Ostreopsis cf. ovata (Dinophyceae) Molecular Phylogeny, Morphology, and Detection of Ovatoxins in Strains and Field Samples from Brazil
Abstract
1. Introduction
2. Results
2.1. Morphology
2.2. Molecular Phylogeny
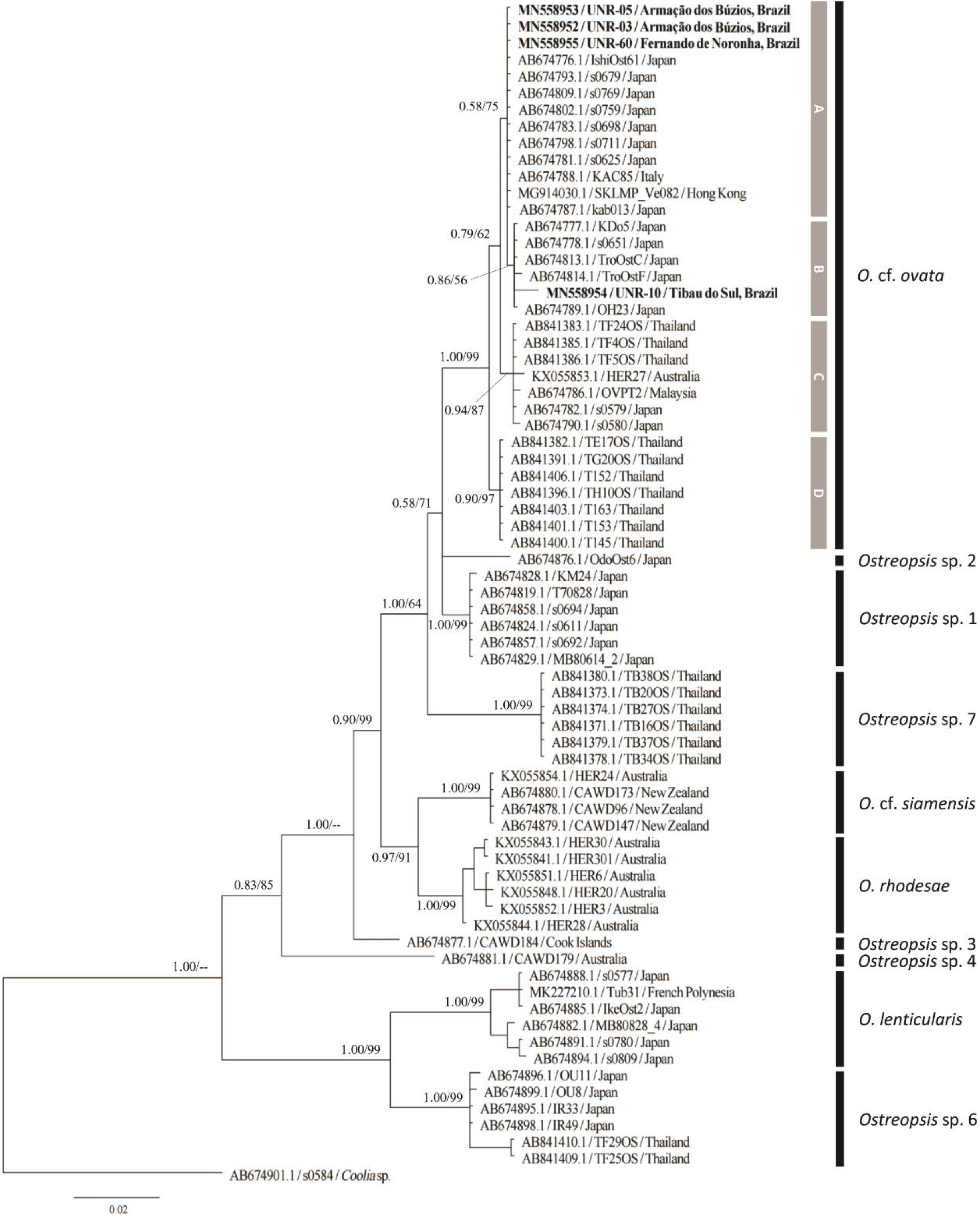
2.2.1. ITS rDNA Phylogeny
2.2.2. LSU rDNA D1–D3 Phylogeny
2.2.3. LSU rDNA D8–D10 Phylogeny
2.3. Toxin Content and Profile
3. Discussion
3.1. Morphology
3.2. Molecular Phylogeny
3.3. Toxins
4. Conclusions
5. Materials and Methods
5.1. Strains Isolations and Cultures Establishment
5.2. Morphological Characterization
5.3. Molecular Characterization of Strains
5.4. Molecular Characterization of Field Specimens
5.5. Toxin Analysis by Liquid Chromatography-Mass Spectrometry
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schmidt, J. Flora of Koh Chang. Contributions to the knowledge of the vegetation in the Gulf of Siam. Bot. Tidsskr. 1901, 24, 212–221. [Google Scholar] [CrossRef]
- Fukuyo, Y. Taxonomical Study on Benthic Dinoflagellates Collected in Coral Reefs. Bull. Jpn. Soc. Sci. Fish. 1981, 47, 967–978. [Google Scholar] [CrossRef]
- Faust, M.A. Three new Ostreopsis species (Dinophyceae): O. marinus sp. nov., O. belizeanus sp. nov., and O. caribbeanus sp. nov. Phycologia 1999, 38, 92–99. [Google Scholar] [CrossRef]
- Rhodes, L.L.; Smith, K.F.; Munday, R.; Selwood, A.I.; McNabb, P.S.; Holland, P.T.; Bottein, M.-Y. Toxic dinoflagellates (Dinophyceae) from Rarotonga, Cook Islands. Toxicon 2010, 56, 751–758. [Google Scholar] [CrossRef]
- Norris, D.; Bomber, J.; Balech, E. Benthic dinoflagellates associated with ciguatera from the Florida Keys. I. Ostreopsis heptagona sp. nov. Toxic Dinoflag. 1985, 40, 39–44. [Google Scholar]
- Quod, J.P. Ostreopsis mascarenensis sp. nov (Dinophyceae) dinoflagellé toxique associéà la ciguatéra dans l’Océan Indien. Cryptogam. Algol. 1994, 15, 243–251. [Google Scholar]
- Faust, M.A.; Morton, S.L. Morphology and ecology of the marine dinoflagellate Ostreopsis labens sp. nov. (Dinophyceae). J. Phycol. 1995, 31, 456–463. [Google Scholar] [CrossRef]
- Accoroni, S.; Romagnoli, T.; Penna, A.; Capellacci, S.; Ciminiello, P.; Dell’Aversano, C.; Tartaglione, L.; Abboud-Abi Saab, M.; Giussani, V.; Asnaghi, V.; et al. Ostreopsis fattorussoi sp. nov. (Dinophyceae), a new benthic toxic Ostreopsis species from the eastern Mediterranean Sea. J. Phycol. 2016, 52, 1064–1084. [Google Scholar] [CrossRef]
- Verma, A.; Hoppenrath, M.; Dorantes-Aranda, J.J.; Harwood, D.T.; Murray, S.A. Molecular and phylogenetic characterization of Ostreopsis (Dinophyceae) and the description of a new species, Ostreopsis rhodesae sp. nov., from a subtropical Australian lagoon. Harmful Algae 2016, 60, 116–130. [Google Scholar] [CrossRef]
- Penna, A.; Vila, M.; Fraga, S.; Giacobbe, M.G.; Francesco, A.; Riobó, P.; Vernesi, C. Characterization of Ostreopsis and Coolia (Dinophyceae) isolates in the western Mediterranean Sea based on morphology, toxicity and internal transcribed spacer 5.8s rDNA sequences. J. Phycol. 2005, 41, 212–225. [Google Scholar] [CrossRef]
- Parsons, M.L.; Aligizaki, K.; Bottein, M.Y.D.; Fraga, S.; Morton, S.L.; Penna, A.; Rhodes, L. Gambierdiscus and Ostreopsis: Reassessment of the state of knowledge of their taxonomy, geography, ecophysiology, and toxicology. Harmful Algae 2012, 14, 107–129. [Google Scholar] [CrossRef]
- David, H.; Laza-Martínez, A.; Miguel, I.; Orive, E. Ostreopsis cf. siamensis and Ostreopsis cf. ovata from the Atlantic Iberian Peninsula: Morphological and phylogenetic characterization. Harmful Algae 2013, 30, 44–55. [Google Scholar] [CrossRef]
- Hoppenrath, M.; Murray, S.A.; Chomérat, N.; Horiguchi, T. Marine Benthic Dinoflagellates-Unveiling their Worldwide Biodiversity; Senckenberg Gesellschaft für Naturkunde: Frankfurt, Germany, 2014; ISBN 978-3-510-61402-8. [Google Scholar]
- Sato, S.; Nishimura, T.; Uehara, K.; Sakanari, H.; Tawong, W.; Hariganeya, N.; Smith, K.; Rhodes, L.; Yasumoto, T.; Taira, Y.; et al. Phylogeography of Ostreopsis along west Pacific coast, with special reference to a novel clade from Japan. PLoS ONE 2011, 6, e27983. [Google Scholar] [CrossRef]
- Tawong, W.; Nishimura, T.; Sakanari, H.; Sato, S.; Yamaguchi, H.; Adachi, M. Distribution and molecular phylogeny of the dinoflagellate genus Ostreopsis in Thailand. Harmful Algae 2014, 37, 160–171. [Google Scholar] [CrossRef]
- Chomérat, N.; Bilien, G.; Derrien, A.; Henry, K.; Ung, A.; Viallon, J.; Darius, H.T.; Mahana iti Gatti, C.; Roué, M.; Hervé, F.; et al. Ostreopsis lenticularis Y. Fukuyo (Dinophyceae, Gonyaulacales) from French Polynesia (South Pacific Ocean): A revisit of its morphology, molecular phylogeny and toxicity. Harmful Algae 2019, 84, 95–111. [Google Scholar] [CrossRef]
- Gleibs, S.; Mebs, D. Distribution and sequestration of palytoxin in coral reef animals. Toxicon 1999, 37, 1521–1527. [Google Scholar] [CrossRef]
- Ukena, T.; Satake, M.; Usami, M.; Oshima, Y.; Naoki, H.; Fujita, T.; Kan, Y.; Yasumoto, T. Structure elucidation of ostreocin D, a palytoxin analog isolated from the dinoflagellate Ostreopsis siamensis. Biosci. Biotechnol. Biochem. 2001, 65, 2585–2588. [Google Scholar] [CrossRef]
- Thakur, L.K.; Jha, K.K. Palytoxin-induced acute respiratory failure. Respir. Med. Case Rep. 2017, 20, 4–6. [Google Scholar] [CrossRef]
- Onuma, Y.; Satake, M.; Ukena, T.; Roux, J.; Chanteau, S.; Rasolofonirina, N.; Ratsimaloto, M.; Naoki, H.; Yasumoto, T. Identification of putative palytoxin as the cause of clupeotoxism. Toxicon 1999, 37, 55–65. [Google Scholar] [CrossRef]
- Ciminiello, P.; Dell’Aversano, C.; Fattorusso, E.; Forino, M.; Magno, G.S.; Tartaglione, L.; Grillo, C.; Melchiorre, N. The Genoa 2005 Outbreak. Determination of Putative Palytoxin in Mediterranean Ostreopsis ovata by a New Liquid Chromatography Tandem Mass Spectrometry Method. Anal. Chem. 2006, 78, 6153–6159. [Google Scholar] [CrossRef]
- Tartaglione, L.; Dell’Aversano, C.; Mazzeo, A.; Forino, M.; Wieringa, A.; Ciminiello, P. Determination of Palytoxins in Soft Coral and Seawater from a Home Aquarium. Comparison between Palythoa—and Ostreopsis -Related Inhalatory Poisonings. Environ. Sci. Technol. 2016, 50, 1023–1030. [Google Scholar] [CrossRef]
- Sansoni, G.; Borghini, B.; Camici, G.; Casotti, M.; Righini, P.; Rustighi, C. Fioriture algali di Ostreopsis ovata (Gonyaulacales: Dinophyceae): Un problema emergente. Biol. Ambient. 2003, 17, 17–23. [Google Scholar]
- Illoul, H.; Hernández, F.R.; Vila, M.; Adjas, N.; Younes, A.A.; Bournissa, M.; Koroghli, A.; Marouf, N.; Rabia, S.; Ameur, F.L.K. The Genus Ostreopsis along the Algerian Coastal Waters (SW Mediterranean Sea) Associated with a Human Respiratory Intoxication Episode. Cryptogam. Algol. 2012, 33, 209–216. [Google Scholar] [CrossRef]
- Ferreira, C.E.L. Sea Urchins Killed by Toxic Algae. JMBA Glob. Mar. Environ. 2006, 3, 22–23. [Google Scholar]
- Nascimento, S.M.; Corrêa, E. V.; Menezes, M.; Varela, D.; Paredes, J.; Morris, S. Growth and toxin profile of Ostreopsis cf. ovata (Dinophyta) from Rio de Janeiro, Brazil. Harmful Algae 2012, 13, 1–9. [Google Scholar] [CrossRef]
- Shears, N.T.; Ross, P.M. Blooms of benthic dinoflagellates of the genus Ostreopsis; an increasing and ecologically important phenomenon on temperate reefs in New Zealand and worldwide. Harmful Algae 2009, 8, 916–925. [Google Scholar] [CrossRef]
- Rossini, G.P.; Hess, P. Phycotoxins: Chemistry, mechanisms of action and shellfish poisoning. In Molecular, Clinical and Environmental Toxicology; Clinical Toxicology; Luch, A., Ed.; Birkhäuser: Basel, Switzerland, 2010; Volume 2, pp. 65–122. [Google Scholar] [CrossRef]
- Nascimento, S.M.; França, J.V.; Gonçalves, J.E.A.A.; Ferreira, C.E.L.L. Ostreopsis cf. ovata (Dinophyta) bloom in an equatorial island of the Atlantic Ocean. Mar. Pollut. Bull. 2012, 64, 1074–1078. [Google Scholar] [CrossRef]
- Tibiriçá, C.E.J.A.; Leite, I.P.; Batista, T.V. V.; Fernandes, L.F.; Chomérat, N.; Herve, F.; Hess, P.; Mafra, L.L. Ostreopsis cf. ovata Bloom in Currais, Brazil: Phylogeny, Toxin Profile and Contamination of Mussels and Marine Plastic Litter. Toxins (Basel) 2019, 11, 446. [Google Scholar] [CrossRef]
- De’Carli, G.d.A.e.L. Distribuição e Abundância de Dinoflagelados Epi-Bentônicos na Costa Nordeste do Brasil. Master’s Thesis, Universidade Federal do Estado do Rio de Janeiro, Rio de Janeiro, Brazil, 2014. [Google Scholar]
- Besada, E.G.; Loeblich, L.A.; Loeblich, A.R., III. Observations on Tropical, Benthic Dinoflagellates from Ciguatera-Endemic Areas: Coolia, Gambierdiscus, and Ostreopsis. Bull. Mar. Sci. 1982, 32, 723–735. [Google Scholar]
- Zhang, H.; Lu, S.; Li, Y.; Cen, J.; Wang, H.; Li, Q.; Nie, X. Morphology and molecular phylogeny of Ostreopsis cf. ovata and O. lenticularis (Dinophyceae) from Hainan Island, South China Sea. Phycol. Res. 2018, 66, 3–14. [Google Scholar] [CrossRef]
- Penna, A.; Fraga, S.; Battocchi, C.; Casabianca, S.; Giacobbe, M.G.; Riobó, P.; Vernesi, C. A phylogeographical study of the toxic benthic dinoflagellate genus Ostreopsis Schmidt. J. Biogeogr. 2010, 37, 830–841. [Google Scholar] [CrossRef]
- Gómez, F.; Qiu, D.; Lopes, R.M.; Lin, S. Morphological and molecular characterization of the toxic dinoflagellate Ostreopsis cf. ovata (Dinophyta) from Brazil (South Atlantic Ocean). Rev. Biol. Trop. 2017, 65, 1022–1032. [Google Scholar] [CrossRef][Green Version]
- Abdennadher, M.; Zouari, A.B.; Sahnoun, W.F.; Alverca, E.; Penna, A.; Hamza, A. Ostreopsis cf. ovata in the Gulf of Gabès (south-eastern Mediterranean Sea): Morphological, molecular and ecological characterization. Harmful Algae 2017, 63, 56–67. [Google Scholar] [CrossRef]
- Carnicer, O.; Guallar, C.; Andree, K.B.; Diogène, J.; Fernández-Tejedor, M. Ostreopsis cf. ovata dynamics in the NW Mediterranean Sea in relation to biotic and abiotic factors. Environ. Res. 2015. [Google Scholar] [CrossRef]
- Scalco, E.; Brunet, C.; Marino, F.; Rossi, R.; Soprano, V.; Zingone, A.; Montresor, M. Growth and toxicity responses of Mediterranean Ostreopsis cf. ovata to seasonal irradiance and temperature conditions. Harmful Algae 2012, 17, 25–34. [Google Scholar] [CrossRef]
- Accoroni, S.; Romagnoli, T.; Pichierri, S.; Colombo, F.; Totti, C. Morphometric analysis of Ostreopsis cf. ovata cells in relation to environmental conditions and bloom phases. Harmful Algae 2012. [Google Scholar] [CrossRef]
- Selina, M.S.; Levchenko, E.V. Species composition and morphology of dinoflagellates (Dinophyta) of epiphytic assemblages of Peter the Great Bay in the Sea of Japan. Russ. J. Mar. Biol. 2011, 37, 23–32. [Google Scholar] [CrossRef]
- Honsell, G.; De Bortoli, M.; Boscolo, S.; Dell’Aversano, C.; Battocchi, C.; Fontanive, G.; Penna, A.; Berti, F.; Sosa, S.; Yasumoto, T.; et al. Harmful dinoflagellate Ostreopsis cf. ovata Fukuyo: Detection of ovatoxins in field samples and cell immunolocalization using antipalytoxin antibodies. Environ. Sci. Technol. 2011, 45, 7051–7059. [Google Scholar] [CrossRef]
- Monti, M.; Minocci, M.; Beran, A.; Iveša, L. First record of Ostreopsis cf. ovata on macroalgae in the Northern Adriatic Sea. Mar. Pollut. Bull. 2007, 54, 598–601. [Google Scholar] [CrossRef]
- Aligizaki, K.; Nikolaidis, G. The presence of the potentially toxic genera Ostreopsis and Coolia (Dinophyceae) in the North Aegean Sea, Greece. Harmful Algae 2006, 5, 717–730. [Google Scholar] [CrossRef]
- Leaw, C.P.; Lim, P.T.; Ahmad, A.; Usup, G. Genetic diversity of Ostreopsis ovata (Dinophyceae) from Malaysia. Mar. Biotechnol. 2001, 3, 246–255. [Google Scholar] [CrossRef]
- Chang, F.H.; Shimizu, Y.; Hay, B.; Stewart, R.; Mackay, G.; Tasker, R. Three recently recorded Ostreopsis spp. (Dinophyceae) in New Zealand: Temporal and regional distribution in the upper North Island from 1995 to 1997. N. Z. J. Mar. Freshw. Res. 2000, 34, 29–39. [Google Scholar] [CrossRef]
- Tognetto, L.; Bellato, S.; Moro, I.; Andreoli, C. Occurrence of Ostreopsis ovata (Dinophyceae) in the Tyrrhenian Sea during Summer 1994. Bot. Mar. 1995, 38, 291–295. [Google Scholar] [CrossRef]
- Mendes, M.C.Q.; Nunes, J.M.C.; Menezes, M.; Fraga, S.; Rodríguez, F.; Vázquez, J.A.; Blanco, J.; Franco, J.M.; Riobó, P. Toxin production, growth kinetics and molecular characterization of Ostreopsis cf. ovata isolated from Todos os Santos Bay, tropical southwestern Atlantic. Toxicon 2017, 138, 18–30. [Google Scholar] [CrossRef]
- Tartaglione, L.; Dello Iacovo, E.; Mazzeo, A.; Casabianca, S.; Ciminiello, P.; Penna, A.; Dell’Aversano, C. Variability in Toxin Profiles of the Mediterranean Ostreopsis cf. ovata and in Structural Features of the Produced Ovatoxins. Environ. Sci. Technol. 2017, 51, 13920–13928. [Google Scholar] [CrossRef]
- Pelin, M.; Forino, M.; Brovedani, V.; Tartaglione, L.; Dell’Aversano, C.; Pistocchi, R.; Poli, M.; Sosa, S.; Florio, C.; Ciminiello, P.; et al. Ovatoxin-a, A Palytoxin Analogue Isolated from Ostreopsis cf. ovata Fukuyo: Cytotoxic Activity and ELISA Detection. Environ. Sci. Technol. 2016, 50, 1544–1551. [Google Scholar] [CrossRef]
- Brissard, C.; Herrenknecht, C.; Séchet, V.; Hervé, F.; Pisapia, F.; Harcouet, J.; Lémée, R.; Chomérat, N.; Hess, P.; Amzil, Z. Complex toxin profile of French Mediterranean Ostreopsis cf. ovata strains, seafood accumulation and ovatoxins prepurification. Mar. Drugs 2014, 12, 2851–2876. [Google Scholar] [CrossRef]
- García-Altares, M.; Tartaglione, L.; Dell’Aversano, C.; Carnicer, O.; de la Iglesia, P.; Forino, M.; Diogène, J.; Ciminiello, P. The novel ovatoxin-g and isobaric palytoxin (so far referred to as putative palytoxin) from Ostreopsis cf. ovata (NW Mediterranean Sea): Structural insights by LC-high resolution MS(n.). Anal. Bioanal. Chem. 2015, 407, 1191–1204. [Google Scholar] [CrossRef]
- Pezzolesi, L.; Pistocchi, R.; Fratangeli, F.; Dell’Aversano, C.; Dello Iacovo, E.; Tartaglione, L. Growth dynamics in relation to the production of the main cellular components in the toxic dinoflagellate Ostreopsis cf. ovata. Harmful Algae 2014, 36, 1–10. [Google Scholar] [CrossRef]
- Vila, M.; Abós-Herràndiz, R.; Isern-Fontanet, J.; Àlvarez, J.; Berdalet, E. Establishing the link between Ostreopsis cf. ovata blooms and human health impacts using ecology and epidemiology. Sci. Mar. 2016, 80, 107–115. [Google Scholar] [CrossRef]
- Guillard, R.R.J. Culture methods. In Manual on Harmful Marine Microalgae; Hallegraeff, G.M., Anderson, D.M., Cembella, A., Eds.; UNESCO: Paris, France, 1995; pp. 45–56. [Google Scholar]
- Kofoid, C.A. On Peridinium steini Jorgensen, with a note on the nomenclature of the skeleton of the Peridinidae. Arch. Protistenk. 1909, 16, 25–47. [Google Scholar]
- Scholin, C.A.; Anderson, D.M. Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). I. RFLP analysis of SSU rRNA genes. J. Phycol. 1994, 30, 744–754. [Google Scholar] [CrossRef]
- Litaker, R.W.; Vandersea, M.W.; Kibler, S.R.; Reece, K.S.; Stokes, N.A.; Steidinger, K.A.; Millie, D.F.; Bendis, B.J.; Pigg, R.J.; Tester, P.A. Identification of Pfiesteria piscicida (Dinophyceae) and Pfiesteria-like organisms using internal transcribed spacer-specific PCR assays. J. Phycol. 2003, 39, 754–761. [Google Scholar] [CrossRef]
- Chinain, M.; Faust, M.A.; Pauillac, S. Morphology and molecular analyses of three toxic species of Gambierdiscus (Dinophyceae): G. pacificus, sp. nov., G. australes, sp. nov., and G. polynesiensis, sp. nov. J. Phycol. 1999, 35, 1282–1296. [Google Scholar] [CrossRef]
- Hall, T.A. BIOEDIT: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Woodman, M.E.; Savage, C.R.; Arnold, W.K.; Stevenson, B. Direct PCR of intact bacteria (colony PCR). Curr. Protoc. Microbiol. 2016, 42, A.3D.1–A.3D.7. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]







| Sample Location | DV | W | DV/W | Origin | Reference |
|---|---|---|---|---|---|
| Rio Grande do Norte, Brazil (strain UNR–10) | 47.4–64.0 (53.7 ± 5.7) | 37.4–57.7 (42.3 ± 7.8) | 1.1–1.4 (1.3 ± 0.1) | Culture | Current study |
| Fernando de Noronha Island, Brazil | 30.0–57.5 (45.1 ± 6.6) | 17.5–37.5 (26.3 ± 4.8) | 1.3–2.3 (1.7 ± 0.2) | Field | Current study |
| Forte, Bahia, Brazil | 36.6–59.5 (49.0 ± 5.2) | 21.0–42.6 (29.3 ± 4.5) | 1.4–2.1 (1.7 ± 0.1) | Field | Current study |
| Penha, Bahia, Brazil | 48.4–85.1 (63.7 ± 7.3) | 31.3–60.7 (45.7 ± 6.1) | 1.2–1.7 (1.4 ± 0.1) | Field | Current study |
| Trindade Island, Brazil | 30.0–82.5 (58.7 ± 11.2) | 12.5–58.8 (29.0 ± 9.2) | 1.2–3.2 (2.1 ± 0.4) | Field | Current study |
| Armação de Búzios, Rio de Janeiro, Brazil | 39.2–65.0 (53.6 ± 6.2) | 24.1–58.8 (31.8 ± 5.8) | 0.9–2.0 (1.7 ± 0.2) | Field | Current study |
| Armação de Búzios, Rio de Janeiro, Brazil | 28.6–62.3 (49.7 ± 6.0) | 16.6–41.6 (29.8 ± 4.9) | 1.4–2.2 (1.7 ± 0.1) | Field | Current study |
| Armação de Búzios, Rio de Janeiro, Brazil (strain UNR–05) | 29.8–61.4 (45.0 ± 6.4) | 21.1–45.2 (32.5 ± 5.1) | 1.2–1.7 (1.4 ± 0.1) | Culture | Current study |
| Arraial do Cabo, Rio de Janeiro, Brazil (strain LCA–B7) | 33.0–68.0 (53.0 ± 10.8) | 18.0–45.0 (31.1 ± 8.2) | 1.4–2.5 (1.7 ± 0.2) | Culture | Nascimento et al. [26] |
| Arraial do Cabo, Rio de Janeiro, Brazil (strain LCAE7) | 40.0–62.0 (52.6 ± 5.8) | 20.0–48.0 (34.4 ± 6.1) | 1.2–2.0 (1.6 ± 0.2) | Culture | Nascimento et al. [26] |
| Arraial do Cabo, Rio de Janeiro, Brazil | 40.0–65.0 (51.3 ± 7.5) | 18.0–45.0 (28.3 ± 6.7) | 1.4–2.5 (1.9 ± 0.3) | Field | Nascimento et al. [26] |
| Saint Paul’s Rocks, Brazil | 45.9–65.6 (55.4 ± 4.1) | 27.5–45.6 (35.7 ± 3.9) | 1.3–1.9 (1.6 ± 0.1) | Field | Nascimento et al. [29] |
| Currais Archipelago, Paraná, Brazil (strains LM062, LM086, LM129, LM130) | 23.7–60.1 (40.8 ± 8.7) | 15.4–48.9 (31 ± 7.5) | 1.04–1.68 (1.33 ± 0.13) | Culture | Tibiriçá et al. [30] |
| Currais Archipelago, Paraná, Brazil | 29.9–65.9 (49.9 ± 6.6) | 17.1–45.9 (32.6 ± 5.9 | 1.31–1.79 (1.53 ± 0.12) | Field | Tibiriçá et al. [30] |
| Ubatuba, São Paulo, Brazil | 35–65 (55.1) | 20–40 (32.6) | (1.69) | Field | Gómez et al. [35] |
| Hainan Island, China (strains 1S1D2, 1S1D4, 1S1D6) | 39.9–56.4 (47.5 ± 3.1) | 30.4–47.4 (37.1 ± 3.3) | 1.1–1.4 (1.3 ± 0.1) | Culture | Zhang et al. [33] |
| Gulf of Gabès, Mediterranean Sea (strains Oso.1, Oso.2, Oso.3, Oso.4, Oso.5, Oso.6, Oso.7) | 27–65 | 19–57 | – | Culture | Abdennadher et al. [36] |
| Heron Island, Australia (strain HER27) | 30.2–48.3 (37.7 ± 4.3) | 21.9–37.5 (28.7 ± 3.7) | 1.1–1.8 (1.3) | Culture | Verma et al. [9] |
| Ebro Delta, Spain, NW Mediterranean Sea | 21.24–76.9 (54.50 ± 6.80) | 15.57–51.0 (33.05 ± 5.51) | – | Field | Carnicer et al. [37] |
| Iberian Peninsula, Spain, Atlantic Ocean | 54.8–84.3 (69.6 ± 7) | 30.3–62.0 (44.7 ± 6.3) | 1.2–1.9 (1.6 ± 0.1) | Field | David et al. [12] |
| Gulf of Naples, Tyrrhenian Sea, Mediterranean (strain D483) | (49.5 ± 5.1) | (33.3 ± 4.7) | – | Culture | Scalco et al. [38] |
| Portonovo, Adriatic Sea, Mediterranean (strain CBA–T) | (49.2 ± 4.1) | (33.7 ± 4.8) | – | Culture | Scalco et al. [38] |
| Gulf of Trieste, Adriatic Sea, Mediterranean (strain OS2T) | (46.8 ± 6.1) | (32.8 ± 5.4) | – | Culture | Scalco et al. [38] |
| Conero Riviera, Adriatic Sea, Mediterranean | 18.7–75.0 | 12.5–60.0 | – | Field | Accoroni et al. [39] |
| Ussuriiskii Bay (Peter the Great Bay) Sea of Japan | 36–60 (49.4 ± 6.4) | 24–45 (29.6 ± 5.3) | – | Field | Selina and Levchenko [40] |
| Gulf of Trieste, Italy, Adriatic Sea, Mediterranean | 48–65 | 31–46 | 1.33–1.74 | Field | Honsell et al. [41] |
| Subogata, Otsuki Town, Kochi, Japan (strain s0726) | (28.1 ± 2.6) | (21.2 ± 2.8) | – | Culture | Sato et al. [14] |
| Gulf of Trieste, Italy, Adriatic Sea, Mediterranean | 29.6–70.8 (55.3 ± 8.0) | 18.5–53.1 (36.4 ± 6.4) | – | Field | Monti et al. [42] |
| Rovinj, Croatia, Adriatic Sea, Mediterranean | 33.3–66.6 (54.8 ± 7.1) | 18.5–44.4 (34.3 ± 4.7) | – | Field | Monti et al. [42] |
| North Aegen Sea, Mediterranean | 26.2–61.9 | 13.1–47.6 | – | Field | Aligizaki and Nikolaidis [43] |
| Port Dickson, Malaysia (strains OvPD04, OvPD06,OvPD07) | 33–41 | 24–34 | – | Culture | Leaw et al. [44] |
| Kota Kinabalu, Malaysia (strains OvSA02, OvSA04, OvSA06, OvSA09, OvSA10) | 32–55 | 22–39 | – | Culture | Leaw et al. [44] |
| Pulau Redang, Malaysia (strains OvPR01, OvPR02, OvPR03, OvPR04) | 44–48 | 33–37 | – | Culture | Leaw et al. [44] |
| Rangaunu Harbour, New Zealand | 38–50 | 25–35 | – | Field | Chang et al. [45] |
| Civitavecchia, Italy, Tyrrhenian Sea, Mediterranean | 34.2–66.6 (55.7 ± 6.1) | 25.2–39.6 (31.9 ± 4.1) | – | Field | Tognetto et al. [46] |
| French Polynesia, New Caledonia and Ryukyu Islands | 50–56 | 25–35 | – | Field | Fukuyo [2] |
| Samples | OVTX-a | OVTX-b | OVTX-c | OVTX-d | OVTX-e |
|---|---|---|---|---|---|
| Strain UNR-03 | 58.5 (20.9) | 40.0 (14.3) | 0.5 (0.2) | 0.4 (0.1) | 0.6 (0.2) |
| Strain UNR-05 | 68.2 (20.0) | 31.7 (9.3) | 0.03 (<0.1) | 0.02 (<0.1) | 0.05 (<0.1) |
| Armação dos Búzios bloom | 61.0 | 26.7 | 4.1 | 2.5 | 5.7 |
| Arraial do Cabo bloom | 64.7 | 28.3 | 1.3 | 2.2 | 3.4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nascimento, S.M.; Neves, R.A.F.; De’Carli, G.A.L.; Borsato, G.T.; da Silva, R.A.F.; Melo, G.A.; de Morais, A.M.; Cockell, T.C.; Fraga, S.; Menezes-Salgueiro, A.D.; et al. Ostreopsis cf. ovata (Dinophyceae) Molecular Phylogeny, Morphology, and Detection of Ovatoxins in Strains and Field Samples from Brazil. Toxins 2020, 12, 70. https://doi.org/10.3390/toxins12020070
Nascimento SM, Neves RAF, De’Carli GAL, Borsato GT, da Silva RAF, Melo GA, de Morais AM, Cockell TC, Fraga S, Menezes-Salgueiro AD, et al. Ostreopsis cf. ovata (Dinophyceae) Molecular Phylogeny, Morphology, and Detection of Ovatoxins in Strains and Field Samples from Brazil. Toxins. 2020; 12(2):70. https://doi.org/10.3390/toxins12020070
Chicago/Turabian StyleNascimento, Silvia M., Raquel A. F. Neves, Gabriela A. L. De’Carli, Geovanna T. Borsato, Rodrigo A. F. da Silva, Guilherme A. Melo, Agatha M. de Morais, Thais C. Cockell, Santiago Fraga, Adriana D. Menezes-Salgueiro, and et al. 2020. "Ostreopsis cf. ovata (Dinophyceae) Molecular Phylogeny, Morphology, and Detection of Ovatoxins in Strains and Field Samples from Brazil" Toxins 12, no. 2: 70. https://doi.org/10.3390/toxins12020070
APA StyleNascimento, S. M., Neves, R. A. F., De’Carli, G. A. L., Borsato, G. T., da Silva, R. A. F., Melo, G. A., de Morais, A. M., Cockell, T. C., Fraga, S., Menezes-Salgueiro, A. D., Mafra, L. L., Hess, P., & Salgueiro, F. (2020). Ostreopsis cf. ovata (Dinophyceae) Molecular Phylogeny, Morphology, and Detection of Ovatoxins in Strains and Field Samples from Brazil. Toxins, 12(2), 70. https://doi.org/10.3390/toxins12020070







