Human Leukemia T-Cell Lines as Alternatives to Animal Use for Detecting Biologically Active Staphylococcal Enterotoxin Type B
Abstract
1. Introduction
2. Results
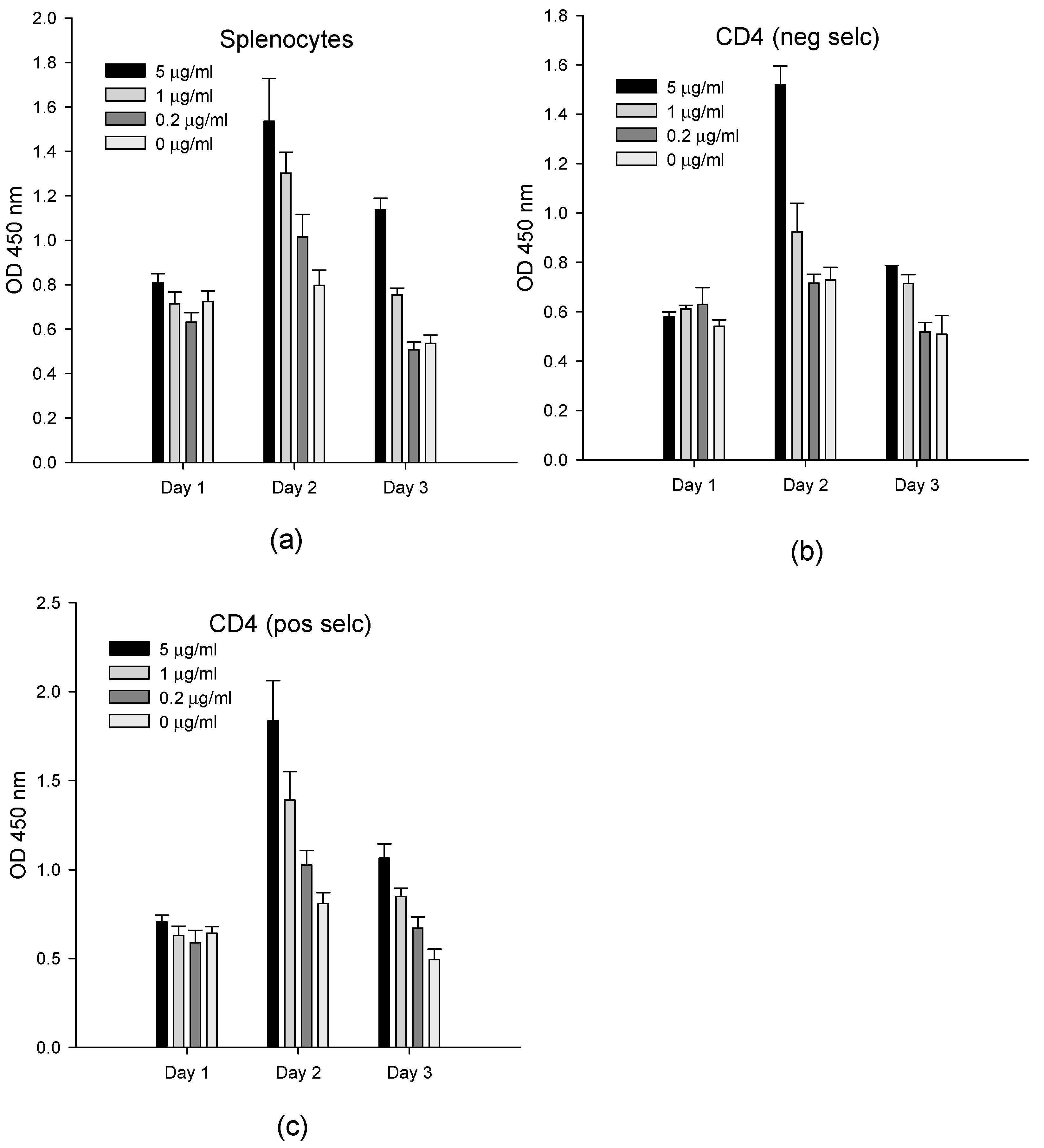
2.1. Ex Vivo Cell-Based Assay
2.2. In Vitro Cell-Based Assay
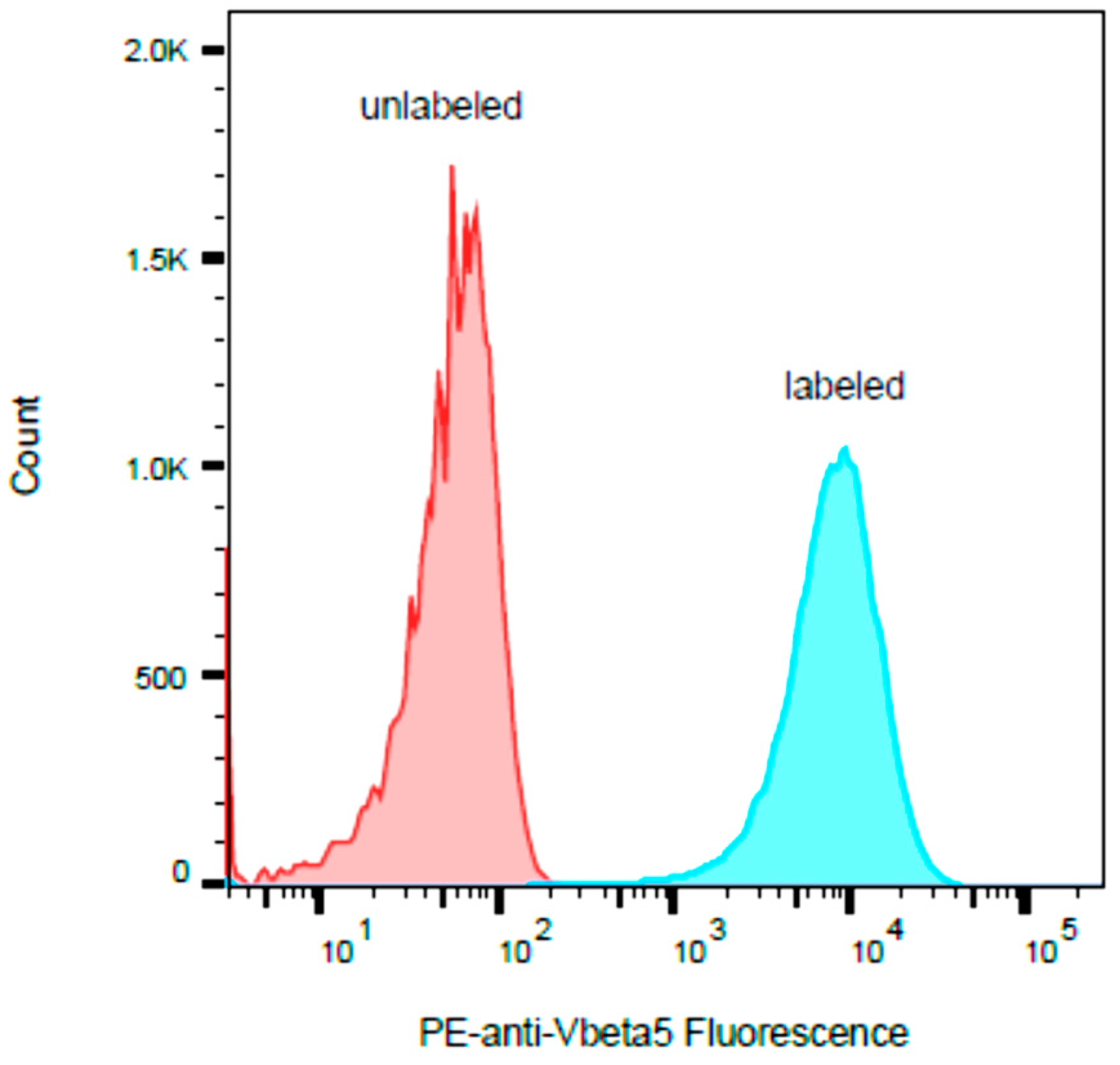
2.3. HPB-ALL Cells Express Vβ5.3
2.4. Limit of Detection
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Media
4.3. Splenocyte Isolation and Human Cell Lines
4.4. Positive or Negative Isolation of Murine CD4+ T-Cells
4.5. Quantitative Determinations of SEs by Cytokine Secretion
4.6. Measurement of Bromodeoxyuridine (BrdU)
4.7. Flow Cytometry
4.8. Statistical Analysis
4.9. Ethics Statement
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Ahanotu, E.; Alvelo-Ceron, D.; Ravita, T.; Gaunt, E. Staphylococcal Enterotoxin B as a Biological Weapon: Recognition, Management, and Surveillance of Staphylococcal Enterotoxin. Appl. Biosaf. 2006, 11, 120–126. [Google Scholar] [CrossRef]
- Pinchuk, I.V.; Beswick, E.J.; Reyes, V.E. Staphylococcal enterotoxins. Toxins 2010, 2, 2177–2197. [Google Scholar] [CrossRef]
- Rasooly, R.; Do, P.; He, X.; Hernlem, B. T cell Receptor Vbeta9 in Method for Rapidly Quantifying Active Staphylococcal Enterotoxin Type-A without Live Animals. Toxins 2019, 11, 399. [Google Scholar] [CrossRef]
- Mourenza, Á.; Gil, J.A.; Mateos, L.M.; Letek, M. Novel Treatments and Preventative Strategies Against Food-Poisoning Caused by Staphylococcal Species. Pathogens 2021, 10, 91. [Google Scholar] [CrossRef]
- Krakauer, T.; Pradhan, K.; Stiles, B.G. Staphylococcal Superantigens Spark Host-Mediated Danger Signals. Front. Immunol. 2016, 7, 23. [Google Scholar] [CrossRef]
- Krakauer, T.; Stiles, B.G. The staphylococcal enterotoxin (SE) family: SEB and siblings. Virulence 2013, 4, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.L.; Omoe, K.; Sashinami, H.; Shinagawa, K.; Nakane, A. Immunization with a nontoxic mutant of staphylococcal enterotoxin A, SEAD227A, protects against enterotoxin-induced emesis in house musk shrews. J. Infect. Dis. 2009, 199, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Hui, J.; Cao, Y.; Xiao, F.; Zhang, J.; Li, H.; Hu, F. Staphylococcus aureus enterotoxin C2 mutants: Biological activity assay in vitro. J. Ind. Microbiol. Biotechnol. 2008, 35, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Atanassova, V.; Meindl, A.; Ring, C. Prevalence of Staphylococcus aureus and staphylococcal enterotoxins in raw pork and uncooked smoked ham--a comparison of classical culturing detection and RFLP-PCR. Int. J. Food Microbiol. 2001, 68, 105–113. [Google Scholar] [CrossRef]
- Zeaki, N.; Johler, S.; Skandamis, P.N.; Schelin, J. The Role of Regulatory Mechanisms and Environmental Parameters in Staphylococcal Food Poisoning and Resulting Challenges to Risk Assessment. Front. Microbiol. 2019, 10, 1307. [Google Scholar] [CrossRef]
- Omoe, K.; Ishikawa, M.; Shimoda, Y.; Hu, D.L.; Ueda, S.; Shinagawa, K. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates Harboring seg, seh, or sei genes. J. Clin. Microbiol. 2002, 40, 857–862. [Google Scholar] [CrossRef]
- Bennett, R.W. Staphylococcal enterotoxin and its rapid identification in foods by enzyme-linked immunosorbent assay-based methodology. J. Food Prot. 2005, 68, 1264–1270. [Google Scholar] [CrossRef]
- Bergdoll, M.S. Monkey feeding test for staphylococcal enterotoxin. Methods Enzymol. 1988, 165, 324–333. [Google Scholar]
- Wu, S.; Duan, N.; Gu, H.; Hao, L.; Ye, H.; Gong, W.; Wang, Z. A Review of the Methods for Detection of Staphylococcus aureus Enterotoxins. Toxins 2016, 8, 176. [Google Scholar] [CrossRef]
- Bergdoll, M.S.; Borja, C.R.; Robbins, R.N.; Weiss, K.F. Identification of enterotoxin E. Infect. Immun. 1971, 4, 593–595. [Google Scholar] [CrossRef]
- Fujikawa, H.; Igarashi, H. Rapid latex agglutination test for detection of staphylococcal enterotoxins A to E that uses high-density latex particles. Appl. Environ. Microbiol. 1988, 54, 2345–2348. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M.; Rasooly, R.; Do, P.M.; Henika, P.R. The olive compound 4-hydroxytyrosol inactivates Staphylococcus aureus bacteria and Staphylococcal Enterotoxin A (SEA). J. Food Sci. 2011, 76, M558–M563. [Google Scholar] [CrossRef] [PubMed]
- Rasooly, R.; Do, P.M.; Friedman, M. Inhibition of biological activity of staphylococcal enterotoxin A (SEA) by apple juice and apple polyphenols. J. Agric. Food Chem. 2010, 58, 5421–5426. [Google Scholar] [CrossRef] [PubMed]
- Rasooly, R.; Do, P.M. In vitro cell-based assay for activity analysis of staphylococcal enterotoxin A in food. FEMS Immunol. Med. Microbiol. 2009, 56, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Rajkovic, A.; Tomasevic, I.; De Meulenaer, B.; Devlieghere, F. The effect of pulsed UV light on Escherichia coli O157:H7, Listeria monocytogenes, Salmonella Typhimurium, Staphylococcus aureus and staphylococcal enterotoxin A on sliced fermented salami and its chemical quality. Food Control 2017, 73, 829–837. [Google Scholar] [CrossRef]
- Rasooly, R.; Do, P.; Hernlem, B.J. Interleukin 2 Secretion by T Cells for Detection of Biologically Active Staphylococcal Enterotoxin Type E. J. Food Prot. 2017, 1857–1862. [Google Scholar] [CrossRef]
- Rasooly, R.; Do, P.; Hernlem, B.J. Low cost bioluminescence imaging as an alternative to in vivo bioassays for quantifying biologically active staphylococcal enterotoxin type E. Sens. Actuators B Chem. 2018, 259, 387–393. [Google Scholar] [CrossRef]
- Rasooly, R.; Hernlem, B.J. Quantitative analysis of staphylococcus enterotoxin A by differential expression of IFN-gamma in splenocyte and CD4+ T-cells. Sensors 2014, 14, 8869–8876. [Google Scholar] [CrossRef]
- Rasooly, R.; Hernlem, B. TNF as biomarker for rapid quantification of active Staphylococcus enterotoxin A in food. Sensors 2012, 12, 5978–5985. [Google Scholar] [CrossRef]
- Rasooly, R.; Hernlem, B.J. CD154 as a potential early molecular biomarker for rapid quantification analysis of active Staphylococcus enterotoxin A. FEMS Immunol. Med. Microbiol. 2012, 64, 169–174. [Google Scholar] [CrossRef]
- Rasooly, R.; Do, P.M.; Hernlem, B.J. Rapid Cell-Based Assay for Detection and Quantification of Active Staphylococcal Enterotoxin Type D. J. Food Sci. 2017, 82, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Rasooly, R.; Do, P.; He, X.; Hernlem, B. TCR-Vβ8 as Alternative to Animal Testing for Quantifying Active SEE. J. Environ. Anal. Toxicol. 2017, 7. [Google Scholar] [CrossRef]
- Kumar, S.; Menoret, A.; Ngoi, S.M.; Vella, A.T. The systemic and pulmonary immune response to staphylococcal enterotoxins. Toxins 2010, 2, 1898–1912. [Google Scholar] [CrossRef]
- Kuhn, R.; Lohler, J.; Rennick, D.; Rajewsky, K.; Muller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 1993, 75, 263–274. [Google Scholar] [CrossRef]
- Rasooly, R.; Do, P.; He, X.; Hernlem, B. Alternative to Animal Use for Detecting Biologically Active Staphylococcal Enterotoxin Type A. Toxins 2018, 10, 540. [Google Scholar] [CrossRef] [PubMed]
- Rasooly, R.; Do, P.M.; Hernlem, B.J. Auto-presentation of Staphylococcal enterotoxin A by mouse CD4+ T cells. Open J. Immunol. 2011, 1, 8–14. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rasooly, R.; Do, P.; He, X.; Hernlem, B. Human Leukemia T-Cell Lines as Alternatives to Animal Use for Detecting Biologically Active Staphylococcal Enterotoxin Type B. Toxins 2021, 13, 300. https://doi.org/10.3390/toxins13050300
Rasooly R, Do P, He X, Hernlem B. Human Leukemia T-Cell Lines as Alternatives to Animal Use for Detecting Biologically Active Staphylococcal Enterotoxin Type B. Toxins. 2021; 13(5):300. https://doi.org/10.3390/toxins13050300
Chicago/Turabian StyleRasooly, Reuven, Paula Do, Xiaohua He, and Bradley Hernlem. 2021. "Human Leukemia T-Cell Lines as Alternatives to Animal Use for Detecting Biologically Active Staphylococcal Enterotoxin Type B" Toxins 13, no. 5: 300. https://doi.org/10.3390/toxins13050300
APA StyleRasooly, R., Do, P., He, X., & Hernlem, B. (2021). Human Leukemia T-Cell Lines as Alternatives to Animal Use for Detecting Biologically Active Staphylococcal Enterotoxin Type B. Toxins, 13(5), 300. https://doi.org/10.3390/toxins13050300





