Salt Shock Responses of Microcystis Revealed through Physiological, Transcript, and Metabolomic Analyses
Abstract
1. Introduction
2. Results
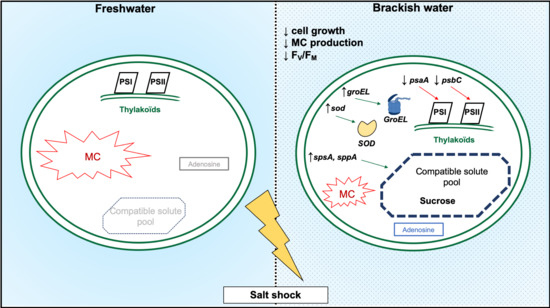
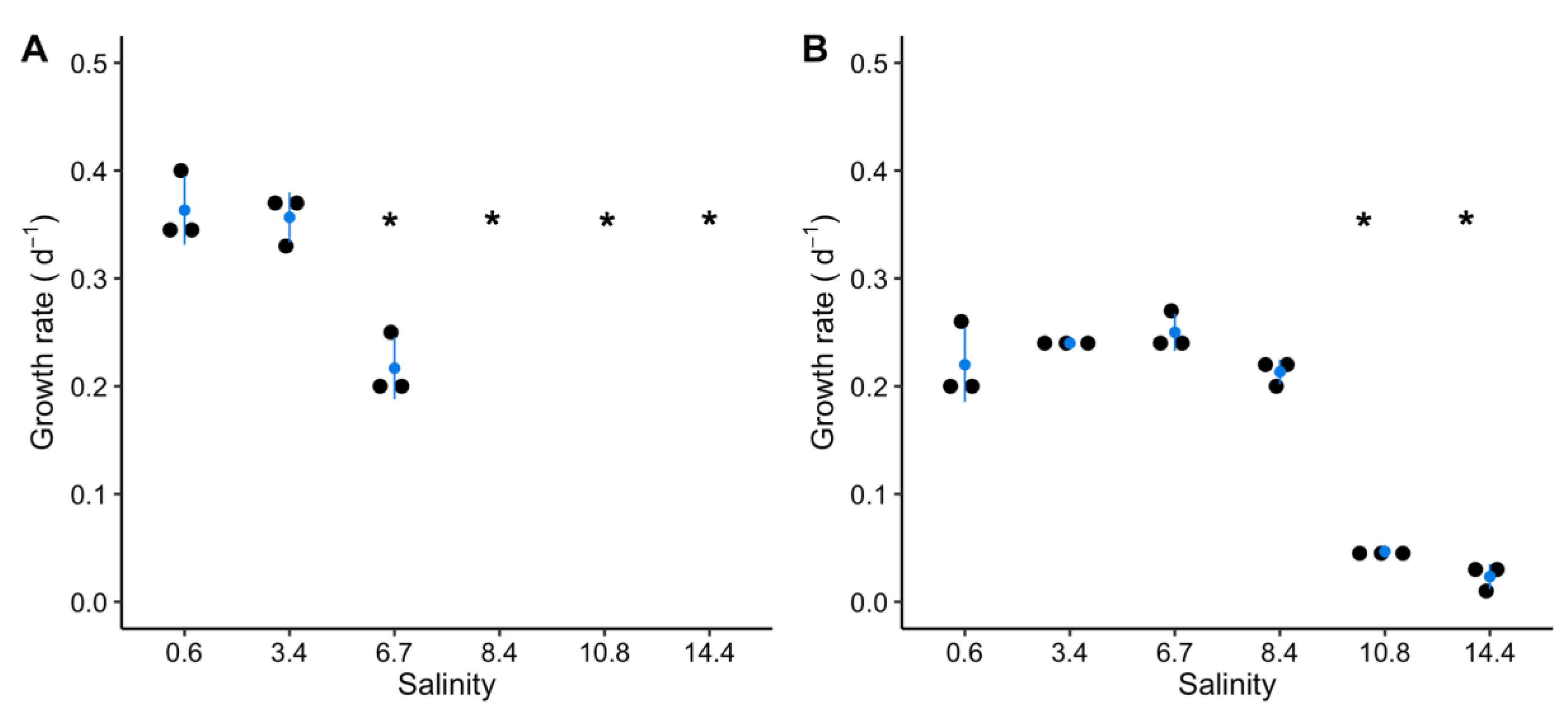
2.1. Growth of M. aeruginosa during a Salt Shock
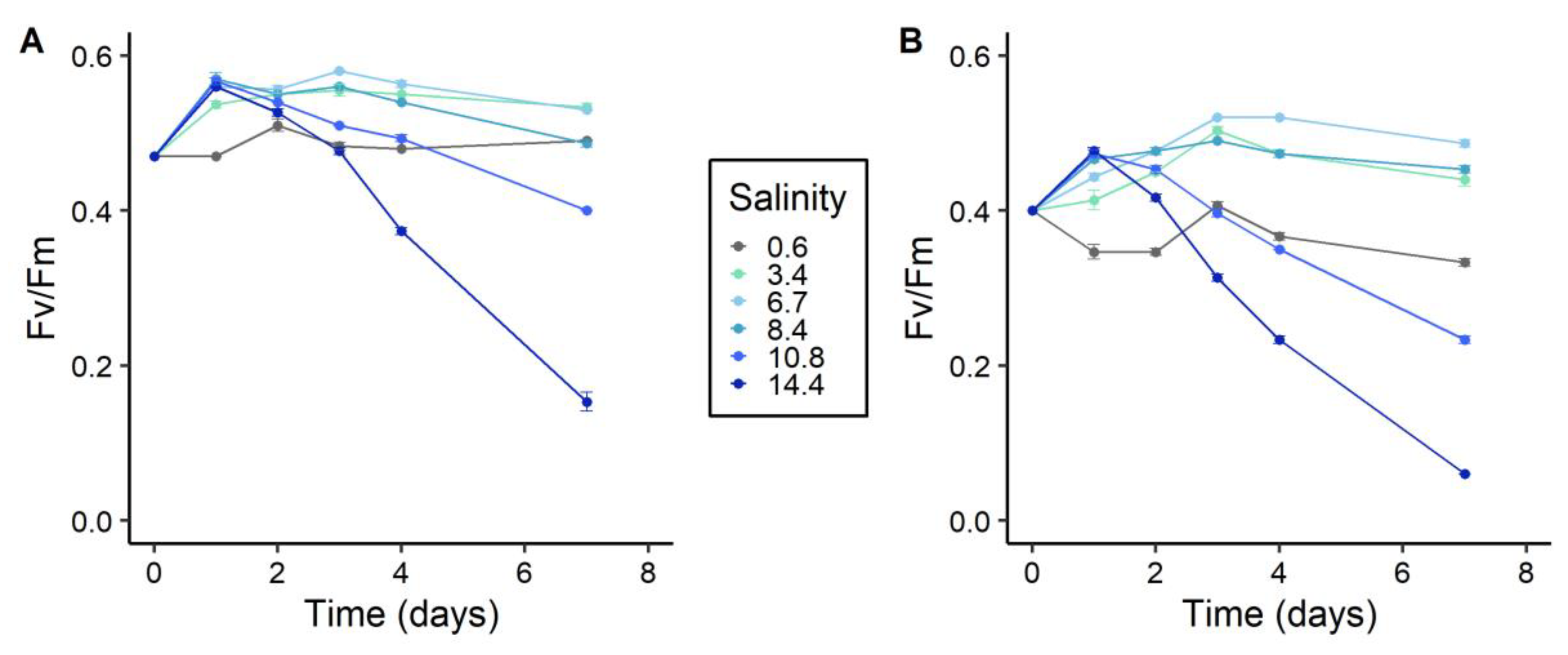
2.2. Photosynthetic Activity
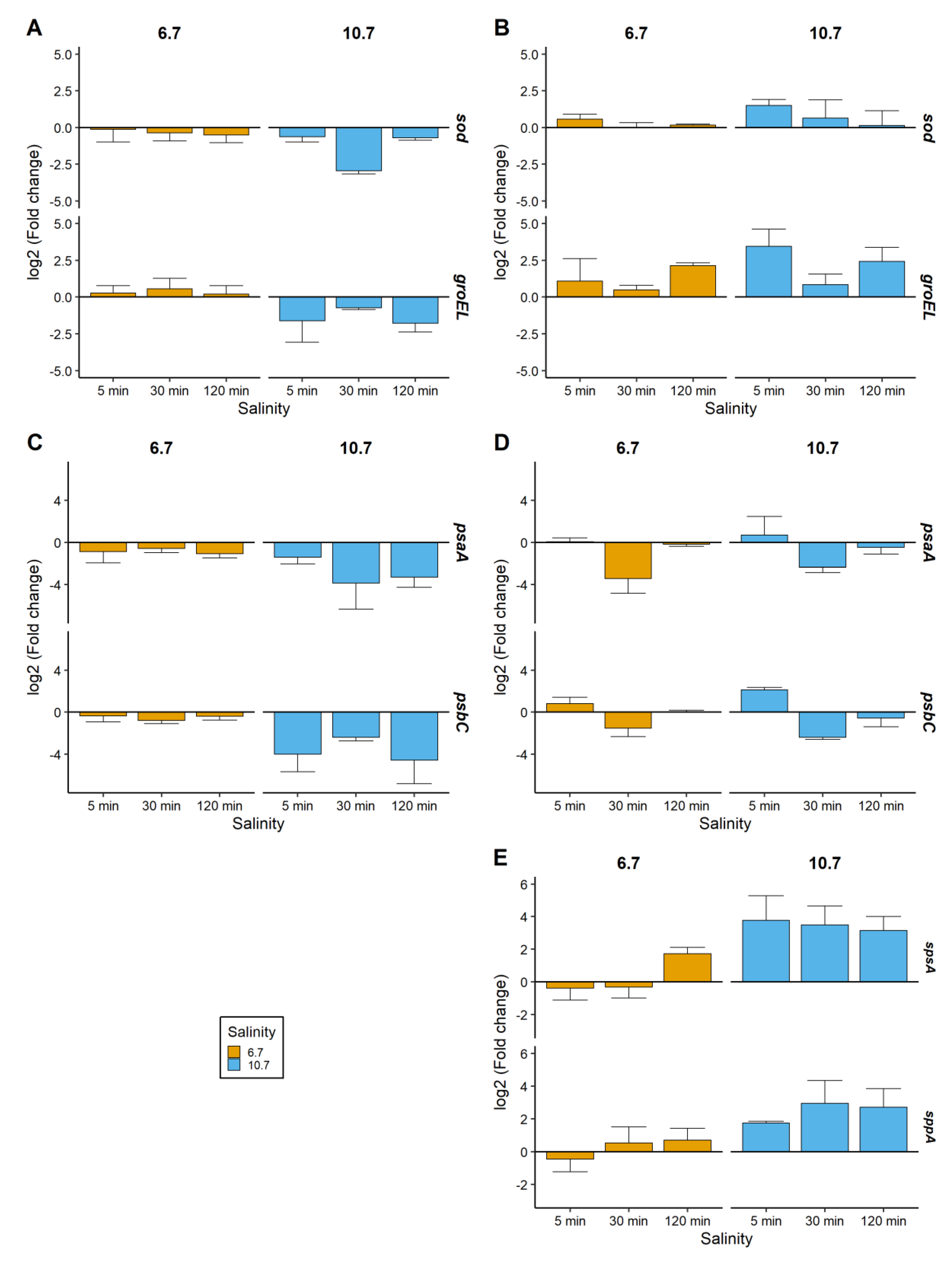
2.3. Relative Gene Expressions
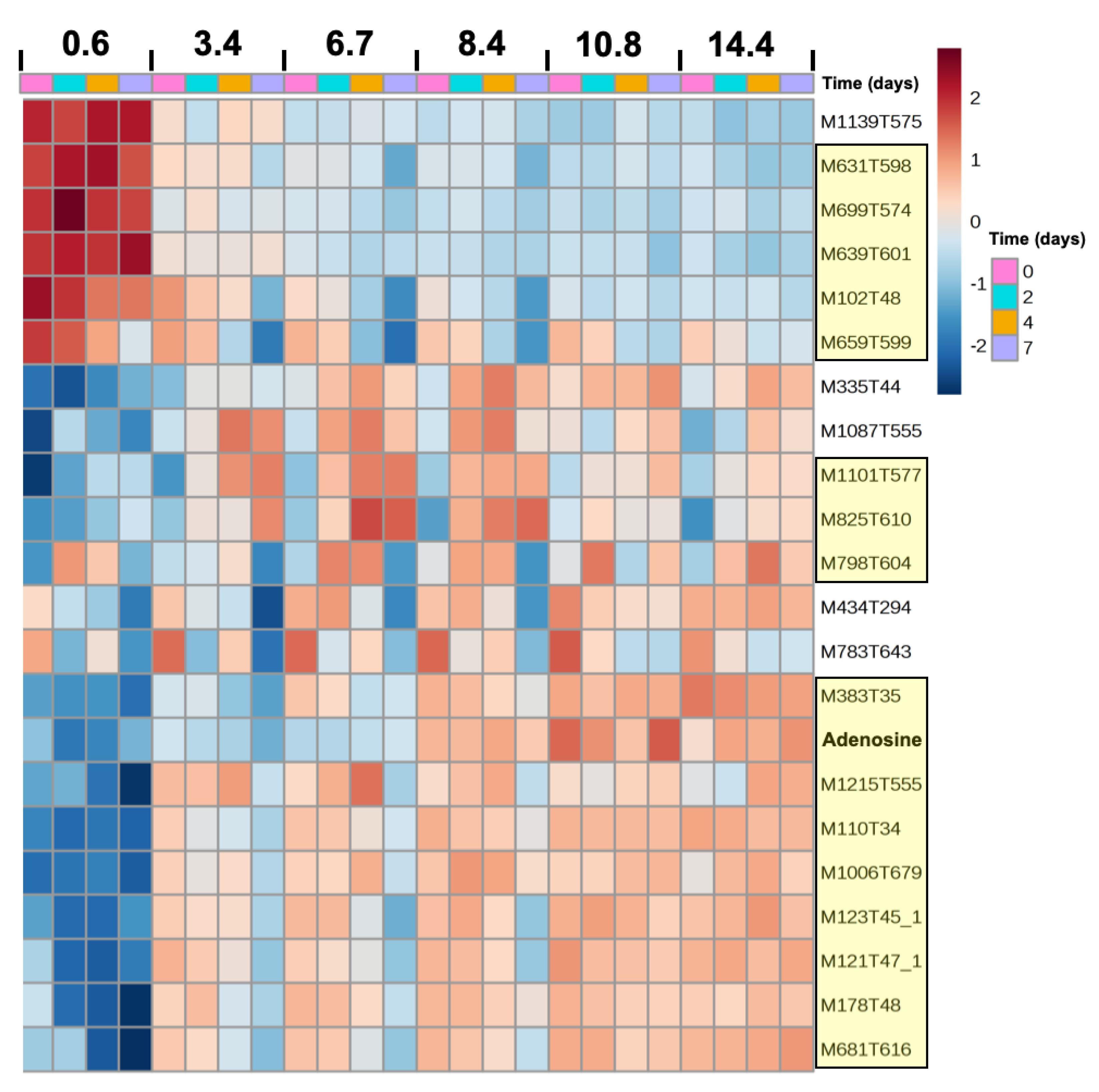
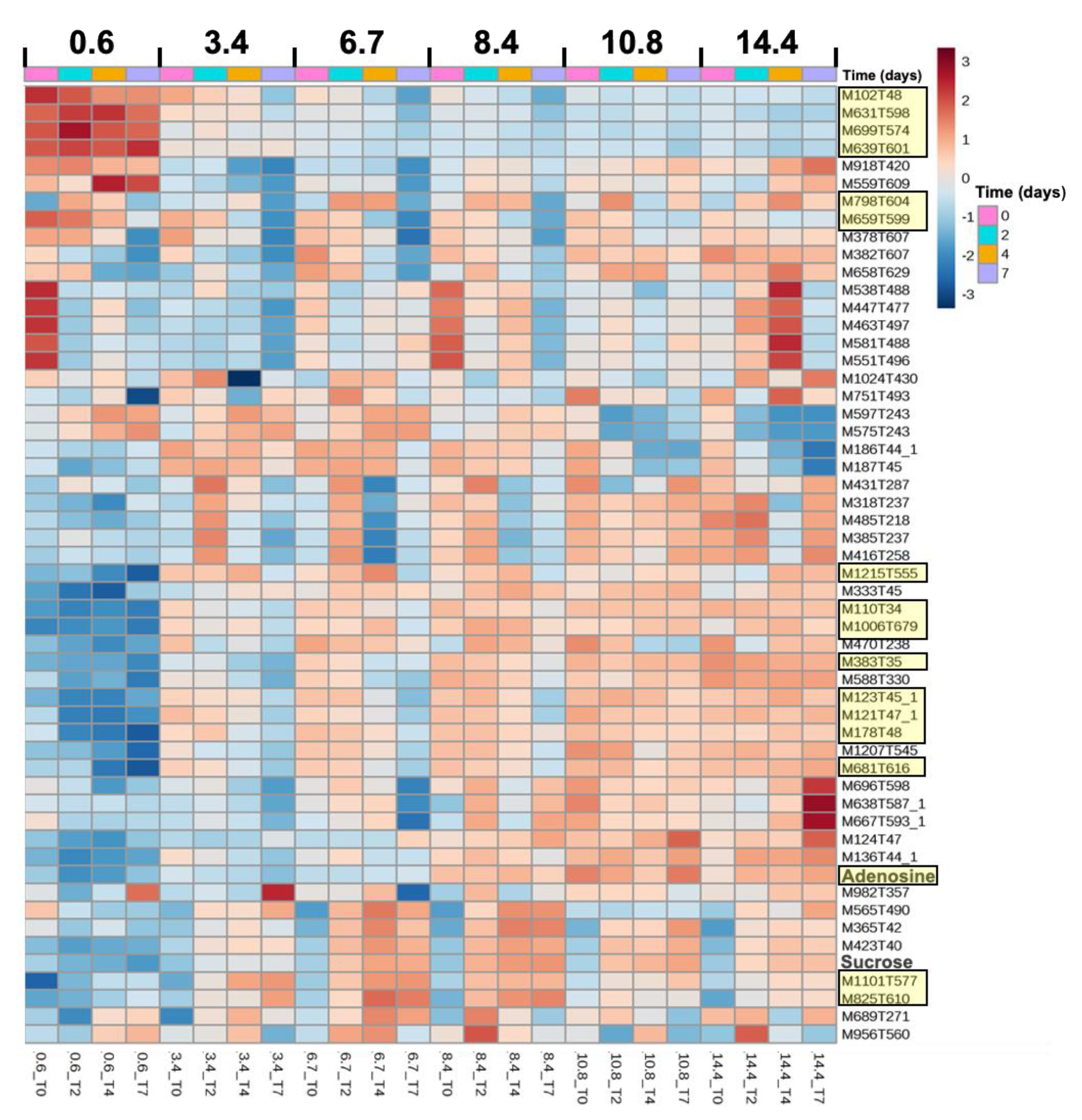
2.4. Metabolome Dynamics
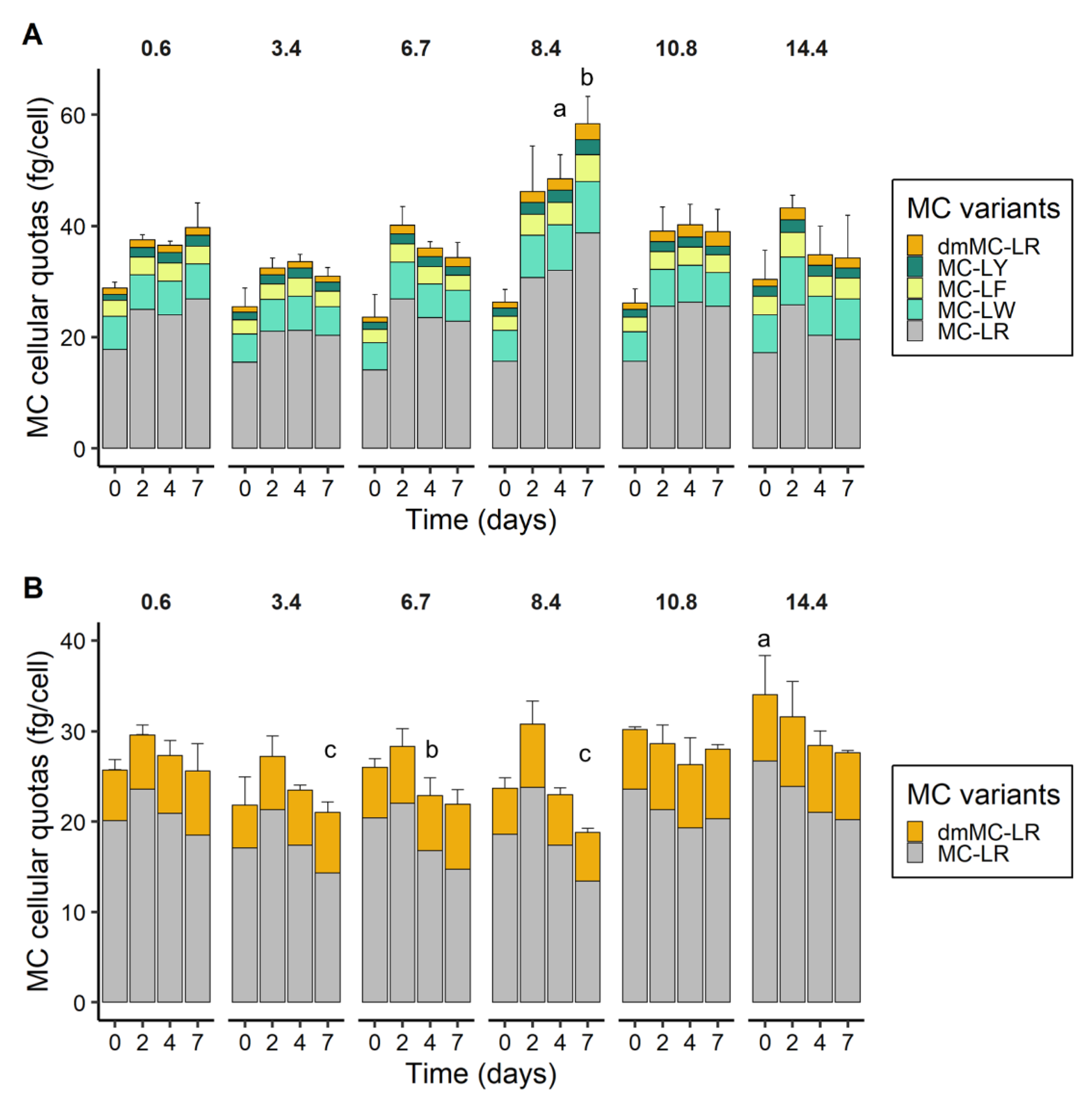
2.5. MC Cellular Quotas
3. Discussion
4. Materials and Methods
4.1. Organisms and Experimental Design
4.2. Cell Enumeration and Photosynthetic Parameter
4.3. LC-MS/MS Analysis of MCs
4.4. RNA Isolation and RT-qPCR
4.5. Metabolomic Analyses
4.6. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harke, M.J.; Steffen, M.M.; Gobler, C.J.; Otten, T.G.; Wilhelm, S.W.; Wood, S.A.; Paerl, H.W. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 2016, 54, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Bouaïcha, N.; Miles, C.O.; Beach, D.G.; Labidi, Z.; Djabri, A.; Benayache, N.Y.; Nguyen-Quang, T. Structural Diversity, Characterization and Toxicology of Microcystins. Toxins 2019, 11, 714. [Google Scholar] [CrossRef] [PubMed]
- Buratti, F.M.; Manganelli, M.; Vichi, S.; Stefanelli, M.; Scardala, S.; Testai, E.; Funari, E. Cyanotoxins: Producing organisms, occurrence, toxicity, mechanism of action and human health toxicological risk evaluation. Arch. Toxicol. 2017, 91, 1049–1130. [Google Scholar] [CrossRef] [PubMed]
- Svirčev, Z.; Lalić, D.; Bojadžija Savić, G.; Tokodi, N.; Drobac Backović, D.; Chen, L.; Meriluoto, J.; Codd, G.A. Global Geographical and Historical Overview of Cyanotoxin Distribution and Cyanobacterial Poisonings; Springer: Berlin/Heidelberg, Germany, 2019; ISBN 0123456789. [Google Scholar]
- Preece, E.P.; Hardy, F.J.; Moore, B.C.; Bryan, M. A review of microcystin detections in Estuarine and Marine waters: Environmental implications and human health risk. Harmful Algae 2017, 61, 31–45. [Google Scholar] [CrossRef]
- Paldavičienė, A.; Mazur-Marzec, H.; Razinkovas, A. Toxic cyanobacteria blooms in the Lithuanian part of the Curonian Lagoon. Oceanologia 2009, 51, 203–216. [Google Scholar] [CrossRef]
- Mazur-Marzec, H.; Forycka, K.; Kobos, J.; Pliński, M.; Browarczyk-Matusiak, G. Morphological, genetic, chemical and ecophysiological characterisation of two Microcystis aeruginosa isolates from the Vistula Lagoon, southern Baltic. Oceanologia 2010, 52, 127–146. [Google Scholar] [CrossRef]
- Chen, D.Z.X.; Boland, M.P.; Smillie, M.A.; Klix, H.; Ptak, C.; Andersen, R.J.; Holmes, C.F.B. Identification of protein phosphatase inhibitors of the microcystin class in the marine environment. Toxicon 1993, 31, 1407–1414. [Google Scholar] [CrossRef]
- Vareli, K.; Jaeger, W.; Touka, A.; Frillingos, S.; Briasoulis, E.; Sainis, I. Hepatotoxic Seafood Poisoning (HSP) due to microcystins: A threat from the ocean? Mar. Drugs 2013, 11, 2751–2768. [Google Scholar] [CrossRef]
- Paerl, H.W.; Otten, T.G.; Kudela, R. Mitigating the expansion of harmful algal blooms across the freshwater-to-marine continuum. Environ. Sci. Technol. 2018, 52, 5519–5529. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Gibble, C.M.; Peacock, M.B.; Kudela, R.M. Evidence of freshwater algal toxins in marine shellfish: Implications for human and aquatic health. Harmful Algae 2016, 59, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Umehara, A.; Takahashi, T.; Komorita, T.; Orita, R.; Choi, J.W.; Takenaka, R.; Mabuchi, R.; Park, H.D.; Tsutsumi, H. Widespread dispersal and bio-accumulation of toxic microcystins in benthic marine ecosystems. Chemosphere 2017, 167, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Atkins, R.; Rose, T.; Brown, R.S.; Robb, M. The Microcystis cyanobacteria bloom in the Swan River - February 2000. Water Sci. Technol. 2001, 43, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Robson, B.J.; Hamilton, D.P. Summer flow event induces a cyanobacterial bloom in a seasonal Western Australian estuary. Mar. Freshw. Res. 2003, 54, 139. [Google Scholar] [CrossRef]
- Lehman, P.W.; Boyer, G.; Satchwell, M.; Waller, S. The influence of environmental conditions on the seasonal variation of Microcystis cell density and microcystins concentration in San Francisco Estuary. Hydrobiologia 2008, 600, 187–204. [Google Scholar] [CrossRef]
- Preece, E.P.; Moore, B.C.; Hardy, F.J. Transfer of microcystin from freshwater lakes to Puget Sound, WA and toxin accumulation in marine mussels (Mytilus trossulus). Ecotoxicol. Environ. Saf. 2015, 122, 98–105. [Google Scholar] [CrossRef]
- Tanabe, Y.; Hodoki, Y.; Sano, T.; Tada, K.; Watanabe, M.M. Adaptation of the freshwater bloom-forming cyanobacterium microcystis aeruginosa to brackish water is driven by recent horizontal transfer of sucrose genes. Front. Microbiol. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Bormans, M.; Amzil, Z.; Mineaud, E.; Brient, L.; Savar, V.; Robert, E.; Lance, E. Demonstrated transfer of cyanobacteria and cyanotoxins along a freshwater-marine continuum in France. Harmful Algae 2019, 87, 101639. [Google Scholar] [CrossRef]
- Kramer, B.J.; Davis, T.W.; Meyer, K.A.; Rosen, B.H.; Goleski, J.A.; Dick, G.J.; Oh, G.; Gobler, C.J. Nitrogen limitation, toxin synthesis potential, and toxicity of cyanobacterial populations in Lake Okeechobee and the St. Lucie River Estuary, Florida, during the 2016 state of emergency event. PLoS ONE 2018, 13. Available online: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0196278 (accessed on 12 March 2020). [CrossRef]
- Kurobe, T.; Lehman, P.W.; Hammock, B.G.; Bolotaolo, M.B.; Lesmeister, S.; Teh, S.J. Biodiversity of cyanobacteria and other aquatic microorganisms across a freshwater to brackish water gradient determined by shotgun metagenomic sequencing analysis in the San Francisco Estuary, USA. PLoS ONE 2018, 13, e0203953. [Google Scholar] [CrossRef]
- Yunes, J.S.; Salomon, P.S.; Matthiensen, A.; Beattie, K.A.; Raggett, S.L.; Codd, G.A. Toxic blooms of cyanobacteria in the Patos Lagoon Estuary, southern Brazil. J. Aquati. Ecosyst. Stress Recover. 1996, 5, 223–229. [Google Scholar] [CrossRef]
- Lehman, P.W.; Kurobe, T.; Lesmeister, S.; Baxa, D.; Tung, A.; Teh, S.J. Impacts of the 2014 severe drought on the Microcystis bloom in San Francisco Estuary. Harmful Algae 2017, 63, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Paul, V.J. Climate change: Links to global expansion of harmful cyanobacteria. Water Res. 2012, 46, 1349–1363. [Google Scholar] [CrossRef]
- Reed, R.H.; Walsby, A.E. Changes in turgor pressure in response to increases in external NaCl concentration in the gas-vacuolate cyanobacterium Microcystis sp. Arch. Microbiol. 1985, 143, 290–296. [Google Scholar] [CrossRef]
- Otsuka, S.; Suda, S.; Li, R.; Watanabe, M.; Oyaizu, H.; Matsumoto, S.; Watanabe, M.M. Characterization of morphospecies and strains of the genus Microcystis (Cyanobacteria) for a reconsideration of species classification. Phycol. Res. 1999, 47, 189–197. [Google Scholar] [CrossRef]
- Tonk, L.; Bosch, K.; Visser, P.; Huisman, J. Salt tolerance of the harmful cyanobacterium Microcystis aeruginosa. Aquati. Microb. Ecol. 2007, 46, 117–123. [Google Scholar] [CrossRef]
- Mitsch, W.J.; Gosselink, J.G. Wetlands; John Wiley & Sons. Inc.: New York, NY, USA, 2000. [Google Scholar]
- Ross, C.; Warhurst, B.C.; Brown, A.; Huff, C.; Ochrietor, J.D. Mesohaline conditions represent the threshold for oxidative stress, cell death and toxin release in the cyanobacterium Microcystis aeruginosa. Aquat. Toxicol. 2019, 206, 203–211. [Google Scholar] [CrossRef]
- Georges des Aulnois, M.; Roux, P.; Caruana, A.; Réveillon, D.; Briand, E.; Hervé, F.; Savar, V.; Bormans, M.; Amzil, Z. Physiological and Metabolic Responses of Freshwater and Brackish-Water Strains of Microcystis aeruginosa Acclimated to a Salinity Gradient: Insight into Salt Tolerance. Appl. Environ. Microbiol. 2019, 85, AEM.01614-19. [Google Scholar] [CrossRef]
- Orr, P.T.; Jones, G.J.; Douglas, G.B. Response of cultured Microcystis aeruginosa from the Swan River, Australia, to elevated salt concentration and consequences for bloom and toxin management in estuaries. Mar. Freshw. Res. 2004, 55, 277. [Google Scholar] [CrossRef]
- Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 2011, 35, 87–123. [Google Scholar] [CrossRef]
- Ross, C.; Santiago-Vázquez, L.; Paul, V. Toxin release in response to oxidative stress and programmed cell death in the cyanobacterium Microcystis aeruginosa. Aquat. Toxicol. 2006, 78, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Vonshak, A. Effects of salinity stress on photosystem II function in cyanobacterial Spirulina platensis cells. Physiol. Plant. 2002, 114, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Mao, F.; Kirumba, G.C.; Jiang, C.; Manefield, M.; He, Y. Changes in metabolites, antioxidant system, and gene expression in Microcystis aeruginosa under sodium chloride stress. Ecotoxicol. Environ. Saf. 2015, 122, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Black, K.; Yilmaz, M.; Phlips, E.J. Growth and Toxin Production by Microcystis Aeruginosa PCC 7806 (Kutzing) Lemmerman at Elevated Salt Concentrations. J. Environ. Prot. 2011, 02, 669–674. [Google Scholar] [CrossRef]
- Hagemann, M.; Erdmann, N.; Wittenburg, E. Synthesis of glucosylglycerol in salt-stressed cells of the cyanobacterium Microcystis firma. Arch. Microbiol. 1987, 148, 275–279. [Google Scholar] [CrossRef]
- Kolman, M.A.; Salerno, G.L. Sucrose in bloom-forming cyanobacteria: Loss and gain of genes involved in its biosynthesis. Environ. Microbiol. 2016, 18, 439–449. [Google Scholar] [CrossRef]
- Kolman, M.; Nishi, C.; Perez-Cenci, M.; Salerno, G. Sucrose in cyanobacteria: From a salt-response Molecule to play a key role in nitrogen fixation. Life 2015, 5, 102–126. [Google Scholar] [CrossRef]
- Nueda, M.J.; Conesa, A.; Westerhuis, J.A.; Hoefsloot, H.C.J.; Smilde, A.K.; Talón, M.; Ferrer, A. Discovering gene expression patterns in time course microarray experiments by ANOVA–SCA. Bioinformatics 2007, 23, 1792–1800. [Google Scholar] [CrossRef]
- Adhikary, S.P. NaCl induced changes in photosystem stoichiometry and photosynthetic activity of the cyanobacterium Microcystis aeruginosa K??tz. Indian J. Exp. Biol. 2005, 43, 1192–1195. [Google Scholar]
- Allakhverdiev, S.I.; Murata, N. Salt stress inhibits photosystems II and I in cyanobacteria. Photosynth. Res. 2008, 98, 529–539. [Google Scholar] [CrossRef]
- Chen, L.; Gin, K.Y.H.; He, Y. Effects of sulfate on microcystin production, photosynthesis, and oxidative stress in Microcystis aeruginosa. Environ. Sci. Pollut. Res. 2016, 23, 3586–3595. [Google Scholar] [CrossRef]
- Sinetova, M.A.; Los, D.A. Systemic analysis of stress transcriptomics of Synechocystis reveals common stress genes and their universal triggers. Mol. BioSyst. 2016, 12, 3254–3258. [Google Scholar] [CrossRef]
- Wei, N.; Hu, L.; Song, L.R.; Gan, N.Q. Microcystin-bound protein patterns in different cultures of Microcystis aeruginosa and field samples. Toxins 2016, 8, 293. [Google Scholar] [CrossRef]
- Schuurmans, J.M.; Brinkmann, B.W.; Makower, A.K.; Dittmann, E.; Huisman, J.; Matthijs, H.C.P. Microcystin interferes with defense against high oxidative stress in harmful cyanobacteria. Harmful Algae 2018, 78, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Sandrini, G.; Cunsolo, S.; Schuurmans, J.M.; Matthijs, H.C.P.; Huisman, J. Changes in gene expression, cell physiology and toxicity of the harmful cyanobacterium Microcystis aeruginosa at elevated CO2. Front. Microbiol. 2015, 6, 401. [Google Scholar] [CrossRef] [PubMed]
- Guljamow, A.; Jenke-Kodama, H.; Saumweber, H.; Quillardet, P.; Frangeul, L.; Castets, A.M.; Bouchier, C.; Tandeau de Marsac, N.; Dittmann, E. Horizontal gene transfer of two cytoskeletal elements from a eukaryote to a cyanobacterium. Curr. Biol. 2007, 17, 757–759. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, Y.; Yamaguchi, H.; Sano, T.; Kawachi, M. A novel salt-tolerant genotype illuminates the sucrose gene evolution in freshwater bloom-forming cyanobacterium Microcystis aeruginosa. FEMS Microbiol. Lett. 2019, 111, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Steffen, M.M.; Dearth, S.P.; Dill, B.D.; Li, Z.; Larsen, K.M.; Campagna, S.R.; Wilhelm, S.W. Nutrients drive transcriptional changes that maintain metabolic homeostasis but alter genome architecture in Microcystis. ISME J. 2014, 8, 2080–2092. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Song, Y.; Li, L.; Gan, N.; Brand, J.J.; Song, L. The highly heterogeneous methylated genomes and diverse restriction-modification systems of bloom-forming Microcystis. Harmful Algae 2018, 75, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.; Orf, I.; Kopka, J.; Hagemann, M. Recent applications of metabolomics toward cyanobacteria. Metabolites 2013, 3, 72–100. [Google Scholar] [CrossRef]
- Rohrlack, T.; Hyenstrand, P. Fate of intracellular microcystins in the cyanobacterium Microcystis aeruginosa (Chroococcales, Cyanophyceae). Phycologia 2007, 46, 277–283. [Google Scholar] [CrossRef]
- Orr, P.T.; Willis, A.; Burford, M.A. Application of first order rate kinetics to explain changes in bloom toxicity—the importance of understanding cell toxin quotas. J. Oceanol. Limnol. 2018, 36, 1063–1074. [Google Scholar] [CrossRef]
- Zilliges, Y.; Kehr, J.C.; Meissner, S.; Ishida, K.; Mikkat, S.; Hagemann, M.; Kaplan, A.; Börner, T.; Dittmann, E. The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS ONE 2011, 6, e17615. [Google Scholar] [CrossRef] [PubMed]
- Meissner, S.; Fastner, J.; Dittmann, E. Microcystin production revisited: Conjugate formation makes a major contribution. Environ. Microbiol. 2013, 15, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.A.; Borges, H.; Puddick, J.; Biessy, L.; Atalah, J.; Hawes, I.; Dietrich, D.R.; Hamilton, D.P. Contrasting cyanobacterial communities and microcystin concentrations in summers with extreme weather events: Insights into potential effects of climate change. Hydrobiologia 2017, 785, 71–89. [Google Scholar] [CrossRef]
- Briand, E.; Escoffier, N.; Straub, C.; Sabart, M.; Quiblier, C.; Humbert, J.F. Spatiotemporal changes in the genetic diversity of a bloom-forming Microcystis aeruginosa (cyanobacteria) population. ISME J. 2009, 3, 419–429. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Bail, A.L.; Charrier, B. Plant Organogenesis; Methods in Molecular Biology; De Smet, I., Ed.; Humana Press: Totowa, NJ, USA, 2013; Volume 959, ISBN 978-1-62703-220-9. [Google Scholar]
- International Association for the Physical Sciences of the Ocean. Working Group on Symbols, Units and Nomenclature in Physical Oceanography. The International System of Units (SI) in Oceanography. In Technical Paper in Marine Science; UNESCO: Paris, France, 1985; Available online: https://unesdoc.unesco.org/ark:/48223/pf0000065031 (accessed on 12 March 2020).
- Wood, A.M.; Everroad, R.C.; Wingard, L.M. Measuring growth rates in microalgal cultures. Algal Cult. Tech. 2005, 18, 269–288. [Google Scholar]
- Ogawa, T.; Misumi, M.; Sonoike, K. Estimation of photosynthesis in cyanobacteria by pulse-amplitude modulation chlorophyll fluorescence: Problems and solutions. Photosynth. Res. 2017, 133, 63–73. [Google Scholar] [CrossRef]
- Makower, A.K.; Schuurmans, J.M.; Groth, D.; Zilliges, Y.; Matthijs, H.C.P.; Dittmann, E. Transcriptomics-aided dissection of the intracellular and extracellular roles of microcystin in Microcystis aeruginosa PCC 7806. Appl. Environ. Microbiol. 2015, 81, 544–554. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Gil-de-la-Fuente, A.; Godzien, J.; Saugar, S.; Garcia-Carmona, R.; Badran, H.; Wishart, D.S.; Barbas, C.; Otero, A. CEU Mass Mediator 3.0: A Metabolite Annotation Tool. J. Proteome Res. 2019, 18, 797–802. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Guitton, Y.; Tremblay-Franco, M.; Le Corguillé, G.; Martin, J.F.; Pétéra, M.; Roger-Mele, P.; Delabrière, A.; Goulitquer, S.; Monsoor, M.; Duperier, C.; et al. Create, run, share, publish, and reference your LC–MS, FIA–MS, GC–MS, and NMR data analysis workflows with the Workflow4Metabolomics 3.0 Galaxy online infrastructure for metabolomics. Int. J. Biochem. Cell Biol. 2017, 93, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- van der Kloet, F.M.; Bobeldijk, I.; Verheij, E.R.; Jellema, R.H. Analytical Error Reduction Using Single Point Calibration for Accurate and Precise Metabolomic Phenotyping. J. Proteome Res. 2009, 8, 5132–5141. [Google Scholar] [CrossRef]
- Adams, M.A.; Chen, Z.; Landman, P.; Colmer, T.D. Simultaneous Determination by Capillary Gas Chromatography of Organic Acids, Sugars, and Sugar Alcohols in Plant Tissue Extracts as Their Trimethylsilyl Derivatives. Anal. Biochem. 1999, 266, 77–84. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical 2017. Available online: https://www.r-project.org (accessed on 12 March 2020).
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Smilde, A.K.; Jansen, J.J.; Hoefsloot, H.C.J.; Lamers, R.J.A.N.; van der Greef, J.; Timmerman, M.E. ANOVA-simultaneous component analysis (ASCA): A new tool for analyzing designed metabolomics data. Bioinformatics 2005, 21, 3043–3048. [Google Scholar] [CrossRef] [PubMed]
- Vis, D.J.; Westerhuis, J.A.; Smilde, A.K.; van der Greef, J. Statistical validation of megavariate effects in ASCA. BMC Bioinform. 2007, 8, 322. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georges des Aulnois, M.; Réveillon, D.; Robert, E.; Caruana, A.; Briand, E.; Guljamow, A.; Dittmann, E.; Amzil, Z.; Bormans, M. Salt Shock Responses of Microcystis Revealed through Physiological, Transcript, and Metabolomic Analyses. Toxins 2020, 12, 192. https://doi.org/10.3390/toxins12030192
Georges des Aulnois M, Réveillon D, Robert E, Caruana A, Briand E, Guljamow A, Dittmann E, Amzil Z, Bormans M. Salt Shock Responses of Microcystis Revealed through Physiological, Transcript, and Metabolomic Analyses. Toxins. 2020; 12(3):192. https://doi.org/10.3390/toxins12030192
Chicago/Turabian StyleGeorges des Aulnois, Maxime, Damien Réveillon, Elise Robert, Amandine Caruana, Enora Briand, Arthur Guljamow, Elke Dittmann, Zouher Amzil, and Myriam Bormans. 2020. "Salt Shock Responses of Microcystis Revealed through Physiological, Transcript, and Metabolomic Analyses" Toxins 12, no. 3: 192. https://doi.org/10.3390/toxins12030192
APA StyleGeorges des Aulnois, M., Réveillon, D., Robert, E., Caruana, A., Briand, E., Guljamow, A., Dittmann, E., Amzil, Z., & Bormans, M. (2020). Salt Shock Responses of Microcystis Revealed through Physiological, Transcript, and Metabolomic Analyses. Toxins, 12(3), 192. https://doi.org/10.3390/toxins12030192