Reduced Membrane-Bound Alkaline Phosphatase Does Not Affect Binding of Vip3Aa in a Heliothis virescens Resistant Colony
Abstract
1. Introduction
2. Results
2.1. Vip3Aa Binding to Midgut Brush Border Membrane Vesicles (BBMV)
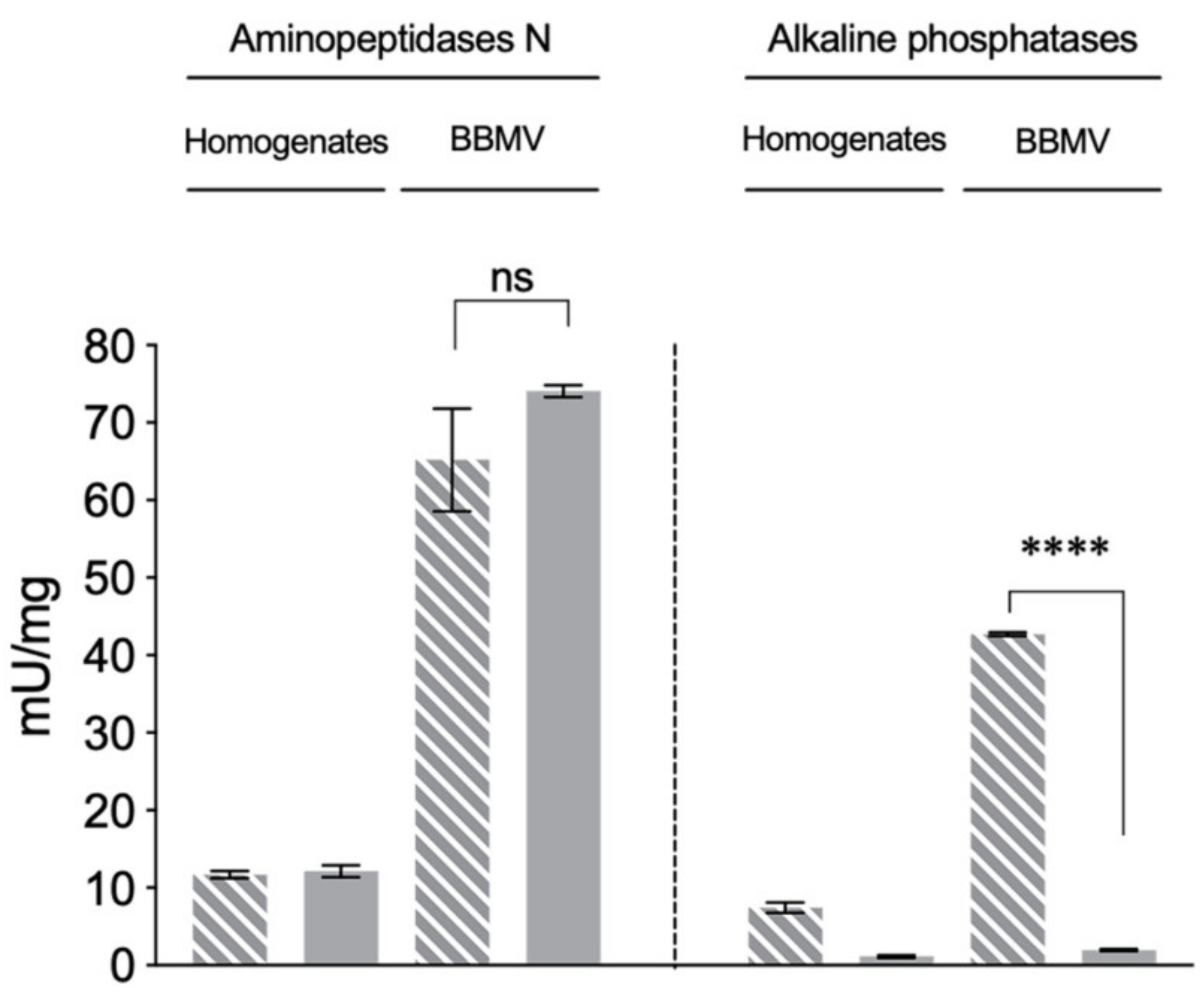
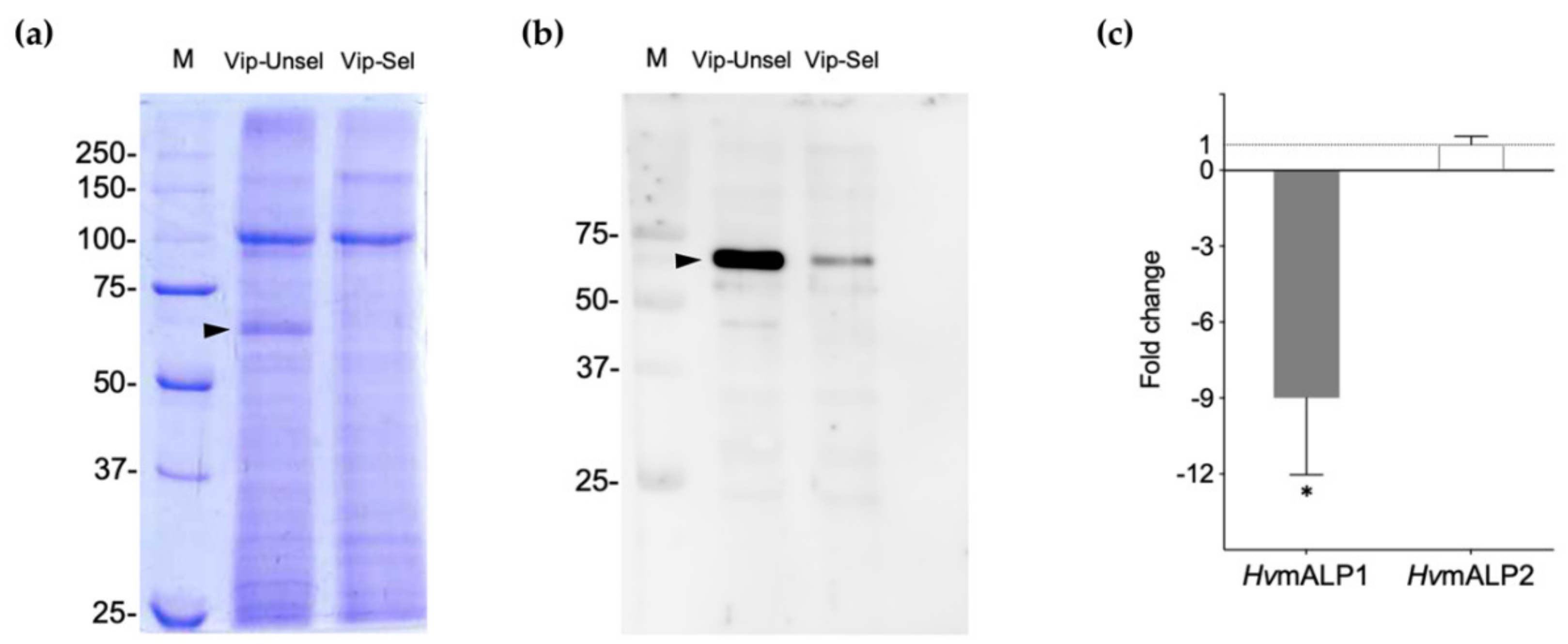
2.2. Reduced ALP Levels in the Vip3Aa-Resistant H. virescens Colony
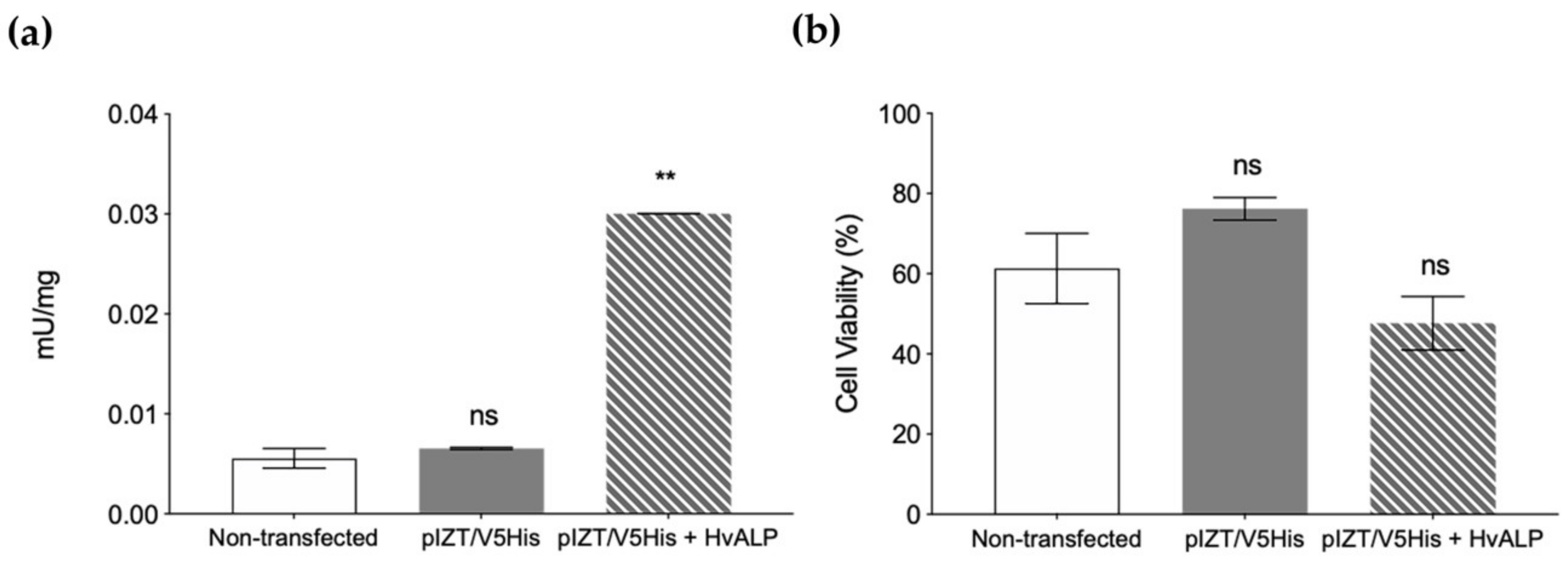
2.3. Functional Role of HvmALP1 in Vip3Aa Binding
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Insects
5.2. BBMV Preparation and Enzyme Activity Assays
5.3. Vip3Aa Protein Expression and Purification
5.4. Vip3Aa Labeling and Binding Experiments
5.5. Western and Ligand Blotting
5.6. Proteomic Analysis
5.7. RT-qPCR
5.8. Expression Vector Construction
5.9. Transient Expression of HvmALP1 in Sf21 Cells
5.10. Cell Viability Assays
Author Contributions
Funding
Conflicts of Interest
References
- Wolfenbarger, D.A.; Bodegas, P.R.; Flores, R. Development of resistance in Heliothis spp. in the Americas, Australia, Africa, and Asia. Bull. Entomol. Soc. Am. 1981, 27, 181–185. [Google Scholar] [CrossRef]
- Terán-Vargas, A.P. Insecticide resistance of tobacco budworm in Southern Tamaulipas Mexico. In Proceedings of the Beltwide Cotton Conference, Nashville, TN, USA, 9–12 January 1996; National Cotton Council of America: Memphis, TN, USA; pp. 784–786. [Google Scholar]
- Tabashnik, B.E.; Carrière, Y. Global patterns of resistance to Bt crops highlighting Pink Bollworm in the United States, China, and India. J. Econ. Entomol. 2019, 112, 2513–2523. [Google Scholar] [CrossRef]
- Roush, R.T. Bt-transgenic crops: Just another pretty insecticide or a chance for a new start in resistance management? Pestic. Sci. 1997, 51, 328–334. [Google Scholar] [CrossRef]
- Moar, W.J.; Anilkumar, K.J. Plant science: The power of the pyramid. Science 2007, 318, 1561–1562. [Google Scholar] [CrossRef]
- Carrière, Y.; Crickmore, N.; Tabashnik, B.E. Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat. Biotechnol. 2015, 33, 161–168. [Google Scholar] [CrossRef]
- Adang, M.; Crickmore, N.; Jurat-Fuentes, J.L. Diversity of Bacillus thuringiensis crystal toxins and mechanism of action. In Advances in Insect Physiology; Dhadialla, T.S., Gill, S., Eds.; Academic Press: San Diego, CA, USA, 2014; Volume 47, pp. 39–87. [Google Scholar] [CrossRef]
- Lee, M.K.; Miles, P.; Chen, J. Brush border membrane binding properties of Bacillus thuringiensis Vip3A toxin to Heliothis virescens and Helicoverpa zea midguts. Biochem. Biophys. Res. Commun. 2006, 339, 1043–1047. [Google Scholar] [CrossRef]
- Sena, J.A.D.; Hernández-Rodríguez, C.S.; Ferré, J. Interaction of Bacillus thuringiensis Cry1 and Vip3A proteins with Spodoptera frugiperda midgut binding sites. Appl. Environ. Microbiol. 2009, 75, 2236–2237. [Google Scholar] [CrossRef]
- Gouffon, C.; Van Vliet, A.; Van Rie, J.; Jansens, S.; Jurat-Fuentes, J.L. Binding sites for Bacillus thuringiensis Cry2Ae toxin on heliothine brush border membrane vesicles are not shared with Cry1A, Cry1F, or Vip3A toxin. Appl. Environ. Microbiol. 2011, 77, 3182–3188. [Google Scholar] [CrossRef]
- Chakroun, M.; Ferré, J. In vivo and in vitro binding of Vip3Aa to Spodoptera frugiperda midgut and characterization of binding sites by 125I radiolabeling. Appl. Environ. Microbiol. 2014, 80, 6258–6265. [Google Scholar] [CrossRef]
- Pickett, B. Studies on Resistance to Vegetative (Vip3A) and Crystal (Cry1A) Insecticide Toxin of Bacillus thuringiensis in Heliothis virescens (Fabricius). Ph.D. Thesis, Imperial College London, London, UK, 2009. [Google Scholar]
- Barkhade, U.P.; Thakare, A.S. Protease mediated resistance mechanism to Cry1C and Vip3A in Spodoptera litura. Egypt Acad. J. Biol. Sci. 2010, 3, 43–50. [Google Scholar] [CrossRef]
- Mahon, R.J.; Downes, S.J.; James, B. Vip3A resistance alleles exist at high levels in Australian targets before release of cotton expressing this toxin. PLoS ONE 2012, 7, e39192. [Google Scholar] [CrossRef]
- Bernardi, O.; Bernardi, D.; Horikoshi, R.J.; Okuma, D.M.; Miraldo, L.L.; Fatoretto, J.; Medeiros, F.C.; Burd, T.; Omoto, C. Selection and characterization of resistance to the Vip3Aa20 protein from Bacillus thuringiensis in Spodoptera frugiperda. Pest. Manag. Sci. 2016, 72, 1794–1802. [Google Scholar] [CrossRef]
- Yang, F.; Morsello, S.; Head, G.P.; Sansone, C.; Huang, F.; Gilreath, R.T.; Kerns, D.L. F2 screen, inheritance and cross-resistance of field-derived Vip3A resistance in Spodoptera frugiperda (Lepidoptera: Noctuidae) collected from Louisiana, USA. Pest. Manag. Sci. 2018, 74, 1769–1778. [Google Scholar] [CrossRef]
- Yang, F.; Santiago-González, J.C.; Little, N.; Reisig, D.; Payne, G.; Ferreira Dos Santos, R.; Jurat-Fuentes, J.L.; Kurtz, R.; Kerns, D.L. First documentation of major Vip3Aa resistance alleles in field populations of Helicoverpa zea (Boddie) (Lepidoptera: Noctuidae) in Texas, USA. Sci. Rep. 2020, 10, 5867. [Google Scholar] [CrossRef]
- Chakroun, M.; Banyuls, N.; Walsh, T.; Downes, S.; James, B.; Ferré, J. Characterization of the resistance to Vip3Aa in Helicoverpa armigera from Australia and the role of midgut processing and receptor binding. Sci. Rep. 2016, 6, 24311. [Google Scholar] [CrossRef]
- Pickett, B.R.; Gulzar, A.; Ferré, J.; Wright, D.J. Bacillus thuringiensis Vip3Aa toxin resistance in Heliothis virescens (Lepidoptera: Noctuidae). Appl. Environ. Microbiol. 2017, 83, e03506–e03516. [Google Scholar] [CrossRef]
- Ayra-Pardo, C.; Ochagavía, M.E.; Raymond, B.; Gulzar, A.; Rodríguez-Cabrera, L.; Rodríguez de la Nova, C.; Morán-Bertot, I.; Terauchi, R.; Yoshida, K.; Matsumura, H.; et al. HT-SuperSAGE of the gut tissue of a Vip3Aa-resistant Heliothis virescens (Lepidoptera: Noctuidae) strain provides insights into the basis of resistance. Insect Sci. 2019, 26, 479–498. [Google Scholar] [CrossRef]
- Caccia, S.; Moar, W.J.; Chandrashekhar, J.; Oppert, C.; Anilkumar, K.J.; Jurat-Fuentes, J.L.; Ferré, J. Association of Cry1Ac toxin resistance in Helicoverpa zea (Boddie) with increased alkaline phosphatase levels in the midgut lumen. Appl. Environ. Microbiol. 2012, 78, 5690–5698. [Google Scholar] [CrossRef]
- Guo, Z.; Kang, S.; Chen, D.; Wu, Q.; Wang, S.; Xie, W.; Zhu, X.; Baxter, S.W.; Zhou, X.; Jurat-Fuentes, J.L.; et al. MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS Genet. 2015, 11, e1005124. [Google Scholar] [CrossRef]
- Jakka, S.R.K.; Gong, L.; Hasler, J.; Banerjee, R.; Sheets, J.J.; Narva, K.; Blanco, C.A.; Jurat-Fuentes, J.L. Field-evolved Mode 1 resistance of the fall armyworm to transgenic Cry1Fa-expressing corn associated with reduced Cry1Fa toxin binding and midgut alkaline phosphatase expression. Appl. Environ. Microbiol. 2016, 82, 1023–1034. [Google Scholar] [CrossRef]
- Jurat-Fuentes, J.L.; Karumbaiah, L.; Jakka, S.R.K.; Ning, C.; Liu, C.; Wu, K.; Jackson, J.; Gould, F.; Blanco, C.A.; Portilla, M.; et al. Reduced levels of membrane-bound alkaline phosphatase are common to lepidopteran strains resistant to Cry toxins from Bacillus thuringiensis. PLoS ONE 2011, 6, e17606. [Google Scholar] [CrossRef]
- Jurat-Fuentes, J.L.; Adang, M.J. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur. J. Biochem. 2004, 271, 3127–3135. [Google Scholar] [CrossRef]
- Ferré, J.; Van Rie, J. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 2002, 47, 501–533. [Google Scholar] [CrossRef]
- Ferré, J.; Van Rie, J.; MacIntosh, S.C. Insecticidal genetically modified crops and insect resistance management (IRM). In Integration of Insect-Resistant Genetically Modified Crops Within IPM Programs; Romeis, J., Shelton, A.M., Kennedy, G.G., Eds.; Springer Science Business Media BV: Dordrecht, The Netherlands, 2008; pp. 41–86. [Google Scholar] [CrossRef]
- Peterson, B.; Bezuidenhout, C.C.; Van den Berg, J. An overview of mechanisms of Cry toxin resistance in lepidopteran insects. J. Econ. Entomol. 2017, 110, 362–377. [Google Scholar] [CrossRef]
- Pigott, C.R.; Ellar, D.J. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 2007, 71, 255–281. [Google Scholar] [CrossRef]
- Sato, R.; Adegawa, S.; Li, X.; Tanaka, S.; Endo, H. Function and role of ATP-Binding Cassette Transporters as receptors for 3D-Cry toxins. Toxins 2019, 11, 124. [Google Scholar] [CrossRef]
- Singh, G.; Sachdev, B.; Sharma, N.; Seth, R.; Bhatnagar, R.K. Interaction of Bacillus thuringiensis vegetative insecticidal protein with Ribosomal S2 protein triggers larvicidal activity in Spodoptera frugiperda. Appl. Environ. Microbiol. 2010, 76, 7202–7209. [Google Scholar] [CrossRef]
- Jiang, K.; Hou, X.; Han, L.; Tan, T.; Cao, Z.; Cai, J. Fibroblast growth factor receptor, a novel receptor for vegetative insecticidal protein Vip3Aa. Toxins 2018, 10, 546. [Google Scholar] [CrossRef]
- Jiang, K.; Hou, X.; Tan, T.; Cao, Z.; Mei, S.; Yan, B.; Chang, J.; Han, L.; Zhao, D.; Cai, J. Scavenger receptor-C acts as a receptor for Bacillus thuringiensis vegetative insecticidal protein Vip3Aa and mediates the internalization of Vip3Aa via endocytosis. PLoS Pathog. 2018, 14, e1007347. [Google Scholar] [CrossRef]
- Gong, L.; Wang, H.; Qi, J.; Han, L.; Hu, M.; Jurat-Fuentes, J.L. Homologs to Cry toxin receptor genes in a de novo transcriptome and their altered expression in resistant Spodoptera litura larvae. J. Invertebr. Pathol. 2015, 129, 1–6. [Google Scholar] [CrossRef]
- Tetreau, G.; Bayyareddy, K.; Jones, C.M.; Stalinski, R.; Riaz, M.A.; Paris, M.; David, J.; Adang, M.J.; Després, L. Larval midgut modifications associated with Bti resistance in the yellow fever mosquito using proteomic and transcriptomic apaproaches. BMC Genom. 2012, 13, 248. [Google Scholar] [CrossRef]
- Jakka, S.R.K.; Ferré, J.; Jurat-Fuentes, J.L. Cry toxin binding sites and their use in strategies to delay resistance evolution. In Bt Resistance—Characterization and Strategies for GM Crops Producing Bacillus thuringiensis Toxins; Soberón, M., Gao, Y., Bravo, A., Eds.; CAB International: Boston, MA, USA, 2015; pp. 138–149. [Google Scholar] [CrossRef]
- Tiewsiri, K.; Wang, P. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl. Acad. Sci. USA 2011, 108, 14037–14042. [Google Scholar] [CrossRef]
- Coates, B.S.; Sumerford, D.V.; Siegfried, B.D.; Hellmich, R.L.; Abel, C.A. Unlinked genetic loci control the reduced transcription of aminopeptidase N 1 and 3 in the European corn borer and determine tolerance to Bacillus thuringiensis Cry1Ab toxin. Insect Biochem. Mol. Biol. 2013, 43, 1152–1160. [Google Scholar] [CrossRef]
- Chen, Y.; Li, M.; Islam, I.; You, L.; Wang, Y.; Li, Z.; Ling, L.; Zeng, B.; Xu, J.; Huang, Y.; et al. Allelic-specific expression in relation to Bombyx mori resistance to Bt toxin. Insect Biochem. Mol. Biol. 2014, 54, 53–60. [Google Scholar] [CrossRef]
- Wolfersberger, M.; Luethy, P.; Maurer, A.; Parenti, P.; Sacchi, F.V.; Giordana, B.; Hanozet, G.M. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 1987, 86, 301–308. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Hernández, C.S.; Rodrigo, A.; Ferré, J. Lyophilization of lepidopteran midguts: A preserving method for Bacillus thuringiensis toxin binding studies. J. Invertebr. Pathol. 2004, 85, 182–187. [Google Scholar] [CrossRef]
- Abdelkefi-Mesrati, L.; Rouis, S.; Sellami, S.; Jaoua, S. Prays oleae midgut putative receptor of Bacillus thuringiensis vegetative insecticidal protein Vip3LB differs from that of Cry1Ac toxin. Mol. Biol. 2009, 43, 15–19. [Google Scholar] [CrossRef]
- Munson, P.J.; Rodbard, D. Ligand: A versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 1980, 107, 220–239. [Google Scholar] [CrossRef]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Perera, O.P.; Willis, J.D.; Adang, M.J.; Jurat-Fuentes, J.L. Cloning and characterization of the Cry1Ac-binding alkaline phosphatase (HvALP) from Heliothis virescens. Insect Biochem. Mol. Biol. 2009, 39, 294–302. [Google Scholar] [CrossRef]
- Martínez-Solís, M.; Pinos, D.; Endo, H.; Portugal, L.; Sato, R.; Ferré, J.; Herrero, S.; Hernández-Martínez, P. Role of Bacillus thuringiensis Cry1A toxins domains in the binding to the ABCC2 receptor from Spodoptera exigua. Insect Biochem. Mol. Biol. 2018, 101, 47–56. [Google Scholar] [CrossRef]

| Strain | Mean ± SEM 1 | |
|---|---|---|
| Kd (nM) | Rt (pmol/mg) 2 | |
| Susceptible | 138 ± 18 | 443 ± 66 |
| Resistant | 161 ± 34 | 443 ± 109 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinos, D.; Chakroun, M.; Millán-Leiva, A.; Jurat-Fuentes, J.L.; Wright, D.J.; Hernández-Martínez, P.; Ferré, J. Reduced Membrane-Bound Alkaline Phosphatase Does Not Affect Binding of Vip3Aa in a Heliothis virescens Resistant Colony. Toxins 2020, 12, 409. https://doi.org/10.3390/toxins12060409
Pinos D, Chakroun M, Millán-Leiva A, Jurat-Fuentes JL, Wright DJ, Hernández-Martínez P, Ferré J. Reduced Membrane-Bound Alkaline Phosphatase Does Not Affect Binding of Vip3Aa in a Heliothis virescens Resistant Colony. Toxins. 2020; 12(6):409. https://doi.org/10.3390/toxins12060409
Chicago/Turabian StylePinos, Daniel, Maissa Chakroun, Anabel Millán-Leiva, Juan Luis Jurat-Fuentes, Denis J. Wright, Patricia Hernández-Martínez, and Juan Ferré. 2020. "Reduced Membrane-Bound Alkaline Phosphatase Does Not Affect Binding of Vip3Aa in a Heliothis virescens Resistant Colony" Toxins 12, no. 6: 409. https://doi.org/10.3390/toxins12060409
APA StylePinos, D., Chakroun, M., Millán-Leiva, A., Jurat-Fuentes, J. L., Wright, D. J., Hernández-Martínez, P., & Ferré, J. (2020). Reduced Membrane-Bound Alkaline Phosphatase Does Not Affect Binding of Vip3Aa in a Heliothis virescens Resistant Colony. Toxins, 12(6), 409. https://doi.org/10.3390/toxins12060409







