Genomic Analysis of Clostridium perfringens BEC/CPILE-Positive, Toxinotype D and E Strains Isolated from Healthy Children
Abstract
1. Introduction
2. Results
2.1. Genomic Description of Newly Sequenced C. perfringens Isolates
2.2. Comparisons of Toxin Gene Sequences
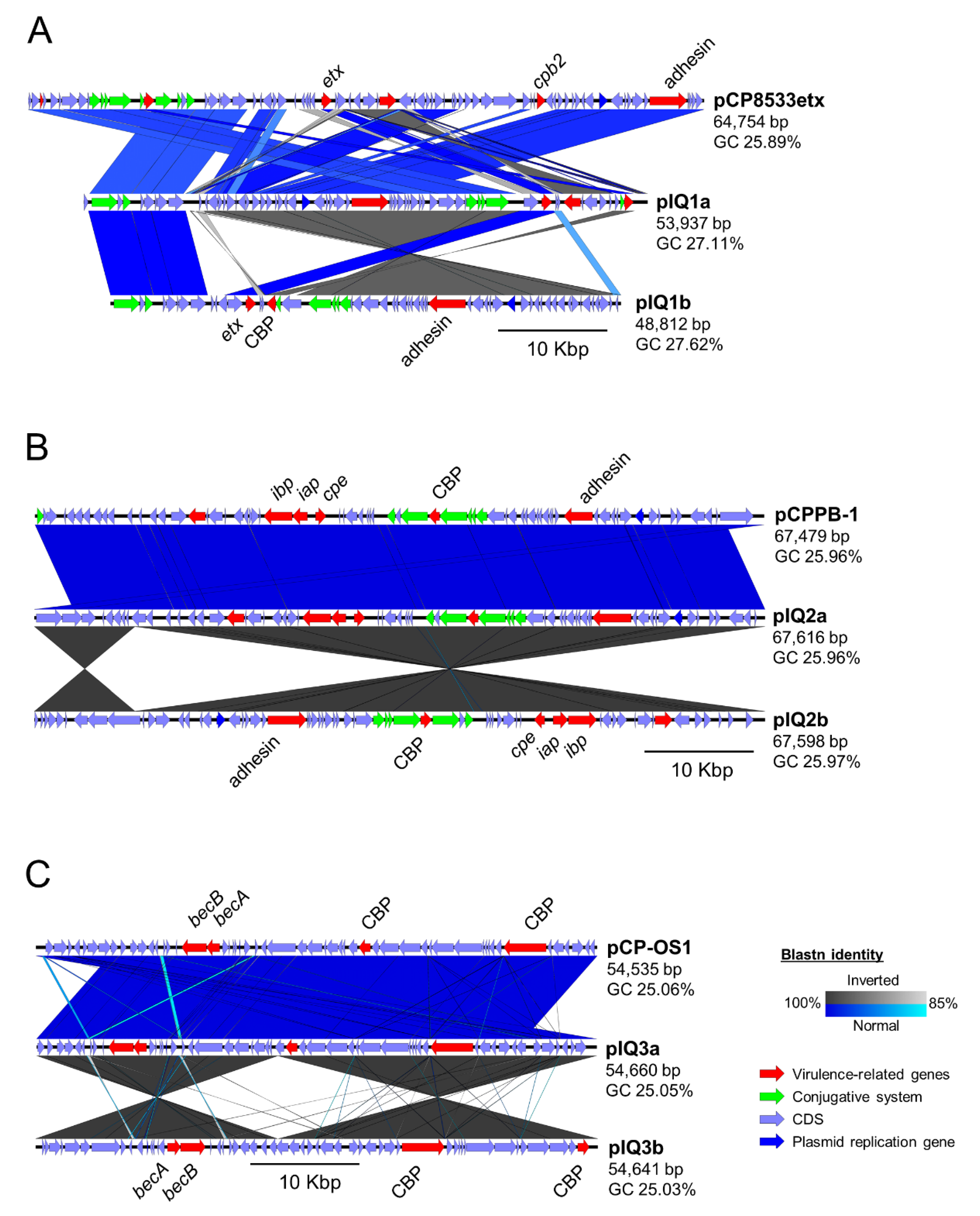
2.3. Comparative Study of Toxin-Encoding Plasmids
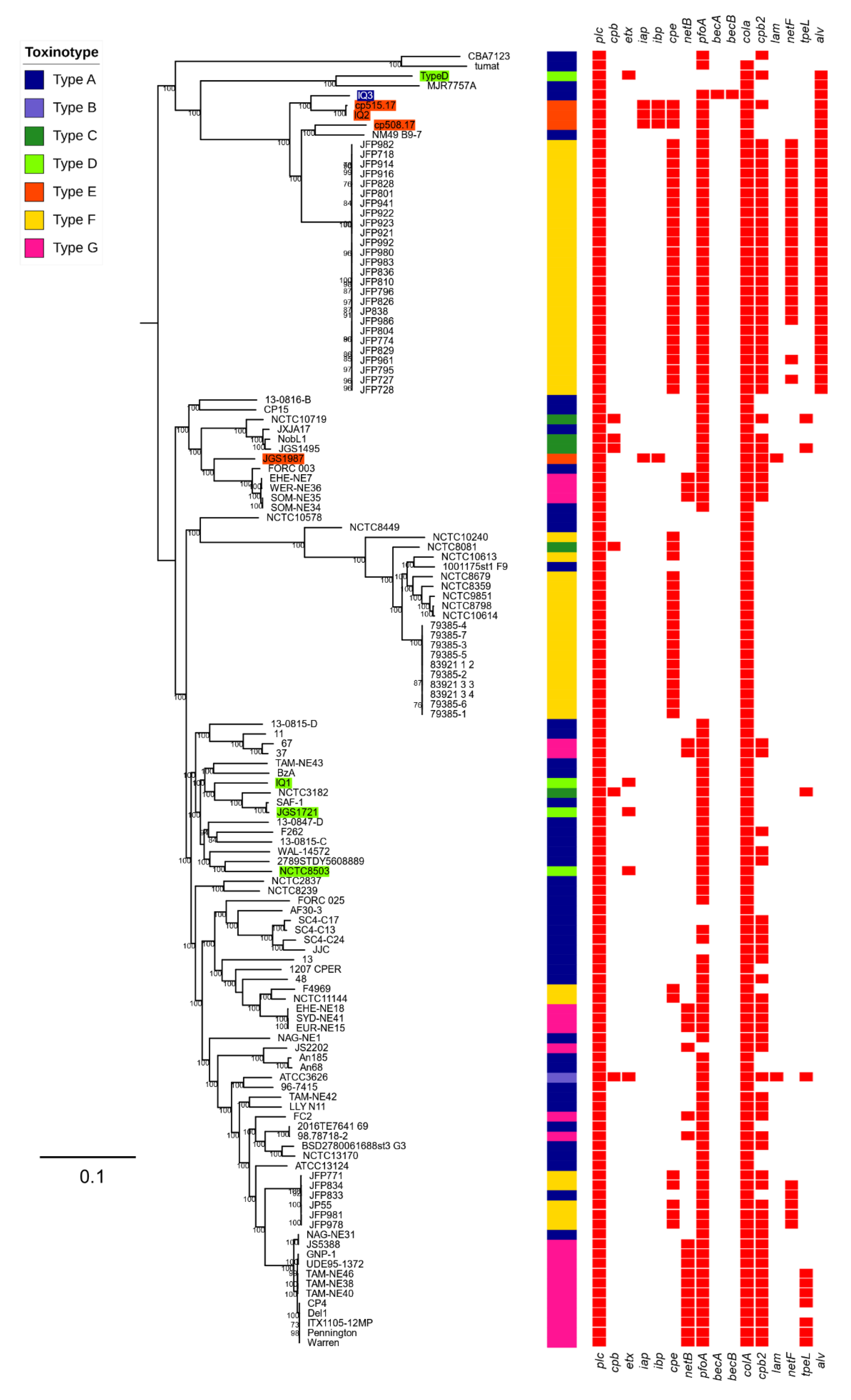
2.4. Phylogenetic Analysis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Ethics Declaration
5.2. Sample Collection
5.3. Bacterial Isolation, Identification and Storage
5.4. Genomic DNA Extraction and Whole Genome Sequencing
5.5. Genome Assembly and Annotation
5.6. Genomic Analysis
5.7. Phylogenetic Analysis
5.8. Data Deposition
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kiu, R.; Hall, L.J. An update on the human and animal enteric pathogen Clostridium perfringens. Emerg. Microbes Infect. 2018, 7, 141. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, M.; Mainil, J.G.; Linden, A. Cattle enterotoxaemia and Clostridium perfringens: Description, diagnosis and prophylaxis. Vet. Rec. 2010, 167, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Prescott, J.F.; Parreira, V.R.; Mehdizadeh Gohari, I.; Lepp, D.; Gong, J. The pathogenesis of necrotic enteritis in chickens: What we know and what we need to know: A review. Avian Pathol. 2016, 45, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Awad, M.M.; Ellemor, D.M.; Boyd, R.L.; Emmins, J.J.; Rood, J.I. Synergistic effects of alpha-toxin and perfringolysin O in Clostridium perfringens-mediated gas gangrene. Infect. Immun. 2001, 69, 7904–7910. [Google Scholar] [CrossRef] [PubMed]
- Rood, J.I.; Adams, V.; Lacey, J.; Lyras, D.; McClane, B.A.; Melville, S.B.; Moore, R.J.; Popoff, M.R.; Sarker, M.R.; Songer, J.G. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe 2018. [Google Scholar] [CrossRef] [PubMed]
- Petit, L.; Gibert, M.; Popoff, M. Clostridium perfringens: Toxinotype and genotype. Trends Microbiol. 1999, 7, 104–110. [Google Scholar] [CrossRef]
- Layana, J.E.; Fernandez Miyakawa, M.E.; Uzal, F.A. Evaluation of different fluids for detection of Clostridium perfringens type D epsilon toxin in sheep with experimental enterotoxemia. Anaerobe 2006, 12, 204–206. [Google Scholar] [CrossRef]
- Filho, E.J.; Carvalho, A.U.; Assis, R.A.; Lobato, F.F.; Rachid, M.A.; Carvalho, A.A.; Ferreira, P.M.; Nascimento, R.A.; Fernandes, A.A.; Vidal, J.E.; et al. Clinicopathologic features of experimental Clostridium perfringens type D enterotoxemia in cattle. Vet. Pathol. 2009, 46, 1213–1220. [Google Scholar] [CrossRef]
- Muth, O.H.; Morrill, D.R. Control of enterotoxemia (pulpy kidney disease) in lambs by the use of alum precipitated toxoid. Am. J. Vet. Res. 1946, 7, 355–357. [Google Scholar]
- Fernandez-Miyakawa, M.E.; Sayeed, S.; Fisher, D.J.; Poon, R.; Adams, V.; Rood, J.I.; McClane, B.A.; Saputo, J.; Uzal, F.A. Development and application of an oral challenge mouse model for studying Clostridium perfringens type D infection. Infect. Immun. 2007, 75, 4282–4288. [Google Scholar] [CrossRef]
- Baker, I.; Van Dreumel, A.; PALMER, N. Chapter I, The Alimentary System. Pathol. Domest. Anim. 1993, 2, 141–144. [Google Scholar]
- Finnie, J.W.; Blumbergs, P.C.; Manavis, J. Neuronal damage produced in rat brains by Clostridium perfringens type D epsilon toxin. J. Comp. Pathol. 1999, 120, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Tamai, E.; Ishida, T.; Miyata, S.; Matsushita, O.; Suda, H.; Kobayashi, S.; Sonobe, H.; Okabe, A. Accumulation of Clostridium perfringens epsilon-toxin in the mouse kidney and its possible biological significance. Infect. Immun. 2003, 71, 5371–5375. [Google Scholar] [CrossRef] [PubMed]
- Smedley, J.G., 3rd; Fisher, D.J.; Sayeed, S.; Chakrabarti, G.; McClane, B.A. The enteric toxins of Clostridium perfringens. Rev. Physiol. Biochem. Pharmacol. 2004, 152, 183–204. [Google Scholar] [CrossRef] [PubMed]
- Sayeed, S.; Li, J.; McClane, B.A. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect. Immun. 2007, 75, 2391–2398. [Google Scholar] [CrossRef]
- McClane, B.A.; Uzal, F.A.; Miyakawa, M.E.F.; Lyerly, D.; Wilkins, T. The enterotoxic Clostridia. In The Prokaryotes: A Handbook on the Biology of Bacteria; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebandt, E., Eds.; Springer: Singapore, 2006; Volume 4, pp. 698–752. [Google Scholar]
- Uzal, F.A.; Vidal, J.E.; McClane, B.A.; Gurjar, A.A. Clostridium Perfringens Toxins Involved in Mammalian Veterinary Diseases. Open Toxinol. J. 2010, 2, 24–42. [Google Scholar] [CrossRef]
- Li, J.; Miyamoto, K.; McClane, B.A. Comparison of virulence plasmids among Clostridium perfringens type E isolates. Infect. Immun. 2007, 75, 1811–1819. [Google Scholar] [CrossRef]
- Yonogi, S.; Matsuda, S.; Kawai, T.; Yoda, T.; Harada, T.; Kumeda, Y.; Gotoh, K.; Hiyoshi, H.; Nakamura, S.; Kodama, T.; et al. BEC, a novel enterotoxin of Clostridium perfringens found in human clinical isolates from acute gastroenteritis outbreaks. Infect. Immun. 2014, 82, 2390–2399. [Google Scholar] [CrossRef]
- Matsuda, A.; Aung, M.S.; Urushibara, N.; Kawaguchiya, M.; Sumi, A.; Nakamura, M.; Horino, Y.; Ito, M.; Habadera, S.; Kobayashi, N. Prevalence and Genetic Diversity of Toxin Genes in Clinical Isolates of Clostridium perfringens: Coexistence of Alpha-Toxin Variant and Binary Enterotoxin Genes (bec/cpile). Toxins 2019, 11, 326. [Google Scholar] [CrossRef]
- Diancourt, L.; Sautereau, J.; Criscuolo, A.; Popoff, M.R. Two Clostridium perfringens Type E Isolates in France. Toxins 2019, 11, 138. [Google Scholar] [CrossRef]
- Geoffroy, C.; Mengaud, J.; Alouf, J.E.; Cossart, P. Alveolysin, the thiol-activated toxin of Bacillus alvei, is homologous to listeriolysin O, perfringolysin O, pneumolysin, and streptolysin O and contains a single cysteine. J. Bacteriol. 1990, 172, 7301–7305. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Adams, V.; Bannam, T.L.; Miyamoto, K.; Garcia, J.P.; Uzal, F.A.; Rood, J.I.; McClane, B.A. Toxin plasmids of Clostridium perfringens. Microbiol. Mol. Biol. Rev. 2013, 77, 208–233. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Yumine, N.; Mimura, K.; Nagahama, M.; Li, J.; McClane, B.A.; Akimoto, S. Identification of novel Clostridium perfringens type E strains that carry an iota toxin plasmid with a functional enterotoxin gene. PLoS ONE 2011, 6, e20376. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Li, J.; Sayeed, S.; Akimoto, S.; McClane, B.A. Sequencing and diversity analyses reveal extensive similarities between some epsilon-toxin-encoding plasmids and the pCPF5603 Clostridium perfringens enterotoxin plasmid. J. Bacteriol. 2008, 190, 7178–7188. [Google Scholar] [CrossRef] [PubMed]
- Gohari, I.M.; Kropinski, A.M.; Weese, S.J.; Whitehead, A.E.; Parreira, V.R.; Boerlin, P.; Prescott, J.F. NetF-producing Clostridium perfringens: Clonality and plasmid pathogenicity loci analysis. Infect. Genet. Evol. 2017, 49, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Brynestad, S.; Sarker, M.R.; McClane, B.A.; Granum, P.E.; Rood, J.I. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 2001, 69, 3483–3487. [Google Scholar] [CrossRef] [PubMed]
- Thelestam, M.; Alouf, J.E.; Geoffroy, C.; Mollby, R. Membrane-damaging action of alveolysin from Bacillus alvei. Infect. Immun. 1981, 32, 1187–1192. [Google Scholar] [PubMed]
- Rossjohn, J.; Gilbert, R.J.; Crane, D.; Morgan, P.J.; Mitchell, T.J.; Rowe, A.J.; Andrew, P.W.; Paton, J.C.; Tweten, R.K.; Parker, M.W. The molecular mechanism of pneumolysin, a virulence factor from Streptococcus pneumoniae. J. Mol. Biol. 1998, 284, 449–461. [Google Scholar] [CrossRef]
- Sim, K.; Shaw, A.G.; Randell, P.; Cox, M.J.; McClure, Z.E.; Li, M.S.; Haddad, M.; Langford, P.R.; Cookson, W.O.; Moffatt, M.F.; et al. Dysbiosis anticipating necrotizing enterocolitis in very premature infants. Clin. Infect. Dis. 2015, 60, 389–397. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Nunez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Lawley, T.D.; Walker, A.W. Intestinal colonization resistance. Immunology 2013, 138, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kiu, R.; Caim, S.; Alcon-Giner, C.; Belteki, G.; Clarke, P.; Pickard, D.; Dougan, G.; Hall, L.J. Preterm Infant-Associated Clostridium tertium, Clostridium cadaveris, and Clostridium paraputrificum Strains: Genomic and Evolutionary Insights. Genome Biol. Evol. 2017, 9, 2707–2714. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Chalita, M.; Ha, S.M.; Na, S.I.; Yoon, S.H.; Chun, J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez, R.L.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Goker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinforma. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Hunt, M.; Silva, N.D.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; Mather, A.E.; Sanchez-Buso, L.; Page, A.J.; Parkhill, J.; Keane, J.A.; Harris, S.R. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom 2017, 3, e000131. [Google Scholar] [CrossRef]
- Kiu, R.; Caim, S.; Alexander, S.; Pachori, P.; Hall, L.J. Probing Genomic Aspects of the Multi-Host Pathogen Clostridium perfringens Reveals Significant Pangenome Diversity, and a Diverse Array of Virulence Factors. Front. Microbiol. 2017, 8, 2485. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. ABRicate: Mass screening of contigs for antimicrobial and virulence genes. Available online: https://github.com/tseemann/abricate (accessed on 2 August 2019).
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Seemann, T.; Klotzl, F.; Page, A.J. Snp-dists: Pairwise SNP distance matrix from a FASTA sequence alignment. Available online: https://github.com/tseemann/snp-dists (accessed on 2 August 2019).
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef]
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016, 2, e000056. [Google Scholar] [CrossRef]
| Strain/Genome | C. perfringens IQ1 | C. perfringens IQ2 | C. perfringens IQ3 |
|---|---|---|---|
| Genome size (bp) | 3,563,782 | 3,438,840 | 3,518,141 |
| Sequencing coverage | 195× | 216× | 212× |
| Sequencing platform | Illumina HiSeq 2500 | Illumina HiSeq 2500 | Illumina HiSeq 2500 |
| No. of contigs | 41 | 15 | 20 |
| Genes | 3320 | 3199 | 3299 |
| CDS | 3219 | 3097 | 3196 |
| tRNAs | 91 | 92 | 92 |
| N50 length (bp) | 452,560 | 2,192,929 | 2,217,274 |
| GC (%) | 27.98 | 28.03 | 28.00 |
| ANI (%) 1 | 98.35 | 96.89 | 97.01 |
| dDDH (%) 1 | 87.40 | 74.50 | 75.30 |
| Toxinotype | D | E | A (BEC-positive) |
| ENA sample accession | SAMEA5818795 | SAMEA5818796 | SAMEA5818797 |
| ENA assembly accession | GCA_902459455 | GCA_902459435 | GCA_902459425 |
| Strain | Type | SNP | Sequence Similarity (%) | ||||
|---|---|---|---|---|---|---|---|
| Pos.726 | ATCC3626 | JGS1721 | NCTC8503 | IQ1 | Type D | ||
| ATCC3626 | B | A | 100.00 | 99.90 | 99.90 | 99.90 | 100.00 |
| JGS1721 | D | G | 100.00 | 100.00 | 100.00 | 99.90 | |
| NCTC8503 | D | G | 100.00 | 100.00 | 99.90 | ||
| IQ1 | D | G | 100.00 | 99.90 | |||
| Type D | D | A | 100.00 | ||||
| Strain | iap Sequence Similarity (%) | ibp Sequence Similarity (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| cp508.17 | cp515.17 | JGS1987 | IQ2 | cp508.17 | cp515.17 | JGS1987 | IQ2 | |
| cp508.17 | 100.00 | 99.85 | 90.40 | 99.85 | 100.00 | 99.96 | 88.44 | 99.96 |
| cp515.17 | - | 100.00 | 90.40 | 100.00 | - | 100.00 | 88.48 | 100.00 |
| JGS1987 | - | - | 100.00 | 90.40 | - | - | 100.00 | 88.48 |
| IQ2 | - | - | - | 100.00 | - | - | - | 100.00 |
| Strain | Gene | Average Nucleotide Sequence Similarity (%) | |||||
|---|---|---|---|---|---|---|---|
| ATCC13124 | IQ1 | IQ2 | IQ3 | IQ2 | IQ3 | ||
| pfoA | pfoA | pfoA | pfoA | alv | alv | ||
| ATCC13124 | pfoA | 100.00 | 98.87 | 97.54 | 97.34 | 86.62 | 86.82 |
| IQ1 | pfoA | - | 100.00 | 97.34 | 97.14 | 86.62 | 86.62 |
| IQ2 | pfoA | - | - | 100.00 | 99.67 | 86.62 | 86.62 |
| IQ3 | pfoA | - | - | - | 100.00 | 86.36 | 86.56 |
| IQ2 | alv | - | - | - | - | 100.00 | 99.43 |
| IQ3 | alv | - | - | - | - | - | 100.00 |
| Strain | Plasmid | Method | Type | Size (bp) | Contig | GC (%) | CDS | Toxin Gene Encoded |
|---|---|---|---|---|---|---|---|---|
| IQ1 | pIQ1a | RBA | D | 53,937 | 13 | 27.11 | 61 | etx |
| pIQ1b | AG | D | 48,812 | 2 | 27.62 | 58 | etx | |
| NCTC8533 | pCP8533etx | REF | B | 64,753 | 1 | 25.89 | 80 | etx, cpb2 |
| IQ2 | pIQ2a | RBA | E | 67,616 | 1 | 25.96 | 66 | iap, ibp, cpe |
| pIQ2b | AG | E | 67,598 | 1 | 25.97 | 66 | iap, ibp, cpe | |
| PB-1 | pCPPB-1 | REF | E | 67,479 | 1 | 25.96 | 69 | iap, ibp, cpe |
| IQ3 | pIQ3a | RBA | A | 54,460 | 1 | 25.05 | 53 | becA, becB |
| pIQ3b | AG | A | 54,641 | 1 | 25.03 | 53 | becA, becB | |
| OS-1 | pCP_OS1 | REF | A | 54,535 | 1 | 25.06 | 54 | becA, becB |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiu, R.; Sim, K.; Shaw, A.; Cornwell, E.; Pickard, D.; Kroll, J.S.; Hall, L.J. Genomic Analysis of Clostridium perfringens BEC/CPILE-Positive, Toxinotype D and E Strains Isolated from Healthy Children. Toxins 2019, 11, 543. https://doi.org/10.3390/toxins11090543
Kiu R, Sim K, Shaw A, Cornwell E, Pickard D, Kroll JS, Hall LJ. Genomic Analysis of Clostridium perfringens BEC/CPILE-Positive, Toxinotype D and E Strains Isolated from Healthy Children. Toxins. 2019; 11(9):543. https://doi.org/10.3390/toxins11090543
Chicago/Turabian StyleKiu, Raymond, Kathleen Sim, Alex Shaw, Emma Cornwell, Derek Pickard, J. Simon Kroll, and Lindsay J. Hall. 2019. "Genomic Analysis of Clostridium perfringens BEC/CPILE-Positive, Toxinotype D and E Strains Isolated from Healthy Children" Toxins 11, no. 9: 543. https://doi.org/10.3390/toxins11090543
APA StyleKiu, R., Sim, K., Shaw, A., Cornwell, E., Pickard, D., Kroll, J. S., & Hall, L. J. (2019). Genomic Analysis of Clostridium perfringens BEC/CPILE-Positive, Toxinotype D and E Strains Isolated from Healthy Children. Toxins, 11(9), 543. https://doi.org/10.3390/toxins11090543






