A Transcriptomic Approach to the Recruitment of Venom Proteins in a Marine Annelid
Abstract
1. Introduction
2. Results and Discussion
2.1. Discovery of Putative Toxins
2.2. Conserved Domains in Toxin-Like Proteins Reveal the Common Signature of Venoms and Poisons
2.3. Cysteine-Rich Neurotoxins Are a Major Component of Eulalia’s Toxungen
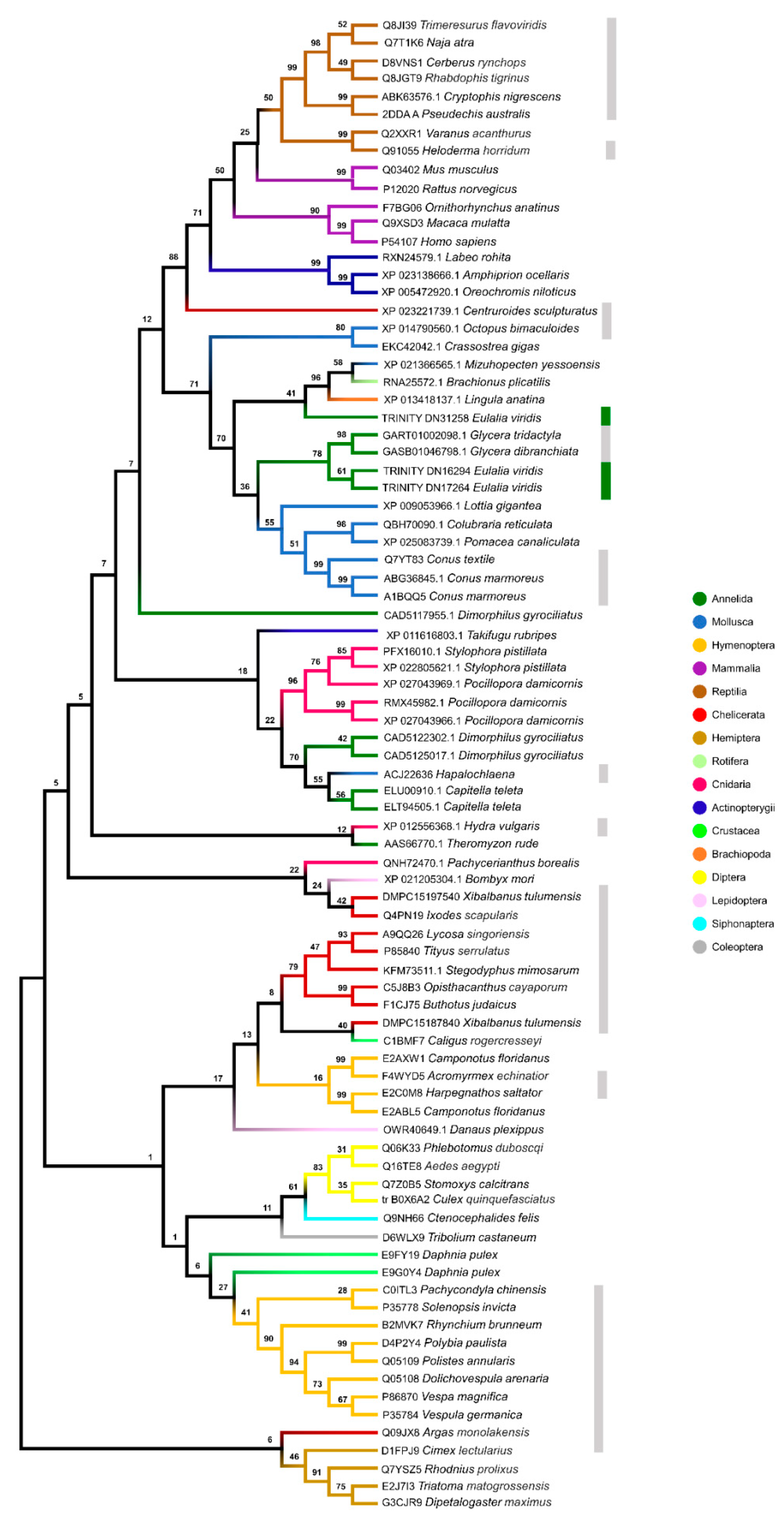
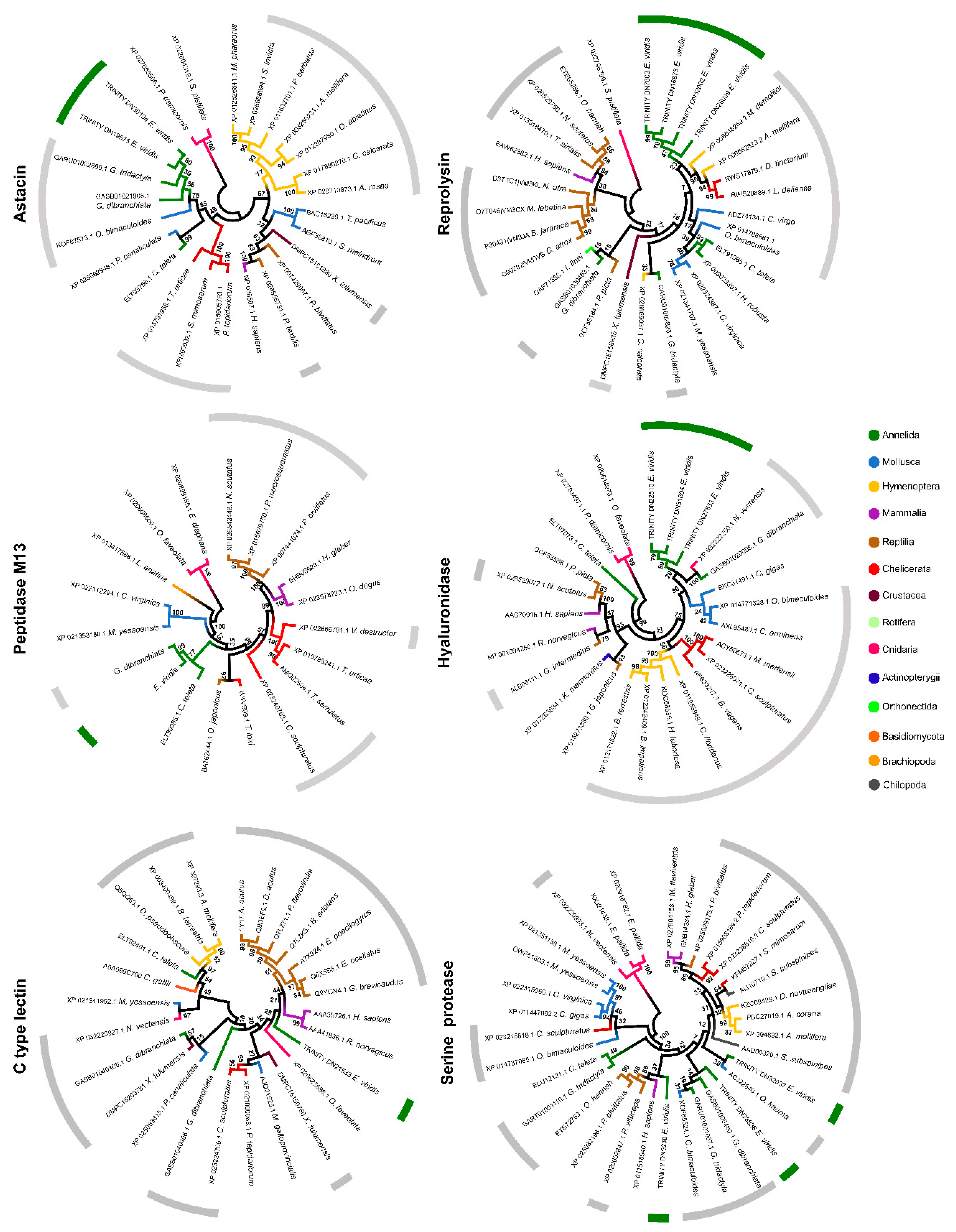
2.4. Phylogenetic Analysis of Individual Components
2.5. Multigene Phylogeny
3. Conclusions
4. Materials and Methods
4.1. Animal Collection
4.2. RNA Extraction and High-Throughput Sequencing (RNA-seq)
4.3. Transcriptome Data Analysis
4.4. Quality Assessment and Validation
4.5. Multigene Phylogenetics
4.6. Microscopy
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Von Reumont, B.M.; Blanke, A.; Richter, S.; Alvarez, F.; Bleidorn, C.; Jenner, R.A. The first venomous crustacean revealed by transcriptomics and functional morphology: Remipede venom glands express a unique toxin cocktail dominated by enzymes and a neurotoxin. Mol. Biol. Evol. 2014, 31, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.M.; Campbell, L.I.; Richter, S.; Hering, L.; Sykes, D.; Hetmank, J.; Jenner, R.A.; Bleidorn, C. A polychaete’s powerful punch: Venom gland transcriptomics of Glycera reveals a complex cocktail of toxin homologs. Genome Biol. Evol. 2014, 6, 2406–2423. [Google Scholar] [CrossRef] [PubMed]
- Schendel, V.; Rash, L.D.; Jenner, R.A.; Undheim, E.A.B. The diversity of venom: The importance of behavior and venom system morphology in understanding its ecology and evolution. Toxins 2019, 11, 666. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Day, M.; Heavner, J.E. Ziconotide: An update and review. Expert Opin. Pharmacother. 2008, 9, 1575–1583. [Google Scholar] [CrossRef]
- Francischetti, I.M.B.; Assumpção, T.C.F.; Ma, D.; Li, Y.; Vicente, E.C.; Uieda, W.; Ribeiro, J.M.C. The “Vampirome”: Transcriptome and proteome analysis of the principal and accessory submaxillary glands of the vampire bat Desmodus rotundus, a vector of human rabies. J. Proteom. 2013, 82, 288–319. [Google Scholar] [CrossRef]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.R.; Kerkkamp, H.M.E.; Vos, R.A.; Guerreiro, I.; Calvete, J.J.; et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 2013, 110, 20651–20656. [Google Scholar] [CrossRef]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.A.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Jackson, T.N.W.; Koludarov, I. How the Toxin got its Toxicity. Front. Pharmacol. 2020, 11, 574925. [Google Scholar] [CrossRef]
- Khalturin, K.; Hemmrich, G.; Fraune, S.; Augustin, R.; Bosch, T.C.G. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet. 2009, 25, 404–413. [Google Scholar] [CrossRef]
- Chang, D.; Duda, T.F. Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol. Biol. Evol. 2012, 29, 2019–2029. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, D.; King, G.F. The venom optimization hypothesis revisited. Toxicon 2013, 63, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M.; Doley, R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Verdes, A.; Simpson, D.; Holford, M. Are fireworms venomous? Evidence for the convergent evolution of toxin homologs in three species of fireworms (Annelida, Amphinomidae). Genome Biol. Evol. 2018, 10, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, N.; Martins, M.; Rodrigo, A.P.; Martins, C.; Costa, P.M. Explorations on the ecological role of toxin secretion and delivery in jawless predatory Polychaeta. Sci. Rep. 2018, 8, 7635. [Google Scholar] [CrossRef]
- Rodrigo, A.P.; Costa, P.M. The hidden biotechnological potential of marine invertebrates: The Polychaeta case study. Environ. Res. 2019, 173, 270–280. [Google Scholar] [CrossRef]
- Whitelaw, B.L.; Strugnell, J.M.; Faou, P.; Da Fonseca, R.R.; Hall, N.E.; Norman, M.; Finn, J.; Cooke, I.R. Combined transcriptomic and proteomic analysis of the posterior salivary gland from the southern blue-ringed octopus and the southern sand octopus. J. Proteome Res. 2016, 15, 3284–3297. [Google Scholar] [CrossRef]
- Nelsen, D.R.; Nisani, Z.; Cooper, A.M.; Fox, G.A.; Gren, E.C.K.; Corbit, A.G.; Hayes, W.K. Poisons, toxungens, and venoms: Redefining and classifying toxic biological secretions and the organisms that employ them. Biol. Rev. 2014, 89, 450–465. [Google Scholar] [CrossRef]
- Rodrigo, A.P.; Martins, C.; Costa, M.H.; Alves de Matos, A.P.; Costa, P.M. A morphoanatomical approach to the adaptive features of the epidermis and proboscis of a marine Polychaeta: Eulalia viridis (Phyllodocida: Phyllodocidae). J. Anat. 2018, 233, 567–579. [Google Scholar] [CrossRef]
- Michel, C. Enzymes digestives intestinales d’ Eulalia viridis (Muller) (Phyllodocidae) et de Glycera convoluta (Keferstein) (Glyceridae) Annelides Polychetes Errantes. Ann. Histochim. 1968, 13, 67–78. [Google Scholar]
- Michel, C. Rôle physiologique de la trompe chez quatre annélides polychètes appartenant aux genres: Eulalia, Phyllodoce, Glycera et Notomastus. Cahiers Biol. Mar. 1970, 11, 209–228. [Google Scholar]
- Michel, C. Ultrastructure et histochimie de la cuticule pharyngienne chez Eulalia viridis Müller (Annélide Polychète Errant, Phyllodocidae). Z. Zellforsch. Mikrosk. Anat. 1969, 98, 54–73. [Google Scholar] [CrossRef] [PubMed]
- Bartolomaeus, T.; Purschke, G.; Hausen, H. Polychaete phylogeny based on morphological data—A comparison of current attempts. In Morphology, Molecules, Evolution and Phylogeny in Polychaeta and Related Taxa; Bartolomaeus, T., Purschke, G., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 341–356. [Google Scholar]
- Struck, T.H.; Paul, C.; Hill, N.; Hartmann, S.; Hösel, C.; Kube, M.; Lieb, B.; Meyer, A.; Tiedemann, R.; Purschke, G.; et al. Phylogenomic analyses unravel annelid evolution. Nature 2011, 471, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Weigert, A.; Bleidorn, C. Current status of annelid phylogeny. Org. Divers. Evol. 2016, 16, 345–362. [Google Scholar] [CrossRef]
- Fauchald, K.; Jumars, P. The diet of worms: A study of polychaete feeding guilds. Oceanogr. Mar. Biol. Annu. Rev. 1979, 17, 193–284. [Google Scholar]
- McHugh, D. Molecular phylogeny of the Annelida. Can. J. Zool. 2000, 78, 1873–1884. [Google Scholar] [CrossRef]
- Ruder, T.; Sunagar, K.; Undheim, E.A.B.; Ali, S.A.; Wai, T.C.; Low, D.H.W.; Jackson, T.N.W.; King, G.F.; Antunes, A.; Fry, B.G. Molecular phylogeny and evolution of the proteins encoded by coleoid (cuttlefish, octopus, and squid) posterior venom glands. J. Mol. Evol. 2013, 76, 192–204. [Google Scholar] [CrossRef]
- Modica, M.V.; Lombardo, F.; Franchini, P.; Oliverio, M. The venomous cocktail of the vampire snail Colubraria reticulata (Mollusca, Gastropoda). BMC Genom. 2015, 16. [Google Scholar] [CrossRef]
- Appella, E.; Weber, I.T.; Blasi, F. Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett. 1988, 231, 1–4. [Google Scholar] [CrossRef]
- Bordon, K.C.F.; Wiezel, G.A.; Amorim, F.G.; Arantes, E.C. Arthropod venom Hyaluronidases: Biochemical properties and potential applications in medicine and biotechnology. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 1–12. [Google Scholar] [CrossRef]
- Winningham, K.M.; Fitch, C.D.; Schmidt, M.; Hoffman, D.R. Hymenoptera venom protease allergens. J. Allergy Clin. Immunol. 2004, 114, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Bork, P.; Beckmann, G. The CUB Domain. A widespread module in developmentally regulated proteins. J. Mol. Biol. 1993, 231, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Font, B.; Eichenberger, D.; Moreau, C.; Ricard-Blum, S.; Hulmes, D.J.S.; Moali, C. Insights into how CUB domains can exert specific functions while sharing a common fold: Conserved and specific features of the CUB1 domain contribute to the molecular basis of procollagen C-proteinase enhancer-1 activity. J. Biol. Chem. 2007, 282, 16924–16933. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, Y.; Morita, T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon 2004, 44, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Scheib, H.; Van der Weerd, L.; Young, B.; McNaughtan, J.; Ramjan, S.F.R.; Vidal, N.; Poelmann, R.E.; Norman, J.A. Evolution of an Arsenal. Mol. Cell. Proteom. 2008, 7, 215–246. [Google Scholar] [CrossRef]
- Kemparaju, K.; Girish, K.S. Snake venom hyaluronidase: A therapeutic target. Cell Biochem. Funct. 2006, 24, 7–12. [Google Scholar] [CrossRef]
- De Oliveira, U.C.; Nishiyama, M.Y.; Dos Santos, M.B.V.; Santos-Da-Silva, A.D.P.; Chalkidis, H.D.M.; Souza-Imberg, A.; Candido, D.M.; Yamanouye, N.; Dorce, V.A.C.; Junqueira-de-Azevedo, I.D.L.M. Proteomic endorsed transcriptomic profiles of venom glands from Tityus obscurus and T. serrulatus scorpions. PLoS ONE 2018, 13, e0193739. [Google Scholar] [CrossRef]
- Smith, A.I.; Rajapakse, N.W.; Kleifeld, O.; Lomonte, B.; Sikanyika, N.L.; Spicer, A.J.; Hodgson, W.C.; Conroy, P.J.; Small, D.H.; Kaye, D.M.; et al. N-terminal domain of Bothrops asper myotoxin II enhances the activity of endothelin converting enzyme-1 and neprilysin. Sci. Rep. 2016, 6, 22413. [Google Scholar] [CrossRef]
- Bordon, K.C.F.; Perino, M.G.; Giglio, J.R.; Arantes, E.C. Isolation, enzymatic characterization and antiedematogenic activity of the first reported rattlesnake hyaluronidase from Crotalus durissus terrificus venom. Biochimie 2012, 94, 2740–2748. [Google Scholar] [CrossRef]
- Bookbinder, L.H.; Hofer, A.; Haller, M.F.; Zepeda, M.L.; Keller, G.A.; Lim, J.E.; Edgington, T.S.; Shepard, H.M.; Patton, J.S.; Frost, G.I. A recombinant human enzyme for enhanced interstitial transport of therapeutics. J. Control. Release 2006, 114, 230–241. [Google Scholar] [CrossRef]
- Fry, B.G.; Wüster, W.; Kini, R.M.; Brusic, V.; Khan, A.; Venkataraman, D.; Rooney, A.P. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 2003, 57, 110–129. [Google Scholar] [CrossRef] [PubMed]
- Sterchi, E.E.; Stöcker, W.; Bond, J.S. Meprins, membrane-bound and secreted astacin metalloproteinases. Mol. Aspects Med. 2009, 29, 309–328. [Google Scholar] [CrossRef] [PubMed]
- Honma, T.; Nagai, H.; Nagashima, Y.; Shiomi, K. Molecular cloning of an epidermal growth factor-like toxin and two sodium channel toxins from the sea anemone Stichodactyla gigantea. Biochim. Biophys. Acta Proteins Proteom. 2003, 1652, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, X.; Shen, D.; Song, J.; Guo, M.; Yan, X. Anticoagulation factor I, a snaclec (snake C-type lectin) from Agkistrodon acutus venom binds to FIX as well as FX: Ca2+ induced binding data. Toxicon 2012, 59, 718–723. [Google Scholar] [CrossRef]
- Nikai, T.; Kato, S.; Komori, Y.; Sugihara, H. Amino acid sequence and biological properties of the lectin from the venom of Trimeresurus okinavensis (Himehabu). Toxicon 2000, 38, 707–711. [Google Scholar] [CrossRef]
- Earl, S.T.H.; Robson, J.; Trabi, M.; De Jersey, J.; Masci, P.P.; Lavin, M.F. Characterisation of a mannose-binding C-type lectin from Oxyuranus scutellatus snake venom. Biochimie 2011, 93, 519–527. [Google Scholar] [CrossRef]
- Havt, A.; Toyama, M.H.; Do Nascimento, N.R.F.; Toyama, D.O.; Nobre, A.C.L.; Martins, A.M.C.; Barbosa, P.S.F.; Novello, J.C.; Boschero, A.C.; Carneiro, E.M.; et al. A new C-type animal lectin isolated from Bothrops pirajai is responsible for the snake venom major effects in the isolated kidney. Int. J. Biochem. Cell Biol. 2005, 37, 130–141. [Google Scholar] [CrossRef]
- Decote-Ricardo, D.; LaRocque-de-Freitas, I.F.; Rocha, J.D.B.; Nascimento, D.O.; Nunes, M.P.; Morrot, A.; Freire-de-Lima, L.; Previato, J.O.; Mendonça-Previato, L.; Freire-de-Lima, C.G. Immunomodulatory role of capsular polysaccharides constituents of Cryptococcus neoformans. Front. Med. 2019, 6, 129. [Google Scholar] [CrossRef]
- Planques, A.; Malem, J.; Parapar, J.; Vervoort, M.; Gazave, E. Morphological, cellular and molecular characterization of posterior regeneration in the marine annelid Platynereis dumerilii. Dev. Biol. 2019, 445, 189–210. [Google Scholar] [CrossRef]
- Sunagar, K.; Moran, Y. The rise and fall of an evolutionary innovation: Contrasting strategies of venom evolutin in ancient and young animals. PLoS Genet. 2015, 11, e1005596. [Google Scholar] [CrossRef]
- Peterson, K.J.; Eernisse, D.J. Animal phylogeny and the ancestry of bilaterians: Inferences from morphology and 18S rDNA gene sequences. Evol. Dev. 2001, 3, 170–205. [Google Scholar] [CrossRef] [PubMed]
- Osipov, A.V.; Levashov, M.Y.; Tsetlin, V.I.; Utkin, Y.N. Cobra venom contains a pool of cysteine-rich secretory proteins. Biochem. Biophys. Res. Commun. 2005, 328, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.S.W.; Belov, K. Venom evolution through gene duplications. Gene 2012, 496, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Struck, T.H.; Schult, N.; Kusen, T.; Hickman, E.; Bleidorn, C.; McHugh, D.; Halanych, K.M. Annelid phylogeny and the status of Sipuncula and Echiura. BMC Evol. Biol. 2007, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Simakov, O.; Marletaz, F.; Cho, S.J.; Edsinger-Gonzales, E.; Havlak, P.; Hellsten, U.; Kuo, D.H.; Larsson, T.; Lv, J.; Arendt, D.; et al. Insights into bilaterian evolution from three spiralian genomes. Nature 2013, 493, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Hedtke, S.M.; Townsend, T.M.; Hillis, D.M. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst. Biol. 2006, 55, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Morgenstern, D.; Reitzel, A.M.; Moran, Y. Ecological venomics: How genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J. Proteom. 2016, 135, 62–72. [Google Scholar] [CrossRef]
- Rodrigo, A.P.; Costa, M.H.; Alves De Matos, A.; Carrapiço, F.; Costa, P.M. A study on the digestive physiology of a marine polychaete (Eulalia viridis) through microanatomical changes of epithelia during the digestive cycle. Microsc. Microanal. 2015, 21, 91–101. [Google Scholar] [CrossRef]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 57. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Suzek, B.E.; Wang, Y.; Huang, H.; McGarvey, P.B.; Wu, C.H. UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef] [PubMed]
- States, D.J.; Gish, W. Combined use of sequence similarity and codon bias for coding region identification. J. Comput. Biol. 1994, 1, 39–50. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 138–141. [Google Scholar] [CrossRef]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009, 205–211. [Google Scholar] [CrossRef]
- Rodrigo, A.P.; Mendes, V.M.; Manadas, B.; Grosso, A.R.; Baptista, P.V.; Costa, P.M.; Fernandes, A.R.; Biology, C.; Caparica, M. De Specific antiproliferative properties of proteinaceous toxin secretions from the marine annelid Eulalia sp. onto ovarian cancer cells. Mar. Drugs 2021, 19, 31. [Google Scholar] [CrossRef]
- Thiel, D.; Hugenschutt, M.; Meyer, H.; Paululat, A.; Quijada-Rodriguez, A.R.; Purschke, G.; Weihrauch, D. Ammonia excretion in the marine polychaete Eurythoe complanata (Annelida). J. Exp. Biol. 2017, 220, 425–436. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Inoué, T.; Osatake, H. A new drying method of biological specimens for scanning method microscopy: The t-butyl alcohol freeze-drying method. Arch. Histol. Cytol. 1988, 51, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, C.; Costa, P.M. Histochemical detection of free sthiols in glandular cells and tissues of different marine Polychaeta. Histochem. Cell Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigo, A.P.; Grosso, A.R.; Baptista, P.V.; Fernandes, A.R.; Costa, P.M. A Transcriptomic Approach to the Recruitment of Venom Proteins in a Marine Annelid. Toxins 2021, 13, 97. https://doi.org/10.3390/toxins13020097
Rodrigo AP, Grosso AR, Baptista PV, Fernandes AR, Costa PM. A Transcriptomic Approach to the Recruitment of Venom Proteins in a Marine Annelid. Toxins. 2021; 13(2):97. https://doi.org/10.3390/toxins13020097
Chicago/Turabian StyleRodrigo, Ana P., Ana R. Grosso, Pedro V. Baptista, Alexandra R. Fernandes, and Pedro M. Costa. 2021. "A Transcriptomic Approach to the Recruitment of Venom Proteins in a Marine Annelid" Toxins 13, no. 2: 97. https://doi.org/10.3390/toxins13020097
APA StyleRodrigo, A. P., Grosso, A. R., Baptista, P. V., Fernandes, A. R., & Costa, P. M. (2021). A Transcriptomic Approach to the Recruitment of Venom Proteins in a Marine Annelid. Toxins, 13(2), 97. https://doi.org/10.3390/toxins13020097






