Genotypes, Enterotoxin Gene Profiles, and Antimicrobial Resistance of Staphylococcus aureus Associated with Foodborne Outbreaks in Hangzhou, China
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Characterization and Identification of S. aureus Isolates
2.2. Antibiotic Resistance
2.3. Molecular Genotyping of S. aureus
2.4. Prevalence of Enterotoxin or Enterotoxin-Like Genes
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Isolates
5.2. Antimicrobial Susceptibility
5.3. Extraction of Genomic DNA
5.4. Detection of 23 SE/SEl Genes
5.5. PFGE
5.6. MLST and Spa Typing
Author Contributions
Funding
Conflicts of Interest
References
- Krishna, S.; Miller, L.S. Host-pathogen interactions between the skin and Staphylococcus aureus. Curr. Opin. Microbiol. 2012, 15, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Liu, J.; Bai, L.; Li, W.; Han, H.; Fu, P.; Ma, X.; Bi, Z.; Yang, X.; Zhang, X.; Zhen, S.; et al. Trends of foodborne diseases in China: Lessons from laboratory-based surveillance since 2011. Front. Med. 2018, 12, 48–57. [Google Scholar] [CrossRef]
- Mead, P.S.; Slutsker, L.; Dietz, V.; McCaig, L.F.; Bresee, J.S.; Shapiro, C.; Griffin, P.M.; Tauxe, R.V. Food-related illness and death in the United States. Emerg. Infect. Dis. 1999, 5, 607–625. [Google Scholar] [CrossRef]
- Johler, S.; Giannini, P.; Jermini, M.; Hummerjohann, J.; Baumgartner, A.; Stephan, R. Further evidence for staphylococcal food poisoning outbreaks caused by egc-encoded enterotoxins. Toxins 2015, 7, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Hennekinne, J.A.; De Buyser, M.L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef] [PubMed]
- Hait, J.; Tallent, S.; Melka, D.; Keys, C.; Bennett, R. Prevalence of enterotoxins and toxin gene profiles of Staphylococcus aureus isolates recovered from a bakery involved in a second staphylococcal food poisoning occurrence. J. Appl. Microbiol. 2014, 117, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Bohach, G.A.; Nair, S.P.; Hiramatsu, K.; Jouvin-Marche, E.; Mariuzza, R.; International Nomenclature Committee for Staphylococcal Superantigens. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 2004, 189, 2334–2336. [Google Scholar] [CrossRef] [PubMed]
- Omoe, K.; Hu, D.L.; Ono, H.K.; Shimizu, S.; Takahashi-Omoe, H.; Nakane, A.; Uchiyama, T.; Shinagawa, K.; Imanishi, K. Emetic potentials of newly identified staphylococcal enterotoxin-like toxins. Infect. Immun. 2013, 81, 3627–3631. [Google Scholar] [CrossRef] [PubMed]
- Nemati, M.; Hermans, K.; Lipinska, U.; Denis, O.; Deplano, A.; Struelens, M.; Devriese, L.A.; Pasmans, F.; Haesebrouck, F. Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: First detection of livestock-associated methicillin-resistant strain ST398. Antimicrob. Agents Chemother. 2008, 52, 3817–3819. [Google Scholar] [CrossRef]
- Vincze, S.; Stamm, I.; Kopp, P.A.; Hermes, J.; Adlhoch, C.; Semmler, T.; Wieler, L.H.; Lubke-Becker, A.; Walther, B. Alarming proportions of methicillin-resistant Staphylococcus aureus (MRSA) in wound samples from companion animals, Germany 2010–2012. PLoS ONE 2014, 9, e85656. [Google Scholar] [CrossRef] [PubMed]
- Haran, K.P.; Godden, S.M.; Boxrud, D.; Jawahir, S.; Bender, J.B.; Sreevatsan, S. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. J. Clin. Microbiol. 2012, 50, 688–695. [Google Scholar] [CrossRef]
- Pu, S.; Wang, F.; Ge, B. Characterization of toxin genes and antimicrobial susceptibility of Staphylococcus aureus isolates from Louisiana retail meats. Foodborne Pathog. Dis. 2011, 8, 299–306. [Google Scholar] [CrossRef]
- Parisi, A.; Caruso, M.; Normanno, G.; Latorre, L.; Sottili, R.; Miccolupo, A.; Fraccalvieri, R.; Santagada, G. Prevalence, antimicrobial susceptibility and molecular typing of Methicillin-Resistant Staphylococcus aureus (MRSA) in bulk tank milk from southern Italy. Food Microbiol. 2016, 58, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Riva, A.; Borghi, E.; Cirasola, D.; Colmegna, S.; Borgo, F.; Amato, E.; Pontello, M.M.; Morace, G. Methicillin-Resistant Staphylococcus aureus in Raw Milk: Prevalence, SCCmec Typing, Enterotoxin Characterization, and Antimicrobial Resistance Patterns. J. Food Prot. 2015, 78, 1142–1146. [Google Scholar] [CrossRef]
- Rhee, C.H.; Woo, G.J. Emergence and characterization of foodborne methicillin-resistant Staphylococcus aureus in Korea. J. Food Prot. 2010, 73, 2285–2290. [Google Scholar] [CrossRef] [PubMed]
- Al-Ashmawy, M.A.; Sallam, K.I.; Abd-Elghany, S.M.; Elhadidy, M.; Tamura, T. Prevalence, Molecular Characterization, and Antimicrobial Susceptibility of Methicillin-Resistant Staphylococcus aureus Isolated from Milk and Dairy Products. Foodborne Pathog. Dis. 2016, 13, 156–162. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [PubMed]
- Cha, J.O.; Lee, J.K.; Jung, Y.H.; Yoo, J.I.; Park, Y.K.; Kim, B.S.; Lee, Y.S. Molecular analysis of Staphylococcus aureus isolates associated with staphylococcal food poisoning in South Korea. J. Appl. Microbiol. 2006, 101, 864–871. [Google Scholar] [CrossRef]
- Sato’o, Y.; Omoe, K.; Naito, I.; Ono, H.K.; Nakane, A.; Sugai, M.; Yamagishi, N.; Hu, D.L. Molecular epidemiology and identification of a Staphylococcus aureus clone causing food poisoning outbreaks in Japan. J. Clin. Microbiol. 2014, 52, 2637–2640. [Google Scholar] [CrossRef]
- Suzuki, Y.; Omoe, K.; Hu, D.L.; Sato’o, Y.; Ono, H.K.; Monma, C.; Arai, T.; Konishi, N.; Kato, R.; Hirai, A.; et al. Molecular epidemiological characterization of Staphylococcus aureus isolates originating from food poisoning outbreaks that occurred in Tokyo, Japan. Microbiol. Immunol. 2014, 58, 570–580. [Google Scholar] [CrossRef]
- Umeda, K.; Nakamura, H.; Yamamoto, K.; Nishina, N.; Yasufuku, K.; Hirai, Y.; Hirayama, T.; Goto, K.; Hase, A.; Ogasawara, J. Molecular and epidemiological characterization of staphylococcal foodborne outbreak of Staphylococcus aureus harboring seg, sei, sem, sen, seo, and selu genes without production of classical enterotoxins. Int. J. Food Microbiol. 2017, 256, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Argudin, M.A.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef]
- Wattinger, L.; Stephan, R.; Layer, F.; Johler, S. Comparison of Staphylococcus aureus isolates associated with food intoxication with isolates from human nasal carriers and human infections. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Wang, B.; Tao, X.; Hu, Q.; Cui, Z.; Zhang, J.; Lin, Y.; You, Y.; Shi, X.; Grundmann, H. Characterization of Staphylococcus aureus stains associated with food poisoning in Shenzhen, China. Appl. Environ. Microbiol. 2012, 78, 6637–6642. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wu, S.; Luo, W.; Su, Y.; Luan, Y.; Wang, X. Staphylococcus aureus ST6-t701 isolates from food-poisoning outbreaks (2006-2013) in Xi’an, China. Foodborne Pathog. Dis. 2015, 12, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Zhu, Z.; Chang, Y.; Shen, X.; Gao, H.; Yang, Y. Prevalence and Characteristics of Enterotoxin B-Producing Staphylococcus aureus Isolated from Food Sources: A Particular Cluster of ST188 Strains was Identified. J. Food Sci. 2016, 81, M715–M718. [Google Scholar] [CrossRef]
- Cui, S.; Li, J.; Hu, C.; Jin, S.; Li, F.; Guo, Y.; Ran, L.; Ma, Y. Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J. Antimicrob. Chemother. 2009, 64, 680–683. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar]
- Xie, Y.; He, Y.; Gehring, A.; Hu, Y.; Li, Q.; Tu, S.I.; Shi, X. Genotypes and toxin gene profiles of Staphylococcus aureus clinical isolates from China. PLoS ONE 2011, 6, e28276. [Google Scholar] [CrossRef]
- Tang, J.; Tang, C.; Chen, J.; Du, Y.; Yang, X.N.; Wang, C.; Zhang, H.; Yue, H. Phenotypic characterization and prevalence of enterotoxin genes in Staphylococcus aureus isolates from outbreaks of illness in Chengdu City. Foodborne Pathog. Dis. 2011, 8, 1317–1320. [Google Scholar] [CrossRef]
- Argudin, M.A.; Mendoza, M.C.; Gonzalez-Hevia, M.A.; Bances, M.; Guerra, B.; Rodicio, M.R. Genotypes, exotoxin gene content, and antimicrobial resistance of Staphylococcus aureus strains recovered from foods and food handlers. Appl. Environ. Microbiol. 2012, 78, 2930–2935. [Google Scholar] [CrossRef] [PubMed]
- Kerouanton, A.; Hennekinne, J.A.; Letertre, C.; Petit, L.; Chesneau, O.; Brisabois, A.; De Buyser, M.L. Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int. J. Food Microbiol. 2007, 115, 369–375. [Google Scholar] [CrossRef]
- Becker, K.; Roth, R.; Peters, G. Rapid and specific detection of toxigenic Staphylococcus aureus: Use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 1998, 36, 2548–2553. [Google Scholar] [PubMed]
- Omoe, K.; Hu, D.L.; Takahashi-Omoe, H.; Nakane, A.; Shinagawa, K. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiol. Lett. 2005, 246, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.J.; Jeon, H.S.; Sung, H.; Kim, M.N.; Hong, S.J. Epidemiological characteristics of methicillin-resistant Staphylococcus aureus isolates from children with eczematous atopic dermatitis lesions. J. Clin. Microbiol. 2008, 46, 991–995. [Google Scholar] [CrossRef]
- Available online: http://saureus.mlst.net/ (accessed on 29 May 2019).
- Available online: http://spaserver.ridom.de/ (accessed on 29 May 2019).
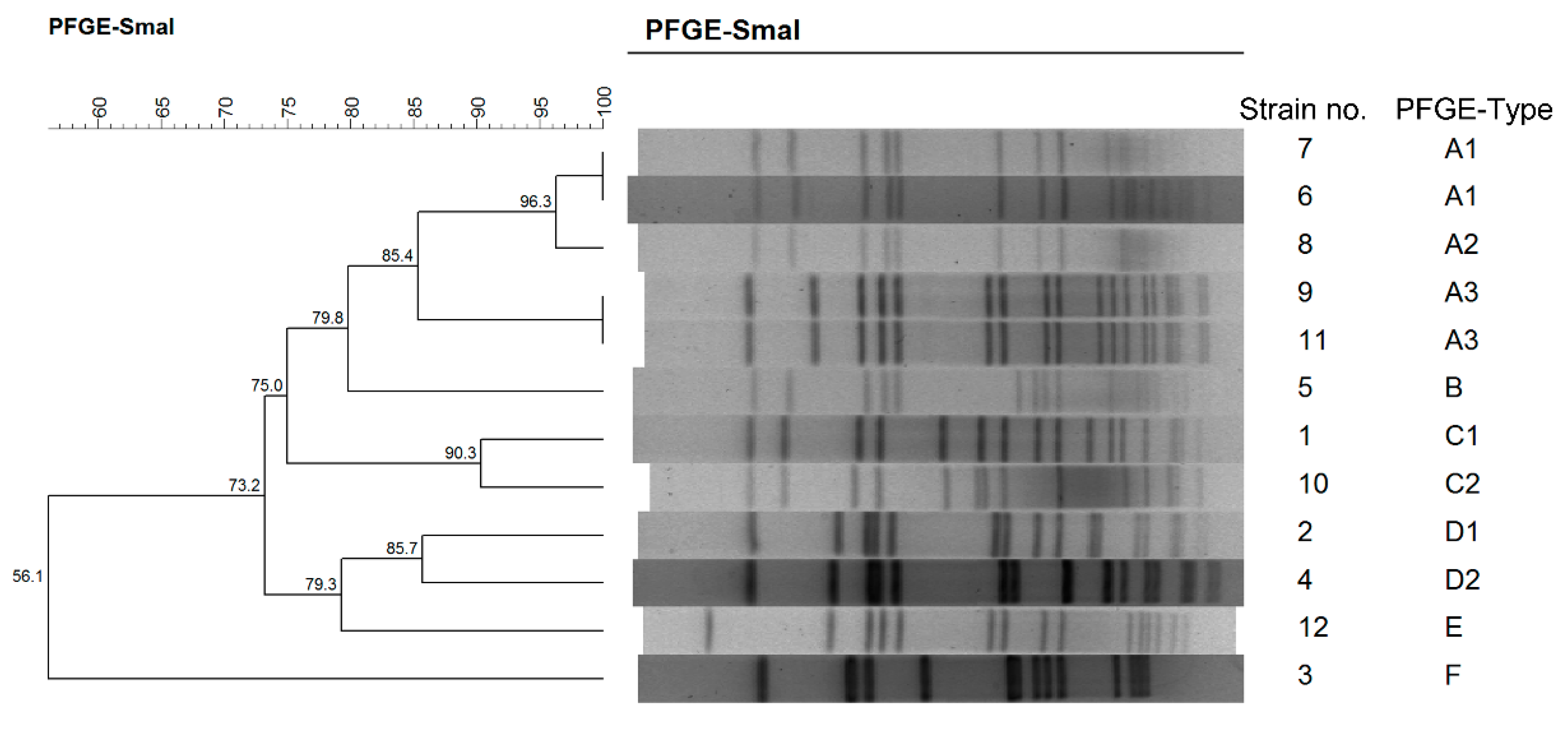
| Outbreaks | Data | District | Origins (no. of Isolates) a | Strains b | MLST | Spa | PFGE | Resistance c |
|---|---|---|---|---|---|---|---|---|
| 1 | October, 2015 | Gongshu | Food (8) Environment (1) Feces (3) | 1 | ST2315 | t11687 | C1 | TE–PEN |
| Feces (1) | 2 | ST188 | t189 | D1 | PEN | |||
| Feces (1) | 3 | ST2250 | t7960 | F | PEN | |||
| Feces (1) | 4 | ST188 | t189 | D2 | PEN | |||
| 2 | June, 2016 | Xiangcheng | Food (1) | 5 | ST3055 | t084 | B | PEN |
| Food (1) | 6 | ST5 | t1228 | A1 | ERY–CD–GEN–SXT–PEN | |||
| Food (1) | 7 | ST5 | t002 | A1 | ERY–CD–GEN–SXT–PEN | |||
| Feces (1) | ||||||||
| Feces (1) | 8 | ST5 | t002 | A2 | ERY–CD–GEN–SXT–PEN | |||
| 3 | September, 2016 | Gongshu | Food (6) | 9 | ST6 | t304 | A3 | PEN |
| Feces (1) | ||||||||
| Feces (1) | 10 | ST573 | t458 | C2 | ERY–PEN | |||
| 4 | October, 2016 | Xihu | Food (5) | 11 | ST6 | t304 | A3 | ERY–PEN |
| Feces (3) | ||||||||
| Feces (1) | 12 | ST72 | t148 | E | ERY |
| Strains | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SE Genes | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| sea | + | + | + | + | + | + | ||||||
| seb | + | |||||||||||
| sec | + | + | + | + | + | + | ||||||
| sed | + | + | + | + | ||||||||
| see | ||||||||||||
| seg | + | + | + | + | + | + | ||||||
| seh | + | |||||||||||
| sei | + | + | + | + | + | + | ||||||
| sej | + | + | + | + | ||||||||
| sek | ||||||||||||
| sel | + | + | + | + | + | + | ||||||
| sem | + | + | + | + | + | + | ||||||
| sen | + | + | + | + | + | + | ||||||
| seo | + | + | + | + | + | + | ||||||
| sep | ||||||||||||
| seq | ||||||||||||
| ser | + | + | + | + | ||||||||
| seu | + | + | + | + | + | + | ||||||
| Total | 9 | 3 | 1 | 1 | 3 | 11 | 11 | 9 | 1 | 9 | 1 | 9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Xie, S. Genotypes, Enterotoxin Gene Profiles, and Antimicrobial Resistance of Staphylococcus aureus Associated with Foodborne Outbreaks in Hangzhou, China. Toxins 2019, 11, 307. https://doi.org/10.3390/toxins11060307
Chen Q, Xie S. Genotypes, Enterotoxin Gene Profiles, and Antimicrobial Resistance of Staphylococcus aureus Associated with Foodborne Outbreaks in Hangzhou, China. Toxins. 2019; 11(6):307. https://doi.org/10.3390/toxins11060307
Chicago/Turabian StyleChen, Qi, and Sangma Xie. 2019. "Genotypes, Enterotoxin Gene Profiles, and Antimicrobial Resistance of Staphylococcus aureus Associated with Foodborne Outbreaks in Hangzhou, China" Toxins 11, no. 6: 307. https://doi.org/10.3390/toxins11060307
APA StyleChen, Q., & Xie, S. (2019). Genotypes, Enterotoxin Gene Profiles, and Antimicrobial Resistance of Staphylococcus aureus Associated with Foodborne Outbreaks in Hangzhou, China. Toxins, 11(6), 307. https://doi.org/10.3390/toxins11060307




