; Toulokhonov, I.; Vassylyev, D.G.; Landick, R.J. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. Mol. Biol. 2010, 401, 708–725. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.E.; Lewis, C.A.; Mooney, R.A.; Kohanski, M.A.; Collins, J.J.; Landick, R.; Walker, G.C. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc. Natl. Acad. Sci. USA 2010, 107, 15517–15522. [Google Scholar] [CrossRef] [PubMed]
- Sato, N. A family of cold-regulated RNA-binding protein genes in the cyanobacterium Anabaena variabilis M3. Nucleic Acids Res. 1995, 23, 2161–2167. [Google Scholar]
- Mori, S.; Castoreno, A.; Mulligan, M.E.; Lammers, P.J. Nitrogen status modulates the expression of RNA-binding proteins in cyanobacteria. FEMS Microbiol. Lett. 2003, 227, 203–210. [Google Scholar] [CrossRef][Green Version]
- Shrivastava, A.K.; Pandey, S.; Yadav, S.; Mishra, Y.; Singh, P.K.; Rai, R.; Rai, L.C. Comparative proteomics of wild type, An+ahpC and An∆ahpC strains of Anabaena sp. PCC7120 demonstrates AhpC mediated augmentation of photosynthesis, N2-fixation and modulation of regulatory network of antioxidative proteins. J. Proteom. 2016, 140, 81–99. [Google Scholar] [CrossRef] [PubMed]
- Lopatovskaya, K.V.; Seliverstov, A.V.; Lyubetsky, V.A. NtcA and NtcB regulons in cyanobacteria and rhodophyta chloroplasts. Mol. Biol. 2011, 45, 522–526. [Google Scholar] [CrossRef]
- Dunlop, R.A.; Cox, P.A.; Banack, S.A.; Rodgers, K.J. The non-protein amino acid BMAA is misincorporated into human proteins in place of L-serine causing protein misfolding and aggregation. PLoS ONE 2013, 8, e75376. [Google Scholar] [CrossRef]
- Han, N.C.; Bullwinkle, T.J.; Loeb, K.F.; Faull, K.F.; Mohler, K.; Rinehart, J.; Ibba, M. The mechanism of β-N-methylamino-l-alanine inhibition of tRNA aminoacylation and its impact on misincorporation. J. Biol. Chem. 2020, 295, 1402–1410. [Google Scholar] [CrossRef]
- Baker, T.A.; Sauer, R.T. ATP-dependent proteases of bacteria: Recognition logic and operating principles. Trends Biochem. Sci. 2006, 31, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Yamamoto, Y. The carboxyl-terminal processing of precursor D1 protein of the photosystem II reaction center. Photosynth. Res. 2007, 94, 203–215. [Google Scholar] [CrossRef]
- Shestakov, S.V.; Anbudurai, P.R.; Stanbekova, G.E.; Gadzhiev, A.; Lind, L.K.; Pakrasi, H.B. Molecular cloning and characterization of the ctpA gene encoding a carboxyl-terminal processing protease: Analysis of a spontaneous photosystem II-deficient mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 1994, 269, 19354–19359. [Google Scholar] [CrossRef]
- Oelmuller, R.; Herrmann, R.G.; Pakrasi, H.B. Molecular studies of CtpA, the carboxyl-terminal processing protease for the D1 protein of the photosystem II reaction center in higher plants. J. Biol. Chem. 1996, 271, 21848–21852. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, L.P.; Sowdhamini, R. Genome-wide survey of prokaryotic serine proteases: Analysis of distribution and domain architectures of five serine protease families in prokaryotes. BMC Genom. 2008, 9, 549. [Google Scholar] [CrossRef] [PubMed]
- Latifi, A.; Ruiz, M.; Zhang, C.-C. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 2009, 33, 258–278. [Google Scholar] [CrossRef]
- Zorina, A.A.; Mironov, K.S.; Stepanchenko, N.S.; Sinetova, M.; Koroban, M.V.; Zinchenko, V.; Kupriyanova, E.V.; Allakherdiev, S.I.; Los, D.A. Regulation systems for stress responses in cyanobacteria. Russ. J. Plant. Physiol. 2011, 58, 749. [Google Scholar] [CrossRef]
- Wase, N.V.; Yen, S.O.; Wright, P.C. A global understanding of light stress in cyanobacteria: Environmental and bioproducts perspectives. In Stress Biology of Cyanobacteria: Molecular Mechanisms to Cellular Responses; Srivastava, A.K., Rai, A.N., Neilan, B.A., Eds.; CRC Press: Boca Raton, FL, USA, 2013; 394p. [Google Scholar]
- Rampon, C.; Volovitch, M.; Joliot, A.; Vriz, S. Hydrogen Peroxide and Redox Regulation of Developments. Antioxidants 2018, 7, 159. [Google Scholar] [CrossRef]
- Pham, K.; Pal, R.; Qu, Y.; Liu, X.; Yu, H.; Shiao, S.L.; Wang, X.; Smith, E.O.; Cui, X.; Rodney, G.G.; et al. Nuclear glutaredoxin 3 is critical for protection against oxidative stress-induced cell death. Free Radic. Biol. Med. 2015, 85, 197–206. [Google Scholar] [CrossRef]
- Alkafeef, S.S.; Lane, S.; Yu, C.; Zhou, T.; Solis, N.V.; Filler, S.G.; Huang, L.; Lui, H. Proteomic profiling of the monothiol glutaredoxin Grx3 reveals its global role in the regulation of iron dependent processes. PLoS Genet. 2020, 16, e1008881. [Google Scholar] [CrossRef]
- Ferreira, F.; Straus, N.A. Iron deprivation in cyanobacteria. J. Appl. Phycol. 1994, 6, 199–210. [Google Scholar] [CrossRef]
- Berman-Frank, I.; Quigg, A.; Finkel, Z.V.; Irwin, A.J.; Haramaty, L. Nitrogen-fixation strategies and Fe requirements in cyanobacteria. Limnol. Oceanogr. 2007, 52, 2260–2269. [Google Scholar] [CrossRef]
- Morrissey, J.; Bowler, C. Iron utilization in marine cyanobacteria and eukaryotic algae. Front. Microbiol. 2012. [Google Scholar] [CrossRef]
- Rubio, L.M.; Ludden, P.W. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 2008, 62, 93–111. [Google Scholar] [CrossRef] [PubMed]
- Pernil, R.; Schleiff, E. Metalloproteins in the Biology of Heterocysts. Life 2019, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, P.C.; Smith, A.D.; Frazzon, J.; Cash, V.L.; Johnson, M.K.; Dean, D.R. Iron-Sulfur Cluster Assembly: NifU-directed activation of the nitrogenase Fe protein. JBC 2004, 279, 19705–19711. [Google Scholar] [CrossRef]
- Florencio, F.J.; Pérez-Pérez, M.E.; López-Maury, L.; Mata-Cabana, A.; Lindahl, M. The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynth. Res. 2006, 89, 157–171. [Google Scholar] [CrossRef]
- Lindahl, M.; Florencio, F.J. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc. Natl. Acad. Sci. USA 2003, 100, 16107–16112. [Google Scholar] [CrossRef]
- Lindahl, M.; Kieselbach, T. Disulphide proteomes and interactions with thioredoxin on the track towards understanding redox regulation in chloroplasts and cyanobacteria. J. Proteomics 2009, 72, 416–438. [Google Scholar] [CrossRef]
- Crawford, N.A.; Sutton, C.W.; Yee, B.C.; Johnson, T.C.; Carlson, D.C.; Buchanan, B.B. Contrasting modes of photosynthetic enzyme regulation in oxygenic and anoxygenic prokaryotes. Arch. Microbiol. 1984, 139, 124–129. [Google Scholar] [CrossRef]
- Ran, L.; Huang, F.; Ekman, M.; Klint, J.; Bergman, B. Proteomic analyses of the photoauto-and diazotrophically grown cyanobacterium Nostoc sp. PCC 73102. Microbiology 2007, 153, 608–618. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Robles-Rengel, R.; Florencio, F.J.; Muro-Pastor, M.I. Redox interference in nitrogen status via oxidative stress is mediated by 2-oxoglutarate in cyanobacteria. New Phytol. 2019, 224, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Issa, A.A.; Abd-Alla, M.H.; Ohyama, T. Nitrogen Fixing Cyanobacteria: Future Prospect, Advances in Biology and Ecology of Nitrogen Fixation. IntechOpen 2014. [Google Scholar] [CrossRef]
- Vergou, Y.; Touraki, M.; Paraskevopoulou, A.; Triantis, T.M.; Hiskia, A.; Gkelis, S. β-Ν-Methylamino-L-alanine interferes with nitrogen assimilation in the cyanobacterium, non-BMAA producer, Synechococcus sp. TAU-MAC 0499. Toxicon 2020. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Arapidi, G.; Osetrova, M.; Ivanova, O.; Butenko, I.; Saveleva, T.; Pavlovich, P.; Anikanov, N.; Ivanov, V.; Govorun, V. Peptidomics dataset: Blood plasma and serum samples of healthy donors fractionated on a set of chromatography sorbents. Data Brief 2018, 18, 1204–1211. [Google Scholar] [CrossRef]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteomics 2014, 13, 2513–2526. [Google Scholar] [CrossRef]







| № | Pathway | Number of Proteins Affected by BMAA | Total Amount | |
|---|---|---|---|---|
| Up-Regulated | Down-Regulated | |||
| 1 | Heterocyst formation and functionality | 3 | 1 | 2 |
| 2 | Photosynthesis | 19 | 1 | 18 |
| 3 | Oxidative phosphorylation | 6 | 0 | 6 |
| 4 | CO2-concentrating mechanism | 2 | 0 | 2 |
| 5 | Carbohydrate metabolism | 14 | 8 | 6 |
| 6 | Transporters | 3 | 1 | 2 |
| 7 | Sulfur metabolism | 1 | 0 | 1 |
| 8 | Secondary metabolites | 4 | 1 | 3 |
| 9 | Proteases | 5 | 4 | 1 |
| 10 | Chaperones | 3 | 2 | 1 |
| 11 | Stress response | 11 | 9 | 2 |
| 12 | SOS-response and DNA repair | 4 | 4 | 0 |
| 13 | Transcription | 2 | 2 | 0 |
| 14 | Translation | 8 | 4 | 4 |
| 15 | Amino acid synthesis and metabolism | 11 | 10 | 1 |
| 16 | Purine and pyrimidine metabolism | 1 | 1 | 0 |
| 17 | Hypothetical proteins | 26 | 20 | 6 |
| Total | 123 | 68 | 55 | |
| № | Protein | Gene | Function | Up Shifted | Down Shifted | p-Value |
|---|---|---|---|---|---|---|
| Nitrogen fixation and heterocyst formation (3 proteins) | ||||||
| 1 | NifK | all1440 # | nifK|nitrogenase molybdenum-iron protein subunit β | 0.199 | 0.0021 *** | |
| 2 | nifD | all1454 # | molybdenum-iron protein subunit α in nitrogenase | 0.59 | 0.0131 ** | |
| 3 | Apb2 | all1939 | transcription regulation of hepA and hepC genes | 1.49 | 0.0286 ** | |
| ABC-transporters and transporters (3 proteins) | ||||||
| 4 | Alr3938 | alr3938 # | ABC transporter iron binding protein (high-affinity iron ion transport) | 0.67 | 0.0771 * | |
| 5 | YidC | alr3415 | inner membrane protein translocase component YidC | 0.81 | 0.0105 ** | |
| 6 | alr4164 | alr4164 # | periplasmic amino acid-binding protein of amino acid ABC transporter | 1.47 | 0.0469 ** | |
| CO2-concentrating mechanism and bicarbonate transport (2 proteins) | ||||||
| 7 | ccmM | all0865 | CcmM, carbon dioxide concentrating mechanism protein | 0.70 | 0.0126 ** | |
| 8 | cmpD | alr2880 | CmpD, bicarbonate transport ATP-binding protein | Found only in control sample | 0.0349 ** | |
| Proteases (5 proteins) | ||||||
| 9 | alr2758 | alr2758 | serine proteinase | 0.67 | 0.0259 ** | |
| 10 | carboxyl-terminal processing protease [EC:3.4.21.102] | all2500 | carboxyl-terminal protease (serine endopeptidase) | 1.43 | 0.0279 ** | |
| 11 | ClpP (subunit 1) [EC:3.4.21.92] | alr1238 | ATP-dependent Clp P protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 1 | 1.59 | 0.0338 ** | |
| 12 | ClpP (subunit 2) [EC:3.4.21.92] | alr3683 | ATP-dependent Clp protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 2 | 1.85 | 0.0167 ** | |
| 13 | ATP-dependent Clp protease, protease subunit [EC:3.4.21.92] | all4358 | ATP-dependent Clp protease-like protein | 1.69 | 0.0287 ** | |
| Photosynthesis (19 proteins) | ||||||
| 14 | photosystem I reaction center subunit IV | psaE asr4319 | photosystem I | 0.65 | 0.0023 *** | |
| 15 | photosystem I reaction center protein subunit XI | psaL all0107 | photosystem I | 0.69 | 0.0372 ** | |
| 16 | psbA1 | alr4866; alr4592; alr3727; #all3572; alr3742 | photosystem II protein D1 | 0.56 | 0.0065 *** | |
| 17 | psbB | all0138 | photosystem II CP47 protein | 0.75 | 0.0034 *** | |
| 18 | psbD | alr4548 # alr4290 | photosystem II protein D2 | 0.62 | 0.0563 * | |
| 19 | psbO | all3854 | manganese-stabilizing protein | 0.81 | 0.0222 ** | |
| 20 | cpcB | alr0528 | phycocyanin β chain | 0.81 | 0.0359 ** | |
| 21 | cpcG1 | alr0534 | phycobilisome rod-core linker protein | 0.81 | 0.0671 * | |
| 22 | cpcG2 | alr0535 | phycobilisome rod-core linker protein | 0.61 | 0.0330 ** | |
| 23 | cpcG4 | alr0537 | phycobilisome rod-core linker protein | 0.69 | 0.0234 ** | |
| 24 | pecB | alr0523 | phycoerythrocyanin β chain | 0.83 | 0.0873 * | |
| 25 | hemC hydroxymethylbilane synthase [EC:2.5.1.61] | alr1878 | Porphyrin and chlorophyll metabolism | 0.47 | 0.0089 *** | |
| 26 | hemH protoporphyrin/coproporphyrin ferrochelatase [EC:4.99.1.1 4.99.1.9] | alr3751 | Porphyrin and chlorophyll metabolism | 0.53 | 0.0075 *** | |
| 27 | protochlorophyllide reductase [EC:1.3.1.33] | all1743 | Porphyrin and chlorophyll metabolism | 0.61 | 0.0161 ** | |
| 28 | magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase [EC:1.14.13.81] | alr3300 | Porphyrin and chlorophyll metabolism | 0.52 | 0.0534 * | |
| 29 | petH | all4121 | Ferredoxin-NADP(+) reductase | 0.72 | 0.0187 ** | |
| 30 | petB | alr3421 | cytochrome b6 | 0.625 | 0.0111 ** | |
| 31 | petC | all2453 | cytochrome b6-f complex iron-sulfur subunit | 0.43 | 0.0844 * | |
| 32 | cytA | alr4251 | cytochrome c6 | 1.69 | 0.0202 ** | |
| Oxidative phosphorylation (6 proteins) | ||||||
| 33 | ndhH | alr3355 | NAD(P)H-quinone oxidoreductase subunit H | 0.60 | 0.0358 ** | |
| 34 | F-type H+-transporting ATPase subunit a | all0010 # | ATP synthase F0F1 subunit A | 0.65 | 0.0232 ** | |
| 35 | atpC | all0004 # | F-type H+-transporting ATPase subunit gamma | 0.63 | 0.0075 *** | |
| 36 | atpD | all0006 # | ATP synthase F0F1 subunit delta | 0.70 | 0.0090 *** | |
| 37 | atpF | all0007 # | ATP synthase F0F1 subunit B | 0.61 | 0.0879 * | |
| 38 | F-type H+/Na+-transporting ATPase subunit β [EC:7.1.2.2 7.2.2.1] | all5039 | ATP synthase F0F1 subunit β | 0.76 | 0.0166 ** | |
| Amino acids biosynthesis and metabolism (11 proteins) | ||||||
| 39 | urease subunit α [EC:3.5.1.5] | alr3670 | Arginine biosynthesis Purine metabolism | 0.63 | 0.0059 *** | |
| 40 | acetolactate synthase I/II/III large subunit [EC:2.2.1.6] | all3555 | Valine, leucine and isoleucine biosynthesis | 1.43 | 0.0211 ** | |
| 41 | valine-pyruvate aminotransferase [EC:2.6.1.66] | alr2811 # | Valine, leucine and isoleucine biosynthesis | 1.49 | 0.0644 * | |
| 42 | glyA glycine hydroxymethyltransferase [EC:2.1.2.1] | alr4806 | Glycine, serine and threonine metabolism | 1.69 | 0.0288 ** | |
| 43 | carbamoyl-phosphate synthase large subunit [EC:6.3.5.5] | alr3809 # | Alanine, aspartate and glutamate metabolism Pyrimidine metabolism | 1.28 | 0.0496 ** | |
| 44 | adenylosuccinate synthase [EC:6.3.4.4] | alr4784 | Alanine, aspartate and glutamate metabolism Purine metabolism | 1.64 | 0.0763 * | |
| 45 | succinate-semialdehyde dehydrogenase/glutarate-semialdehyde dehydrogenase EC:1.2.1.16 1.2.1.79 1.2.1.20 | all3556 | Alanine, aspartate and glutamate metabolism Lysine degradation Tyrosine metabolism | 1.67 | 0.0085 *** | |
| 46 | nodM glutamine-fructose-6-phosphate transaminase (isomerizing) [EC:2.6.1.16] | alr3464 | Alanine, aspartate and glutamate metabolism | 1.85 | 0.0013 *** | |
| 47 | aspartate aminotransferase [EC:2.6.1.1] 2-oxoglutarate-glutamate aminotransferase L-aspartate + 2-oxoglutarate = oxaloacetate + L-glutamate | alr4853 # | Alanine, aspartate and glutamate metabolism Arginine biosynthesis Cysteine and methionine metabolism Arginine and proline metabolism Tyrosine metabolism Phenylalanine metabolism Phenylalanine, tyrosine and tryptophan biosynthesis | 1.61 | 0.0016 *** | |
| 48 | S-adenosylmethionine synthetase [EC:2.5.1.6] | alr4124 | Cysteine and methionine metabolism | 1.54 | 0.0064 *** | |
| 49 | threonine synthase [EC:4.2.3.1] | alr3293 # | Glycine, serine and threonine metabolism Vitamin B6 metabolism | 1.56 | 0.0599 * | |
| Chaperones (3 proteins) | ||||||
| 50 | DnaJ | alr2447 | molecular chaperone DnaJ | 1.35 | 0.0856 * | |
| 51 | DnaK | alr1742 | molecular chaperone DnaK | 1.49 | 0.0176 ** | |
| 52 | GroEL | alr1896 | molecular chaperone GroEL | 0.79 | 0.0402 ** | |
| Stress response (11 proteins) | ||||||
| 53 | glutathione S-transferase | alr3798 | glutathione S-transferase | 1.92 | 0.0014 *** | |
| 54 | Glutaredoxin-3 | asl3860 | glutaredoxin | 4.35 | 0.0032 *** | |
| 55 | gor | all4968 | glutathione reductase | 1.30 | 0.0118 ** | |
| 56 | peroxiredoxin 2 family protein/glutaredoxin | all1541 # | peroxiredoxin 2 family protein/glutaredoxin | 1.41 | 0.0918 * | |
| 57 | peroxiredoxin Q/BCP [EC:1.11.1.15] | alr3183 | Acting on a peroxide as acceptor | 1.69 | 0.0011 *** | |
| 58 | peroxiredoxin | alr4641 | peroxiredoxin | 2.27 | 0.0001 *** | |
| 59 | thioredoxin reductase | all0737 | thioredoxin reductase | 2.17 | 0.0241 ** | |
| 60 | peptidylprolyl isomerase [EC:5.2.1.8] | alr0577 | FKBP-type peptidyl-prolyl cis-trans isomerase | 1.52 | 0.0596 * | |
| 61 | starvation-inducible DNA-binding protein | all4145 | probable DNA-binding stress protein | 1.45 | 0.0015 *** | |
| 62 | trxA|thioredoxin | alr0052 | trxA|thioredoxin | 0.23 | 0.0205 ** | |
| 63 | FMN-dependent NADH-azoreductase [EC:1.7.1.17] | all2105 | The biotransformation and/or detoxification of Nitro aromatic compounds can be possible by microbial azoreductase enzyme. Azoreductase enzyme has an ability to reduce the toxic nitro group to the corresponding amino group. | 0.70 | 0.0508 * | |
| SOS-response and DNA repair (4 proteins) | ||||||
| 64 | recA | all3272 | recA|recombinase A | 4.55 | 0.0002 *** | |
| 65 | DNA gyrase subunit A | all0860 | DNA gyrase subunit A | 1.69 | 0.0096 *** | |
| 66 | gyrB|DNA gyrase subunit B | all5265 | gyrB|DNA gyrase subunit B | 2.04 | 0.0001 *** | |
| 67 | single-stranded DNA-binding protein | alr0088 | single-stranded DNA-binding protein | 2.04 | 0.0968 * | |
| Transcription (2 proteins) | ||||||
| 68 | antitermination protein NusA | alr3829 | transcription termination | 1.85 | 0.0022 *** | |
| 69 | DNA-directed RNA polymerase subunit α [EC:2.7.7.6] | all4191 | rpoA; RNA polymerase α subunit | 1.49 | 0.0486 ** | |
| Translation (8 proteins) | ||||||
| 70 | small subunit ribosomal protein S3 | all4209 | rps3; 30S ribosomal protein S3 | 2.56 | 0.0017 *** | |
| 71 | small subunit ribosomal protein S7 | all4339 | 30S ribosomal protein S7 | 0.63 | 0.0109 ** | |
| 72 | large subunit ribosomal protein L19 | alr5297 | rpl19; 50S ribosomal protein L19 | 0.37 | 0.0097 *** | |
| 73 | rbpD | asl4022 | RNA-binding protein | 0.28 | 0.0097 *** | |
| 74 | fus; translation elongation factor EF-G | all4338 | elongation factor G | 1.19 | 0.0830 * | |
| 75 | glyS, glycyl-tRNA synthetase β chain [EC:6.1.1.14] | alr4111 | glycyl-tRNA synthetase β chain | 1.35 | 0.0063 *** | |
| 76 | aspS, aspartyl-tRNA synthetase [EC:6.1.1.12] | all2436 | aspartate-tRNA ligase | 1.85 | 0.0342 ** | |
| 77 | phenylalanyl-tRNA synthetase β chain [EC:6.1.1.20] | alr4958 | phenylalanyl-tRNA synthetase | 0.77 | 0.0431 ** | |
| Purine and Pyrimidine metabolism (1 protein) | ||||||
| 78 | IMP dehydrogenase [EC:1.1.1.205] | alr0051 | Purine metabolism | 1.45 | 0.0421 ** | |
| Carbohydrate metabolism, Glycolysis and gluconeogenesis, Citrate cycle, Pentose phosphate pathway, Starch and sucrose metabolism (14 proteins) | ||||||
| 79 | 6-phosphogluconate dehydrogenase [EC:1.1.1.44 1.1.1.343] | alr5275 # | Pentose phosphate pathway Glutathione metabolism | 0.67 | 0.0001 *** | |
| 80 | transketolase [EC:2.2.1.1] | alr3344 | Pentose phosphate pathway Carbon fixation | 0.68 | 0.0039 *** | |
| 81 | aconitate hydratase 2/2-methylisocitrate dehydratase [EC:4.2.1.3 4.2.1.99] | all1267 | Citrate cycle, first carbon oxidation, oxaloacetate => 2-oxoglutarate Glyoxylate and dicarboxylate metabolism | 0.65 | 0.0478 ** | |
| 82 | phosphoglycerate kinase [EC:2.7.2.3] | all4131 | Glycolysis/Gluconeogenesis Carbon fixation in photosynthetic organisms | 0.67 | 0.0015 *** | |
| 83 | fructose-bisphosphate aldolase, class II [EC:4.1.2.13] | all4563# | Glycolysis/Gluconeogenesis Pentose phosphate pathway | 0.45 | 0.0073 *** | |
| 84 | fructose-1,6-bisphosphatase II/sedoheptulose-1,7-bisphosphatase [EC:3.1.3.11 3.1.3.37] | alr1041 | Glycolysis/Gluconeogenesis Pentose phosphate pathway | 0.55 | 0.0421 ** | |
| 85 | pyruvate dehydrogenase E1 component beta subunit [EC:1.2.4.1] | all0122 | Glycolysis/Gluconeogenesis Citrate cycle Pyruvate metabolism | 1.19 | 0.0442 ** | |
| 86 | glucose-6-phosphate isomerase [EC:5.3.1.9] | alr1050 | Glycolysis/Gluconeogenesis Pentose phosphate pathway Starch and sucrose metabolism | 1.49 | 0.0085 *** | |
| 87 | Phosphoglucomutase / phosphomannomutase | all5089 | Glycogenolysis and glycogenesis | 1.45 | 0.0561 * | |
| 88 | fructose-1,6-bisphosphatase I [EC:3.1.3.11] | all4021 | Glycolysis/Gluconeogenesis Pentose phosphate pathway Fructose and mannose metabolism | Present only in BMAA treated samples | 0.0048 *** | |
| 89 | glycogen phosphorylase [EC:2.4.1.1] | all1272 | Starch and sucrose metabolism | 2.22 | 0.0862 * | |
| 90 | glgB 1,4-alpha-glucan branching enzyme [EC:2.4.1.18] | all0713 | Starch and sucrose metabolism | 1.67 | 0.0096 *** | |
| 91 | UDP-glucose 6-dehydrogenase [EC:1.1.1.22] | alr0658 | Pentose and glucuronate interconversions Ascorbate and aldarate metabolism | 1.43 | 0.0831 * | |
| 92 | rfbB UDP-glucuronate decarboxylase [EC:4.1.1.35] | alr0657 | Amino sugar and nucleotide sugar metabolism, Starch and sucrose metabolism | 2.5 | 0.0844 * | |
| Sulfur metabolism (1 protein) | ||||||
| 93 | phosphoadenosine phosphosulfate reductase [EC:1.8.4.8 1.8.4.10] | all4464 | Sulfur metabolism | 0.56 | 0.0071 *** | |
| Secondary metabolites (4 proteins) | ||||||
| 94 | (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase [EC:1.17.7.1 1.17.7.3] | all2501 | Terpenoid backbone biosynthesis | 0.53 | 0.0003 *** | |
| 95 | 15-cis-phytoene desaturase [EC:1.3.5.5] | alr1832 | Carotenoid biosynthesis | 0.76 | 0.0102 ** | |
| 96 | carboxymethylenebutenolidase [EC:3.1.1.45] | alr1077 | Dienelactone hydrolase | 0.63 | 0.0479 ** | |
| 97 | NADH dehydrogenase demethylphylloquinone reductase [EC:1.6.5.12] | alr4094 | Ubiquinone and other terpenoid-quinone biosynthesis | 3.33 | 0.0073 *** | |
| № | Gene | Possible Function | Up Shifted | Down Shifted | p-Value |
|---|---|---|---|---|---|
| Hypothetical proteins (29 proteins) | |||||
| 1 | alr4642 | putative thiol-specific antioxidant protein | BTS | 0.0628 * | |
| 2 | alr7504 | ubiquitin-like modifier activating enzyme activity | BTS | 0.0111 ** | |
| 3 | all4387 | Membrane protease subunit, stomatin/prohibitin | BTS | 0.0136 ** | |
| 4 | alr4505 # | May be involved in DNA metabolism and recombination | 20 | 0.0014 *** | |
| 5 | alr4504 # | May be involved in DNA metabolism and recombination. | 4.35 | 0.0309 ** | |
| 6 | all1411 # | Unknown | 5.26 | 0.0335 ** | |
| 7 | alr0740 | stomatin-like protein (uncharacterized) | 3.13 | 0.0011 *** | |
| 8 | alr7502 | Uncharacterized protein with ubiquitin-like domains | 3.03 | 0.0162 ** | |
| 9 | all0646 | Thylakoid formation protein Thf1-like protein | 2.56 | 0.0332 ** | |
| 10 | all2705 | Rho termination factor | 2.17 | 0.0003 *** | |
| 11 | all3984 | Conjugal transfer protein TrbI | 2.08 | 0.0041 *** | |
| 12 | all0459 | Uncharacterized low temperature-induced protein | 2.04 | 0.0248 ** | |
| 13 | alr4995 | Saccharop_dh_N domain-containing protein | 2.00 | 0.0014 *** | |
| 14 | alr1143 | Uncharacterized protein | 1.96 | 0.0239 ** | |
| 15 | asl4547 # | Unknown | 1.92 | 0.00005 *** | |
| 16 | all5026 | Short-chain dehydrogenases/ reductases (SDR) | 1.67 | 0.0831 * | |
| 17 | alr2055 | unknown | 1.54 | 0.0889 * | |
| 18 | alr0114 | Tic22-like family protein involved in the preprotein translocation pore in chloroplasts. | 1.49 | 0.1004 * | |
| 19 | all5218 | PmbA; putative modulator of DNA gyrase | 1.49 | 0.0169 ** | |
| 20 | all3797 | Uncharacterized surface protein containing fasciclin (FAS1) repeats | 1.43 | 0.0232 ** | |
| 21 | all4296 | AAA domain-containing protein belongs to diverse group of enzymes that are able to induce structural changes in a wide range of substrate proteins and protein complexes | Control | 0.0343 ** | |
| 22 | all1351 | Contains region “OxoGdeHyase_C” (2-oxoglutarate dehydrogenase C-terminal) | 0.46 | 0.0534 * | |
| 23 | all7598 | Unknown | 0.53 | 0.0587 * | |
| 24 | all3941 | Unknown | 0.64 | 0.0783 * | |
| 25 | alr1850 | Phosphoketolase region | 0.65 | 0.0280 ** | |
| 26 | all3826 # | Peptidoglycan-binding (PGRP) domain of peptidoglycan hydrolases [Cell wall/membrane/envelope biogenesis | 0.67 | 0.0505 * | |
| Pathways and Cell Processes | Nitrogen Starvation (Previous Study [23]) | Nitrogen Replete Growth (Previous Study [24]) | Diazotrophic Growth (Present Study) | |||
|---|---|---|---|---|---|---|
| Upshifted | Downshifted | Upshifted | Downshifted | Upshifted | Downshifted | |
| Heterocyst formation and functionality | all1454 nifD | all1440 nifK all1454 nifD | ||||
| Nitrogen metabolism | alr0608 nrtA | all2319 PII | all2319 PII | |||
| CO2 fixation | alr1524 RbcL | alr1524 RbcL alr1533 RuBisCO Activase | alr1526 RbcS | |||
| CO2 concentrating mechanism | CcmM | CcmK | CmpA | CmpD | ||
| Carbon metabolism | alr5275 6-phosphogluconate dehydrogenase Pentose phosphate pathway, Glutathione metabolism alr4566 NADH-dependent butanol dehydrogenase | alr5275 6-phosphogluconate dehydrogenase [EC:1.1.1.44 1.1.1.343] all4563 fructose-bisphosphate aldolase, class II [EC:4.1.2.13] | ||||
| Photosynthesis | alr4380 EC:4.2.1.24 delta-aminolevulinic acid dehydratase Porphyrin and chlorophyll metabolism | alr3727, alr3742 psbA photosystem II protein D1 alr4548 psbD photosystem II protein D2 | ||||
| Oxidative phospho- rylation | all3570 inorganic pyrophosphatase [EC:3.6.1.1] | all0010 all0004 all0006 all0007 ATP synthase F0F1 subunits | ||||
| Amino acids metabolism | all2521 cysteine synthase [EC:2.5.1.47] | alr2811 valine-pyruvate aminotransferase [EC:2.6.1.66] all4613 ilvG, acetolactate synthase I/II/III large subunit [EC:2.2.1.6] | alr2811 valine- pyruvate aminotransferase [EC:2.6.1.66] alr3809 carbamoyl-phosphate synthase large subunit [EC:6.3.5.5] alr4853 aspartate aminotransferase [EC:2.6.1.1] alr3293 threonine synthase [EC:4.2.3.1] | |||
| Transporters | alr0140 peptide/nickel transport system substrate- binding protein periplasmic oligopeptide- binding ABC transporter Quorum sensing | alr1554 ATP-binding cassette, subfamily B | alr4164 periplasmic amino acid-binding protein of amino acid ABC transporter | alr3938 ABC transporter iron binding protein high-affinity iron ion transport | ||
| Regulatory proteins Signaling | all4662 cyclic-di-GMP-binding protein | all0089 Uncharacterized conserved protein YggE, contains kinase-interacting SIMPL domain all0129 two-component system, OmpR family, response regulator RpaA | ||||
| Stress response | all1541 peroxi- redoxin 2 family protein/ glutaredoxin | |||||
| Transcription | all5263 sigA|RNA polymerase sigma factor RpoD | |||||
| Translation | all4193 small ribosomal protein S13 | |||||
| Secondary metabolites | alr0599 1-deoxy- xylulose 5-phosphate synthase EC:2.2.1.7 | |||||
| Hypothetical proteins | alr4505 all1411 asl4547 alr2889 asr3294 | all4662 | alr4505 all1411 asr1156 | alr4505 alr4504 all1411 asl4547 | all3826 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koksharova, O.A.; Butenko, I.O.; Pobeguts, O.V.; Safronova, N.A.; Govorun, V.M. β-N-Methylamino-L-Alanine (BMAA) Causes Severe Stress in Nostoc sp. PCC 7120 Cells under Diazotrophic Conditions: A Proteomic Study. Toxins 2021, 13, 325. https://doi.org/10.3390/toxins13050325
Koksharova OA, Butenko IO, Pobeguts OV, Safronova NA, Govorun VM. β-N-Methylamino-L-Alanine (BMAA) Causes Severe Stress in Nostoc sp. PCC 7120 Cells under Diazotrophic Conditions: A Proteomic Study. Toxins. 2021; 13(5):325. https://doi.org/10.3390/toxins13050325
Chicago/Turabian StyleKoksharova, Olga A., Ivan O. Butenko, Olga V. Pobeguts, Nina A. Safronova, and Vadim M. Govorun. 2021. "β-N-Methylamino-L-Alanine (BMAA) Causes Severe Stress in Nostoc sp. PCC 7120 Cells under Diazotrophic Conditions: A Proteomic Study" Toxins 13, no. 5: 325. https://doi.org/10.3390/toxins13050325
APA StyleKoksharova, O. A., Butenko, I. O., Pobeguts, O. V., Safronova, N. A., & Govorun, V. M. (2021). β-N-Methylamino-L-Alanine (BMAA) Causes Severe Stress in Nostoc sp. PCC 7120 Cells under Diazotrophic Conditions: A Proteomic Study. Toxins, 13(5), 325. https://doi.org/10.3390/toxins13050325






