Effects of Destruxin A on Silkworm’s Immunophilins
Abstract
1. Introduction
2. Results
2.1. The Affinity of DA with BmPPI, BmFKBP45, BmFKBP59 Homologue and HsPPIA
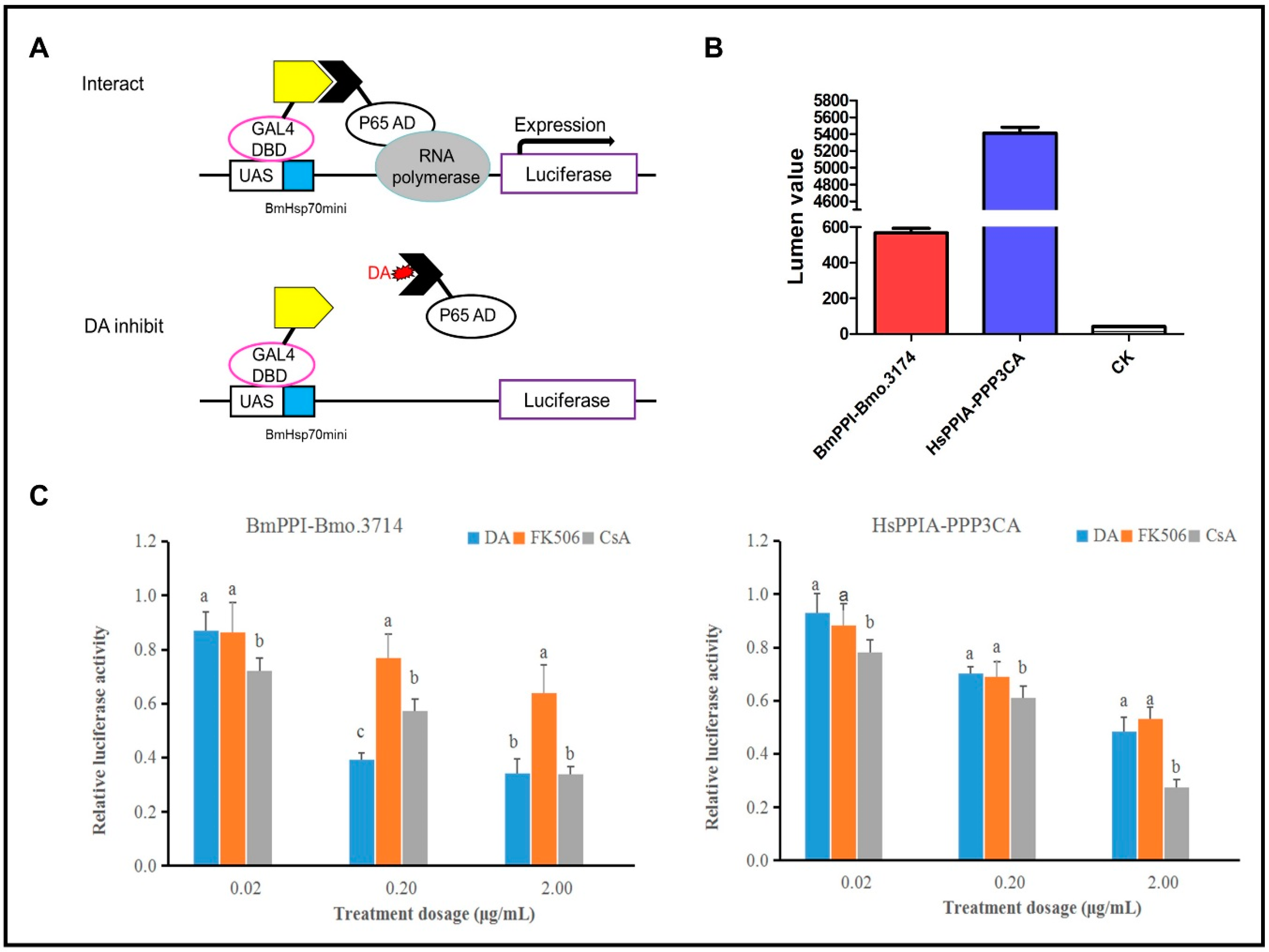
2.2. Influences of DA, CsA, and FK506 on the Interactions of BmPPI–Bmo.3174 and HsPPIA–PPP3CA
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Lines, Culture, and Treatment
5.2. Bio-Layer Interferometry (BLI)
5.3. Insect Two-Hybrid (I2H)
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pedras, M.S.C.; Zaharia, L.I.; Ward, D.E. The destruxins: Synthesis, biosynthesis, biotransformation, and biological activity. Phytochemistry 2002, 59, 579–596. [Google Scholar] [CrossRef]
- Schrank, A.; Vainstein, M.H. Metarhizium anisopliae enzymes and toxins. Toxicon 2010, 56, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Odier, F.V.A.; Bureau, J. In vitro effect of fungal cyclodepsipeptides on leukemic cells: Study of destruxins A, B and E. Biol. Cell 1992, 74, 267–271. [Google Scholar] [CrossRef]
- Yeh, S.F.; Pan, W.; Ong, G.T.; Chiou, A.J.; Chuang, C.C.; Chiou, S.H.; Wu, S.H. Study of Structure–Activity Correlation in Destruxins, a Class of Cyclodepsipeptides Possessing Suppressive Effect on the Generation of Hepatitis B Virus Surface Antigen in Human Hepatoma Cells. Biochem. Biophys. Res. Commun. 1996, 229, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.L.; Tzeng, Y.M. Development and applications of destruxins: A review. Biotechnol. Adv. 2012, 30, 1242–1254. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sanchez, E.; O’Donnell, M.J.J. Effects of the microbial metabolite destruxin A on ion transport by the gut and renal epithelia of Drosophila melanogaster. Arch. Insect Biochem. Physiol. 2012, 80, 109–122. [Google Scholar] [CrossRef]
- Ruiz-Sanchez, E.; Lange, A.B.; Orchard, I. Effects of the mycotoxin destruxin A on Locusta migratoria visceral muscles. Toxicon 2010, 56, 1043–1051. [Google Scholar] [CrossRef]
- Fan, J.Q.; Chen, X.R.; Hu, Q.B. Effects of Destruxin A on Hemocytes Morphology of Bombyx mori. J. Integr. Agric. 2013, 12, 1042–1048. [Google Scholar] [CrossRef]
- Chen, X.R.; Hu, Q.B.; Yu, X.Q.; Ren, S.X. Effects of destruxins on free calcium and hydrogen ions in insect hemocytes. Insect Sci. 2014, 21, 31–38. [Google Scholar] [CrossRef]
- Pal, S.; Leger, R.J.S.; Wu, L.P. Fungal Peptide Destruxin A Plays a Specific Role in Suppressing the Innate Immune Response in Drosophila melanogaster. J. Biol. Chem. 2007, 282, 8969–8977. [Google Scholar] [CrossRef]
- Hu, W.; He, G.; Wang, J.; Hu, Q. The Effects of Destruxin A on Relish and Rel Gene Regulation to the Suspected Immune-Related Genes of Silkworm. Molecules 2016, 22, 41. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, M.; Xu, X.; Xu, J.; Zhu, X.; Li, S.; Zhou, X.; Yu, J.; Xu, X.; Hu, Q.; Yu, X. Identification of immunity-related genes in Plutella xylostella in response to fungal peptide destruxin A: RNA-Seq and DGE analysis. Sci. Rep. 2017, 7, 10966. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Chen, X.; Liu, C.; Jin, F.; Hu, Q. Gene expression profile of Bombyx mori hemocyte under the stress of destruxin A. PLoS ONE 2014, 9, e96170. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Han, P.; Chen, X.; Hu, Q.; Ye, M. Comparative proteomic analysis of Bombyx mori hemocytes treated with Destruxin, A. Arch. Insect Biochem. Physiol. 2014, 86, 33–45. [Google Scholar]
- Lomenick, B.; Hao, R.; Jonai, N.; Chin, R.M.; Aghajan, M.; Warburton, S.; Wohlschlegel, J.A. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 2009, 106, 21984–21989. [Google Scholar] [CrossRef]
- Flaherty, P.T.; Jain, P. Peptidyl Prolyl Isomerase Inhibitors. Annu. Rep. Med. Chem. 2011, 46, 337–349. [Google Scholar]
- Beauchesne, P.R.; Chung, N.S.; Wasan, K.M. Cyclosporine A: A Review of Current Oral and Intravenous Delivery Systems. Drug Dev. Ind. Pharm. 2007, 33, 211–220. [Google Scholar] [CrossRef]
- Fung, J.J. Tacrolimus and transplantation: A decade in review. Transplantation 2004, 77, S41–S43. [Google Scholar] [CrossRef]
- Abdiche, Y.; Dan, M.; Pinkerton, A.; Pons, J.J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 2008, 377, 209–217. [Google Scholar] [CrossRef]
- Mon, H.; Sugahara, R.; Hong, S.M.; Lee, J.M.; Kamachi, Y.; Kawaguchi, Y.; Kusakabe, T.J. Analysis of protein interactions with two-hybrid system in cultured insect cells. Anal. Biochem. 2009, 392, 180–182. [Google Scholar] [CrossRef]
- Wang, J.; Hu, W.; Hu, Q. BmTudor-sn Is a Binding Protein of Destruxin A in Silkworm Bm12 Cells. Toxins 2019, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hu, W.; Xiao, M.; Ou, S.; Hu, Q. Destruxin A induces and binds HSPs in Bombyx mori Bm12 cells. J. Agric. Food Chem. 2017, 65, 9849–9853. [Google Scholar] [CrossRef] [PubMed]
- Ideno, A.; Yoshida, T.; Toshii, D.A., II; Furutani, M.; Maruyama, T. FK506-binding protein of the hyperthermophilic archaeum, Thermococcus sp. KS-1, a cold-shock-inducible peptidyl-prolyl cis–trans isomerase with activities to trap and refold denatured proteins. Biochem. J. 2001, 357, 465–471. [Google Scholar] [PubMed]
- Somarelli, J.A.; Coll, J.L.; Velandia, A.; Martinez, L.; Herrera, R.J. Characterization of immunophilins in the silkmoth Bombyx mori. Arch. Insect Biochem. Physiol. 2010, 65, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Kaur, R.; Akhter, S.; Legerski, R.J. Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep. 2009, 10, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Bertram, K.; Agafonov, D.E.; Liu, W.T.; Dybkov, O.; Will, C.L.; Hartmuth, K.; Lührmann, R. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 2017, 542, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.B.; Ren, S.X.; Wu, J.H.; Chang, J.M.; Musa, P.D. Investigation of destruxin A and B from 80 Metarhizium strains in China, and the optimization of cultural conditions for the strain MaQ10. Toxicon 2006, 48, 491–498. [Google Scholar] [CrossRef] [PubMed]

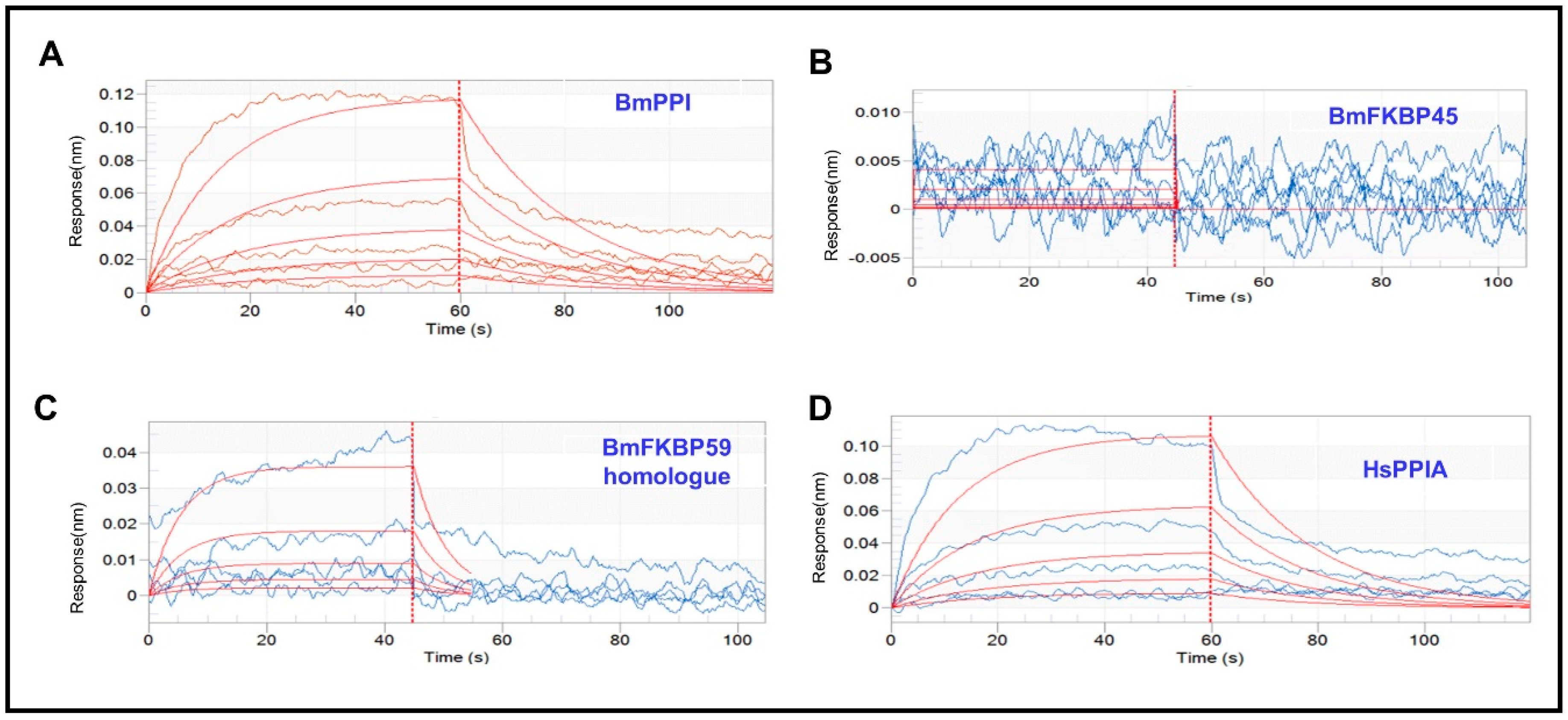
| Protein | DA Conc. (μM) | Response | KD (M) 1 | KON (1/Ms) 2 | KDIS (1/s) 3 |
|---|---|---|---|---|---|
| BmPPI | 62.5 | 0.0045 | 1.98 × 10−3 | 2.38 × 10−1 | 4.70 × 10−2 |
| 125 | 0.0153 | ||||
| 250 | 0.0261 | ||||
| 500 | 0.0547 | ||||
| 1000 | 0.1180 | ||||
| BmFKBP45 | 15.6 | 0.0057 | 8.01 × 10−3 | 5.68 × 10 | 4.55 × 10 |
| 31.3 | 0.0066 | ||||
| 62.5 | 0.0021 | ||||
| 125 | 0.0015 | ||||
| 250 | −0.0001 | ||||
| 500 | −0.0001 | ||||
| 1000 | 0.0058 | ||||
| BmFKBP59 homologue | 62.5 | 0.002 | 1.25 × 10−1 | 1.45 | 0.181 |
| 125 | 0.0083 | ||||
| 250 | 0.0073 | ||||
| 500 | 0.0159 | ||||
| 1000 | 0.0415 | ||||
| HsPPIA | 62.5 | 0.0060 | 2.22 × 10−3 | 2.46 × 10−1 | 5.46 × 10−2 |
| 125 | 0.0077 | ||||
| 250 | 0.0247 | ||||
| 500 | 0.0530 | ||||
| 1000 | 0.1035 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Weng, Q.; Hu, Q. Effects of Destruxin A on Silkworm’s Immunophilins. Toxins 2019, 11, 349. https://doi.org/10.3390/toxins11060349
Wang J, Weng Q, Hu Q. Effects of Destruxin A on Silkworm’s Immunophilins. Toxins. 2019; 11(6):349. https://doi.org/10.3390/toxins11060349
Chicago/Turabian StyleWang, Jingjing, Qunfang Weng, and Qiongbo Hu. 2019. "Effects of Destruxin A on Silkworm’s Immunophilins" Toxins 11, no. 6: 349. https://doi.org/10.3390/toxins11060349
APA StyleWang, J., Weng, Q., & Hu, Q. (2019). Effects of Destruxin A on Silkworm’s Immunophilins. Toxins, 11(6), 349. https://doi.org/10.3390/toxins11060349





